Effects of carbon sources on 17 beta-estradiol degradation by Sphingomonas sp. and the analysis of the involved intracellular metabolomics†
Changwei
Li
,
Xianwang
Kong
,
Lihua
Lan
,
Musa Abubakar
Tadda
and
Dezhao
Liu
 *
*
Institute of Agricultural Bio-Environmental Engineering, College of Biosystems Engineering and Food Science, Zhejiang University, Yuhangtang Road 866, Hangzhou, China. E-mail: dezhaoliu@zju.edu.cn; Tel: +86 13588046533
First published on 3rd December 2019
Abstract
17β-estradiol (E2) ubiquitously exists in various water bodies with long-term endocrine-disrupting and carcinogenic impacts on wildlife even at the trace level of ng L−1. However, it remains unclear how easy-to-degrade carbon sources alter E2 biodegradation patterns. In this study, E2 biodegradation by Sphingomonas sp. MCCC 1A06484 was investigated with regard to alternative carbon sources. Results showed that the bacterium preferentially utilized glucose, sodium succinate and sodium acetate over E2. Interestingly, the presence of these preferred nutrients increased the E2 removal efficiency by 20.1%. Furthermore, a positive relation (p < 0.05) between the utilization of total organic carbon (TOC) and E2 was found. Using intracellular metabolomics by UHPLC-QTOF-MS, 11 up-regulated and 35 down-regulated metabolites (variable importance > 1, p < 0.05) were identified in the bacterium when cultivated with E2 under various carbon and nitrogen backgrounds. The E2 exposure contributed to metabolism changes of lipid, nucleotide, carbohydrate, amino acid and membrane transport, which were considered to play roles in the E2 metabolism. The up-regulated phosphatidylcholine might act as an indicator during the bacterial degradation of E2. Generally, this study contributes to an in-depth understanding of E2 biodegradation in complex environments with multiple carbon and nitrogen sources.
Environmental significanceEstrogens have been listed as group 1 carcinogens by the World Health Organization. In this work, we evaluated how the existence of alternative carbon sources and their concentration levels affected 17β-estradiol (E2) biodegradation by Sphingomonas sp. MCCC 1A06484. Further, the E2 metabolic pathway under various carbon and nitrogen backgrounds was intensively elucidated by the method of intracellular metabolomics. Finally, we found that E2 exposure contributed to metabolism changes of lipid, nucleotide, carbohydrate, amino acid and membrane transport. And the up-regulated phosphatidylcholine might act as an indicator during the bacterial degradation of E2. To the best of our knowledge, there is no published work concerning intracellular estrogen metabolites during the microbial degradation process through the method of metabolomics. The results would expand our understanding of microbial biodegradation of estrogens at the bacterial metabolite level. |
1. Introduction
17β-estradiol (E2) is a typical natural estrogen that is commonly detected in aquatic ecosystems and wastewater treatment facilities worldwide.1 E2 exerts a challenging endocrine disrupting effect on aquatic organisms and human beings even at concentration levels of ng L−1.2–4 E2 could alter sexual developments and physiological activities of aquatic organisms and chronically accumulate in human bodies.5 The issue of E2 occurrence is more alarming since it often appears as a complex conjugate with other co-existing chemicals, which could potentially lead to unexpected synergistic effects.6Biodegradation is in general the predominant mechanism for E2 removal both in engineered and natural ecosystems, and the involved metabolic and co-metabolic pathways critically determine the fate of E2.7–10 Both heterotrophic and autotrophic microorganisms account for the E2 biodegradation. To date, various bacterial strains capable of degrading E2 have been identified and characterized from diverse environmental surroundings.11–14 For example, several bacterial strains affiliated with the genus of Sphingomonas in the literature were found to degrade E2 with high efficiency.15,16 Importantly, Sphingomonas species are also widely distributed in both soil and aquatic environments, and are well known for their distinguished ability to degrade hazardous organic molecules.17
E2 as low as ng L−1 is apparently insufficient for bacterial growth with respect to carbon supply. Thus, additional growth substrates (carbon sources) are essential for the smooth E2 biodegradation.18,19 However, it remains controversial how the additional substrates alter the E2 biodegradation patterns. One study20 recorded that the presence of easily biodegradable organic carbon sources enhanced the estrone (E1, a natural estrogen) biodegradation due to the increased biomass growth, and low influent organic carbon loading was unfavorable for the E1 biodegradation in bioreactors. Besides, it was reported that the utilization of polycyclic aromatic hydrocarbons was accelerated in the presence of additional carbon sources.21 Similarly, C-13 × 102 mineralization rate was found to be positively related to the total organic carbon concentration in sediments (p < 0.01).22 In contrast, it was revealed that23 the substrate present in raw influent slowed down the degradation of E1 and E2 due to the substrate inhibition mechanism, i.e. bacteria would utilize easily biodegradable carbon sources prior to recalcitrant substrates such as estrogens. Correspondingly, E2 degradation by ammonia oxidizers was depressed as a result of a high loading of organic substances.24 Thus, it is necessary to explore the effect of carbon addition on the E2 degradation.
Furthermore, the biochemical mechanisms of the E2 degradation pathway are currently being explored.25 Several studies on E2 biodegradation have applied the methods of systems biology, most of which were based on genomics and proteomics.26–28 However, genomics and proteomics operate at a markedly different timescale, making it difficult to find the underlying causal relationship. Metabolomics solves this problem by monitoring the ultimate outcome without making assumptions about the effect of any single contribution on the results.29 Metabolomics aims to characterize and quantify small molecule metabolites of living organisms as a response to biological stimuli or genetic manipulation. This method is mainly applied in the medical discipline to identify disease markers. Only a few studies have applied metabolomics to monitor the extracellular metabolites of microbial estrogen degradation.30 To the best of our knowledge, there is no published work concerning intracellular metabolites during the process of microbial E2 degradation via metabolomics. Thus, it can potentially bring a new perspective towards complete understanding of the E2 degradation mechanism at the molecular level.
In general, the present study was guided by the following questions: (i) Could the Sphingomonas sp. strain maintain its capacity to degrade E2 in the presence of easy-to-degrade carbon sources? (ii) How does the organic loading affect the E2 biodegradation by the strain? (iii) What are the specific metabolites associated with E2 biodegradation in the strain under various carbon and nitrogen backgrounds?
2. Materials and methods
2.1. Chemicals
17β-estradiol (E2, >98% by mass content) and estrone (E1, >98% by mass content) used in this study were purchased from Aladdin Co. Ltd. (USA) and Macklin Co. Ltd. (USA), respectively. Due to their low solubility in water, stock solutions were prepared in acetone and stored at 4 °C. All organic solvents used were of HPLC grade. Composition details of Reasoner's 2A (R2A) medium (ATCC medium 2258) and nitrate mineral salts medium (NMS, ATCC medium 1306) were accessible on ATCC (https://www.atcc.org). Sodium succinate was obtained from Aladdin Co. Ltd. (USA), while the rest of the carbon sources, glucose and sodium acetate were from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China).2.2. Sphingomonas sp. cultivation
Sphingomonas sp. was purchased from Marine Culture Collection of China (MCCC 1A06484) and maintained on R2A agar medium. Cell growth was assessed over time based on the optical density at 600 nm (OD600) measured using an Agilent Carry 60 UV-Vis spectrophotometer (USA). Cells were harvested by centrifugation (6000 × g, 10 min, 4 °C) and then washed twice with phosphate buffered saline (PBS; 1.80 g L−1 NaH2PO4, 0.30 g L−1 KH2PO4, 0.25 g L−1 KCl, 10.00 g L−1 NaCl; prepared in MilliQwater; pH = 7.4) after shaking cultivation (150 rpm) at 30 °C for three days until the late exponential growth phase was reached for all batch experiments of degradation and metabolomics analysis.2.3. E2 degradation experiment
Harvested cell pellets were cultivated in a series of Erlenmeyer flasks containing 100 mL E2-dissolved (1.8 mg L−1) NMS medium. Prior to the cultivation, acetone in E2 stock solution was removed by heating at 60 °C for 30 min. To elucidate the effects of alternative carbon sources on E2 removal, two types of medium were adopted: (1) NMS (without any carbon source); (2) NMS simultaneously amended with glucose (2 g L−1), sodium succinate (2 g L−1) and sodium acetate (1.167 g L−1) modified from a previous study.15 Two control groups were also set: (1) the second medium (E2 added) with the autoclaved bacterium and (2) the second medium without E2 addition. Due to unsatisfactory growth results, vitamins were supplemented into NMS at 1 mL L−1 to obtain the final concentrations of vitamin B12, biotin and p-aminobenzoic acid as 2 μg L−1, 5 μg L−1, and 10 μg L−1, respectively.31 The TOC concentration of the NMS solution with other carbon sources added after passing through the 0.45 μm filter was determined with a TOC analyzer (Multi N/C 3100, Analytik Jena, Germany).To investigate the effects of carbon concentrations on E2 removal, glucose as a typical carbon source was chosen with concentration gradients of 100, 500 and 5000 mg L−1. These concentrations are in line with the normal chemical oxygen demands (CODs) found in aquaculture wastewater,32 municipal wastewater33 and livestock wastewater,34 respectively.
All degradation experiments were conducted in duplicate in a shaker at 150 rpm, 30 °C. Liquid samples were collected at regular intervals and filtered (0.45 μm) for further measurements.
2.4. Metabolomics analysis in Sphingomonas sp.
The metabolomics analysis of E2 removal by Sphingomonas sp. was conducted under diverse carbon and nitrogen backgrounds. On the one hand, types and concentrations of carbon sources adopted were used in the same way as described in Section 2.3; on the other hand, 1 g L−1 KNO3 (0.1386 g L−1 N−1) in the NMS medium was equally divided into 0.5 g L−1 KNO3 and 0.3267 g L−1 (NH4)2SO4 on the N basis, which served as the experimental group with E2 added (1.8 mg L−1) under diverse carbon and nitrogen backgrounds. Meanwhile, a control group was also set, and the only difference between the control group and the experimental group was the absence of E2 in the former.Metabolic degradation experiments were performed in six replicates for each of the two groups. Cell enrichment was gathered until the late exponential phase. Bacterial cells were centrifuged and washed 5 times, and then quenched in liquid nitrogen (≥3 min) for further metabolomics analysis.
2.5. Analytical methods
Aqueous samples of 20 μL were injected into an HPLC column (Agilent Eclipse XDB-C18, 250 mm × 4.6 mm, 5 μm) for estrogen quantification with UV detection at 200 nm.35 The optimum separation was achieved at a flow rate of 1 mL min−1 with acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (6
water (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v) as the mobile phase.
4, v/v) as the mobile phase.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile
acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (2
water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) with addition of standard substances as the internal standard. Bacterial suspensions were vortexed (30 s), homogenized in a ball mill (45 Hz, 4 min) and ultrasound treated (in ice water, 5 min) thrice. After incubation at −20 °C for another 1 h, the samples were centrifuged for 15 min at 13
1, v/v/v) with addition of standard substances as the internal standard. Bacterial suspensions were vortexed (30 s), homogenized in a ball mill (45 Hz, 4 min) and ultrasound treated (in ice water, 5 min) thrice. After incubation at −20 °C for another 1 h, the samples were centrifuged for 15 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C. The dried residue of the supernatant extracts was soaked in 100 μL of acetonitrile
000 rpm, 4 °C. The dried residue of the supernatant extracts was soaked in 100 μL of acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), reconstituted, vortexed for 30 s and ultrasonically processed for 10 min (4 °C water bath). The supernatant (60 μL) was transferred after centrifugation for further detection.36,37 The extraction method was designed to extract as much water-soluble and fat-soluble substances as possible.
1, v/v), reconstituted, vortexed for 30 s and ultrasonically processed for 10 min (4 °C water bath). The supernatant (60 μL) was transferred after centrifugation for further detection.36,37 The extraction method was designed to extract as much water-soluble and fat-soluble substances as possible.
Determination of intracellular metabolites was performed using an UHPLC (1290, Agilent Technologies, USA) with a UPLC BEH Amide column (2.1 mm × 100 mm × 1.7 μm, Waters, USA) coupled to a Triple TOF 5600 (Q-TOF, AB Sciex, USA). The mobile phase consisting of 25 mM NH4OAc and 25 mM NH4OH in water (pH = 9.75) (A) and acetonitrile (B) was delivered at 0.5 mL min−1 with elution gradient as follows: 0–7 min, A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B = 1
B = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19; 7–9 min, A
19; 7–9 min, A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B = 7
B = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13; 9–9.1 min, A
13; 9–9.1 min, A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B = 4
B = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6; 9.1–12 min, A
6; 9.1–12 min, A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B = 1
B = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19, with an injection volume of 4 μL. Electrospray ionization source (ESI) conditions were set as follows: ion source gas 1 at 60 psi, ion source gas 2 at 60 psi, curtain gas at 35 psi, source temperature at 650 °C and ion spray voltage floating (ISVF) at −4000 V in negative modes. A triple TOF mass spectrometer was used for its ability to acquire MS/MS spectra in an information-dependent basis (IDA) mode.37,38 Differential metabolites were identified using MS/MS acquisition software (Analyst TF 1.7, AB Sciex, USA) and significant changes were defined by variable importance (VIP) >1 and p value <0.05. Metabolic pathway analysis was carried out by searching KEGG (http://www.genome.jp/kegg/) and MetaboAnalyst (http://www.metaboanalyst.ca/) commercial databases based on the identified differential metabolites altered by E2 exposure.
19, with an injection volume of 4 μL. Electrospray ionization source (ESI) conditions were set as follows: ion source gas 1 at 60 psi, ion source gas 2 at 60 psi, curtain gas at 35 psi, source temperature at 650 °C and ion spray voltage floating (ISVF) at −4000 V in negative modes. A triple TOF mass spectrometer was used for its ability to acquire MS/MS spectra in an information-dependent basis (IDA) mode.37,38 Differential metabolites were identified using MS/MS acquisition software (Analyst TF 1.7, AB Sciex, USA) and significant changes were defined by variable importance (VIP) >1 and p value <0.05. Metabolic pathway analysis was carried out by searching KEGG (http://www.genome.jp/kegg/) and MetaboAnalyst (http://www.metaboanalyst.ca/) commercial databases based on the identified differential metabolites altered by E2 exposure.
2.6. Statistical analysis
Significance was assessed by two-way univariate analysis of variance (ANOVA), and a value of p < 0.05 was considered significant. Principal component analysis (PCA) and bubble plots were adopted for metabolite visualization using SIMCA software (V14.1, Sweden).39 The Pearson correlation between the utilization of TOC and E2 was determined using SPSS software (version 22).3. Results
3.1. E2 removal profiles
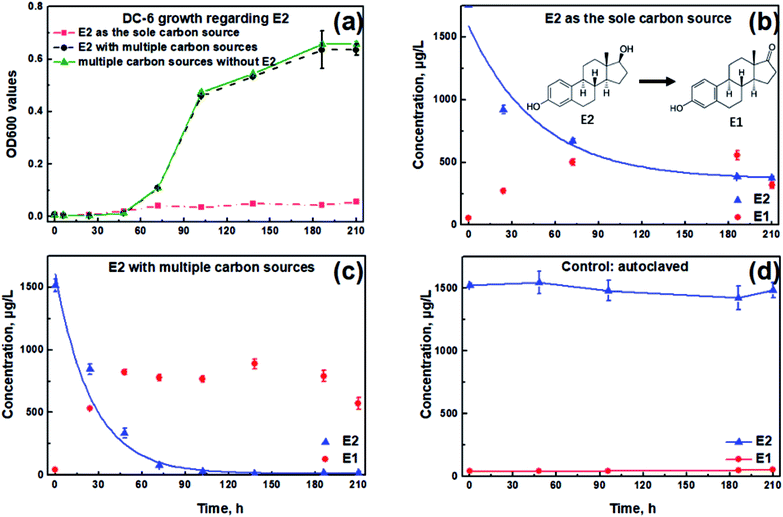
OD600 values remained relatively low throughout the experiment when using E2 as the sole carbon source, indicating that Sphingomonas sp. had little growth (Fig. 1a). However, when supplemented with other carbon sources, OD600 values reached 0.64 at 210 h, one order of magnitude higher than E2 as the sole carbon source. In addition, the OD value in the presence of E2 was slightly lower than the case without E2 at the end of the test (p > 0.05, Fig. 1a).As shown in Fig. 1b, a total E2 removal efficiency of 78.7% was achieved at 210 h in the sole E2 condition. By contrast, up to 98.8% of the spiked E2 could be removed in the presence of additional carbon substrates (Fig. 1c), higher by 20.1% compared with that when E2 existed alone. Among the zero-, first- and second-order kinetics analyses, the first-order exhibited the best fit for the E2 removal without (K = 0.008 h−1, r2 = 0.778) and with (K = 0.027 h−1, r2 = 0.785) carbon additions, respectively. According to the first-order reaction kinetics, the E2 removal constant was 0.008 h−1 when E2 existed alone, which was significantly lower than that in the presence of other carbon sources (0.027 h−1, p < 0.05). Simultaneously, it was observed that E1 accumulated as the intermediate metabolite and then showed a downward trend at the end of the test both in Fig. 1b and c.
To preclude the E2 disappearance due to non-biological processes, Sphingomonas sp. was totally inactivated by autoclave treatment. Concentrations of E2 and E1 remained almost unchanged during the entire degradation period (Fig. 1d). Therefore, it could be concluded that the concentration reduction of E2 in this study was due to the biodegradation process by the bacterium.
E2 removal by Sphingomonas sp. was studied together with TOC changes under the addition of glucose, sodium succinate and sodium acetate to uncover the relationship between TOC utilization and E2 removal. Initially, E2 was biodegraded at a relatively higher rate under a higher TOC supply with a removal rate of 27.9 ± 0.3 μg (L−1 h−1) and 21.3 ± 0.1 μg (L−1 h−1) during the first and second 24 h, respectively (Table S1†). During the third 24 h, the removal rate of E2 decreased to 10.6 μg (L−1 h−1) (Table S1†), only half of that during the first 48 h. From 102 h, E2 degradation slowed down steeply, and only 11.8 μg L−1 E2 in total was removed during the remaining 102 h. Interestingly, similar trends of TOC changes were observed, slowly declining to 0 g L−1 until 210 h with an initially rapid decrease within the first 72 h (Fig. 2). Further, a significant positive correlation via Pearson analysis was found between the utilization of TOC and E2 (r = 0.93, p < 0.05).
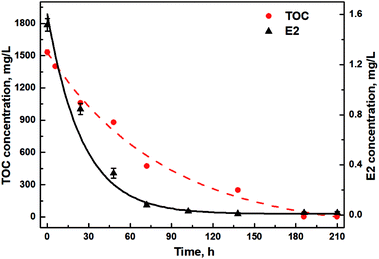 | ||
| Fig. 2 Concentration changes of E2 and TOC in the presence of diverse carbon sources. Lines are exponential fitting curves. | ||
3.2. E2 removal under different glucose concentrations
In view of the strong correlation between the utilization of TOC and E2, degradation experiments by Sphingomonas sp. under glucose concentrations of 100, 500 and 5000 mg L−1 were further conducted to find out the effects of organic carbon concentrations on E2 removal. Glucose was chosen as a typical carbon rather than using multiple carbon sources. Glucose is the most important source of energy metabolism in all organisms, and thus the effect of organic carbon concentration on E2 removal can be easily and clearly revealed. As shown in Fig. 3, E2 removal constants were 0.034 h−1, 0.036 h−1 and 0.034 h−1 for glucose concentrations of 100 mg L−1, 500 mg L−1 and 5000 mg L−1 respectively, without significant differences among them. Additionally, the total E2 removal efficiencies were also similar with no significant differences (99.1%, 99.5% and 99.0% for the three groups, respectively). Meanwhile, E1 accumulated rapidly during 0–48 h and had a slight decline afterwards for all groups.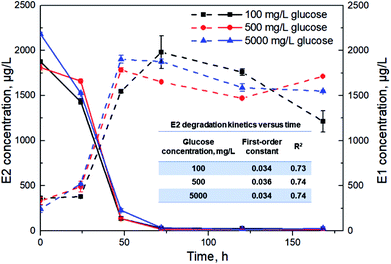 | ||
| Fig. 3 E2 degradation (solid lines) and E1 accumulation (dashed lines) kinetics by Sphingomonas sp. under different glucose concentrations. Error bars represent standard deviation (n = 2). | ||
3.3. Differential intracellular metabolites
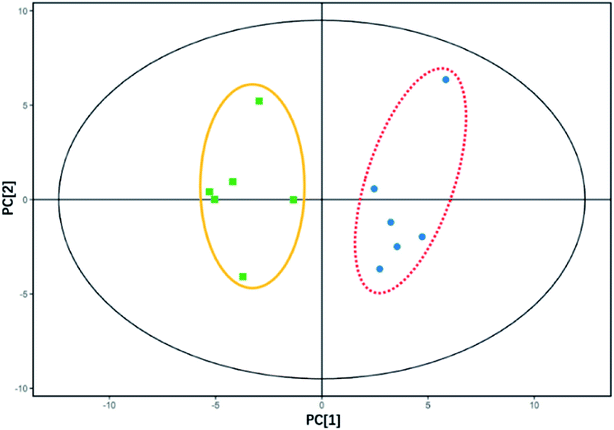
Intracellular metabolites with and without E2 addition under the same carbon and nitrogen backgrounds were clearly separated by PCA where analogous samples gathered (Fig. 4). As can be seen from the figure, an obvious discrimination in intracellular metabolites between samples in the two groups was acquired based on the scores of the first (PC1) and the second (PC2) ranked principal components (p < 0.05), indicating E2 had an effect on the bacterial intracellular metabolites. A total of 46 metabolites can be quantitatively identified in E2-grown Sphingomonas sp. by mapping with the MS/MS database with significant changes (VIP > 1, p < 0.05), 11 up-regulated and 35 down-regulated compared to those without the E2 addition. Up-regulation and down-regulation are processes in which cells increase and decrease the quantity of a cellular component (metabolites in this study), respectively. It is reasonable to assign all of the changes in the production of intracellular metabolites to the E2 addition in the growth solution.3.4. Intracellular metabolomics of E2 degradation
As listed in Table 1, 12 differential expressed metabolites (VIP > 1, p < 0.05) were found to be involved in the bacterial metabolic pathways by searching the MetaboAnalyst database as listed in Table 1. Cross-examination of metabolites (hypoxanthine, deoxyadenosine, adenosine, inosine, cytosine, cytidine and deoxycytidine) involved in purine and pyrimidine metabolism showed that the nucleotide metabolic level was depressed under the exposure to E2. Especially, as shown in Fig. S1,† the metabolism of purine and pyrimidine was most affected by E2 addition. As for lipid metabolism, glycerophospholipid metabolism and biosynthesis of unsaturated fatty acids were down-regulated, while arachidonic acid metabolism was up-regulated. One of the most striking results was the depressed isomaltose and maltose, which are two influential disaccharide substances in starch and sucrose metabolism. It is therefore likely that microbial carbohydrate metabolism was depressed to a certain degree as a response to exposure to E2. Intriguingly, betaine, maltotriose and D-maltose involved in ATP-binding cassette (ABC) transporters (one of the largest known protein families for the membrane transport in environmental information processing) were down-regulated. Meanwhile, choline in glycine, serine and threonine metabolism (assigned to amino acid metabolism) was significantly up-regulated. This was another metabolic pathway that was most affected by E2 in addition to the metabolism of purine and pyrimidine (Fig. S1†).| Metabolite name | KEGG ID | Trends | Pathway description | Function |
|---|---|---|---|---|
| a ABC is the abbreviation of the ATP-binding cassette. Signs of ↑/↓ denote significantly up-regulated/down-regulated metabolites, respectively, compared with that without E2 addition. | ||||
| Hypoxanthine | C00262 | ↓ | Purine metabolism | Nucleotide metabolism |
| Deoxyadenosine | C00559 | ↓ | ||
| Adenosine | C00212 | ↓ | ||
| Inosine | C00294 | ↓ | ||
| Cytosine | C00380 | ↓ | Pyrimidine metabolism | |
| Cytidine | C00475 | ↓ | ||
| Deoxycytidine | C00881 | ↓ | ||
| Triethanolamine | C06771 | ↓ | Glycerophospholipid metabolism | Lipid metabolism |
| Palmitic acid | C00249 | ↓ | Biosynthesis of unsaturated fatty acids | |
| Phosphatidylcholine | C00157 | ↑ | Arachidonic acid metabolism | |
| Arachidonic acid | C00219 | ↑ | ||
| Isomaltose | C00252 | ↓ | Starch and sucrose metabolism | Carbohydrate metabolism |
| D-Maltose/maltose | C00208 | ↓ | ||
| Choline | C00114 | ↑ | Glycine, serine and threonine metabolism | Amino acid metabolism |
| Betaine | C00719 | ↓ | ABC transporters | Membrane transport |
| Maltotriose | C01835 | ↓ | ||
| D-Maltose/maltose | C00208 | ↓ | ||
Given the above, the suppressed nucleotide, carbohydrate and membrane transport metabolisms, the accelerated amino acid metabolism and the changed lipid metabolism were obtained in the presence of E2. All of these were regarded as the response to E2 addition and participated in E2 degradation in the bacterium.
4. Discussion
4.1. Effects of carbon sources on E2 degradation
Sphingomonas species are widely distributed in both soil and aquatic environments with the ability to efficiently degrade organic micropollutants.17 Therefore, the Sphingomonas sp. had strong representativeness as an experimental object in this study. One degradation product of E2 by heterotrophic bacteria was previously found as E1.40,41 The build-up of E1 (Fig. 1b and c) confirmed the biodegradability of E2 by the bacterial strain. Although the decline in E1 concentrations occurred at the end of the test, it should be noted that E1 was continuously generated along with the E2 attenuation during the whole test period. Besides, E1 itself appears to be more recalcitrant to degrade than E2 according to published literature.42 Furthermore, previous studies on isolating estrogen-degrading strains concluded that only a small part of the isolates showed the ability to further degrade E1.11,43 The different abilities of E2-degrading bacteria to breakdown endocrine disrupting chemicals remain to be further studied.E2 was removed by 78.7% as the sole carbon source (Fig. 1b), and in this case OD600 values only changed from 0.008 to 0.057 (Fig. 1a), suggesting that little energy was generated during such a process under the initial E2 concentration up to 1.8 mg L−1, not to mention the much lower concentration of E2 in the environment. Under the condition of sufficient primary carbon sources, the Sphingomonas sp. began to grow rapidly after an initial lag phase of 48 h as expected (Fig. 1a). A more complete degradation of E2 was achieved as Sphingomonas sp. had access to the multiple, non-E2 carbon sources (E2 removal efficiency improved by 20.1%). Besides, a significant increase in E2 removal constant from 0.008 h−1 to 0.027 h−1 was recorded after the carbon addition. These results indicate that an adequate supply of the carbon substrate promoted the degradation of E2. When alternative carbon sources were available, Sphingomonas sp. would have the energy needed to eliminate excess E2 and it did so more efficiently.
It was clear that the E2 removal rate was much faster during the first 24 h than in the subsequent process regardless of the carbon sources (Table S1†), indicating that it was more challenging for Sphingomonas sp. to degrade E2 at lower concentration. One possible reason may be the reduced mass transfer of E2 under lower concentration, since diffusive mass transfer fairly influenced the removal rates of E2.23 Herein, mass transfer represents the net movement of E2 from bulk liquids to the surface of the bacterium. Another explanation might be that exposure to E2 above a certain threshold level likely caused physiological stress on the bacterium. In order to survive under such a harsh environment, strains capable of degrading E2 tended to become stronger in E2 metabolism by reducing E2 sharply down to a relatively “safe” concentration. This was in accordance with the theory of bacterial stress responses, “what doesn't kill them can make them stronger”.44
Interestingly, the E2 removal rate with external carbon during the first 24 h was 27.9 μg (L−1 h−1), significantly lower than that in the absence of other carbon (34.8 μg (L−1 h−1), p < 0.05, as shown in Table S1†). The result indicates that the bacterial strain utilized glucose, sodium succinate and sodium acetate prior to E2. Similarly, a previous study also suggested that heterotrophs preferentially metabolized glucose over E1.24
4.2. E2 removal with regard to carbon concentrations
Under the condition of the presented alternative carbon sources, there existed a significant positive relationship between TOC utilization and E2 degradation by the bacterium (Fig. 2). This indicates that organic carbon utilization can reflect the degradation rate of E2 to a certain degree. In this case, E2 removal can be enhanced by adjusting TOC utilization, which is of great significance for the efficient removal of the estrogen in complex environments. At the beginning of the degradation experiment, easy-to-degrade carbon substrates were exploited rapidly for bacterial maturation and mass expansion. Meanwhile, E2 was eliminated rapidly likely due to the stress responses. Right after the rapid degradation, the decrease in E2 concentration primarily resulted from the increased biomass despite the recalcitrance of E2 for metabolism.8 Later, the bacterial population underwent smooth growth from 96 h based on the OD600 values in Fig. 1a along with the observations of declined TOC and E2 utilization rates (Fig. 2). Importantly, E2 removal seemed to stop at the end of the test along with the scarce additional carbon source (Fig. 2), indicating that E2 under multiple carbon sources was mainly degraded via co-metabolism. It has previously been reported that accelerated E2 degradation occurred in the presence of glucose due to co-metabolism.7,45As can be seen in Fig. 3, E2 was removed at a faster rate at a high glucose level (5000 mg L−1) than the other two conditions (100 and 500 mg L−1) during the first 24 h. This also implies the utilization priority of the easy-to-degrade carbon substrate over E2 by Sphingomonas sp. especially under such an efficient carbon background. However, the total removal efficiency of E2 was not significantly affected by the changes in glucose concentrations, suggesting that the glucose concentration at 100 mg L−1 was probably sufficient to sustain bacterial growth. Higher yields of biomass to promote the E2 degradation might not be expected at higher glucose concentrations (500 mg L−1 or 5000 mg L−1) due to the limited growth space and other nutrients. This result has an important practical significance for the estrogen degradation under different environmental conditions.
4.3. Metabolism analysis
To the best of our knowledge, there are no published papers on intracellular metabolites during microbial E2 degradation using the method of metabolomics. It was found that E2 exposure had a profound and comprehensive effect on the metabolism of Sphingomonas sp. (Table 1). The suppressed nucleotide metabolism suggests a relatively slower degradation of DNA/RNA and clearance of the bacterial population than in the absence of E2. Especially, all differential metabolites detected were most intensively enriched in purine metabolism and pyrimidine metabolism (Fig. S1†), which highlighted the great importance of their involvement in the E2 metabolism. Changes in nucleotide metabolism were also detected previously in other estrogen-spiked samples.46,47 Five possible mechanisms involved in the intracellular action modes of E2, including direct genomic effects, indirect genomic effects, transcription factor activation, ligand-independent effects and rapid non-genomic effects, were previously reported.48,49 The present results thus gave direct evidence of the possible mechanisms since the former four modes ultimately had an impact on gene expression. Furthermore, E2 dosage as high as 1.8 mg L−1 in this study was likely toxic to the tested bacterial species, thus suppressing the nucleotide metabolism. Such toxicity was indicated in previous studies.50,51Moreover, the significantly down-regulated isomaltose and maltose suggested that E2 likely interfered with the carbohydrate metabolism of Sphingomonas sp., although the presence of alternative carbon sources in turn enhanced the E2 degradation (Fig. 1). Exposure to E2 above a certain threshold likely caused physiological stress on Sphingomonas sp., as described in Section 4.1. Carbohydrate metabolism as the way to obtain microbial energy was thus affected to some extent. In addition, the depression of carbohydrate metabolism was consistent with that increasing glucose concentration had no remarkable effect on removal kinetics as observed in Fig. 3. Moreover, it was previously demonstrated that52 the removal rate of sex steroid hormones by a bacterial strain Sphingomonas ZY3 increased to 87% after adding maltose. Yet, few studies have shown the suppressed carbohydrate metabolism caused by estrogen exposure at the bacterial level.47
By contrast, choline relative to amino acid metabolism was up-regulated by E2 exposure, indicating that metabolic end products of E2 might be converted for amino acid synthesis of glycine, serine and threonine in the bacterium. It has been suggested that the intermediate product E1 finally entered the tyrosine metabolism pathway for protein biosynthesis.26,27 The differences of specific amino acid metabolism involved here might result from the biodegradation divergences among different microbial strains. The elevated level of choline in this study was consistent with a previous study53 which reported that estrogen receptor α reprogrammed metabolism upon estrogen stimulation in animal cells including promotion in choline metabolism and phosphatidylcholine synthesis. The up-regulated phosphatidylcholine regarding lipid metabolism was also verified in this study (Table 1). Lipid changes were previously comprehensively detected in fish treated with E2 with phosphatidylcholine included.54 Phosphatidylcholine was proved to bind directly with vitellogenin,55 which was the highly specific biomarker for the contamination of estrogens in aquatic environments.56 Therefore, the up-regulated phosphatidylcholine detected in this study was convincible as an indicator of E2 degradation inside the bacteria. The suggested candidate indicator could be used to monitor the estrogen degradation by a microbial community. Still, further in-depth work needs to be done in this direction.
Microorganisms can alter their intake and transport systems to accommodate different nutritional conditions. It was found that E2 exposure provided the down-regulated ABC transporters for the membrane transport of betaine, maltotriose and D-maltose, which might be direct evidence for explaining why the E2 with reduced bioavailability could be degraded by the bacterium. The down-regulation of ABC transporters relative to the uptake and transportation of maltotriose and D-maltose was consistent with the depressed carbohydrate metabolism. Besides, the down-regulated betaine (the metabolite and derivative of choline) was in accordance with the up-regulated choline, since choline was more used for the amino acid synthesis of glycine, serine and threonine as discussed above. As E2 is hardly soluble in water, it cannot be transported through the water-filled channels. Thus, the specific trans-membrane channels allowing the diffusion of E2 in the outer membrane of Gram-negative bacteria need further in-depth exploration.
Although the concentration of E2 selected in this study differed from the typical concentration levels found in aquatic surroundings (ng L−1 to μg L−1), the present results provided an essential basis and understanding regarding E2 metabolic patterns by the bacterium. Extremely low concentrations of estrogens in aquatic environments usually make quantification difficult. Similarly, a high dosage of estrogen has previously been adopted for batch and continuous tests, ranging from 300 μg L−1 to 5 mg L−1.40,57,58 It should be mentioned that an organic compound could be used via metabolism by microbes as the carbon source to support growth; unless it is insufficient in amount, it may be mainly removed via co-metabolism. In the present study, both the extremely low OD600 with only E2 (Fig. 1a) and the positive relation of utilization of TOC and E2 (Fig. 2) indicate that E2 was mainly removed via co-metabolism, though E2 was spiked as high as 1.8 mg L−1. In this case, the high E2 dosage might not cause changes in the metabolomics analysis in this study. Besides, the unrealistically high E2 spiking concentration of 1.8 mg L−1 might influence its removal rate. It was previously reported that the estrogen degradation rate was slower at spiking concentrations above 100 μg L−1 than at the more environmentally relevant ng L−1 level.59 With information provided in this study, future research can be aimed at evaluating effects of low estrogen concentrations on bacterial samples within a long time span.
5. Conclusions
The results from this study demonstrated that easy-to-degrade carbon sources could preferentially be utilized over E2 by Sphingomonas sp., and the presence of alternative carbon sources accelerated the E2 degradation process. In addition, the co-metabolism of E2 correlated well with the TOC utilization by Sphingomonas sp. (p < 0.05). E2 exposure had a comprehensive effect on the bacterial metabolism, including the altered metabolism of lipid, nucleotide, carbohydrate, amino acid and membrane transport, which were thought to be responsible for E2 stimulation and involved in biodegrading E2. In particular, the up-regulated phosphatidylcholine can be presumably regarded as an indicator of E2 bacterial degradation. The present study could act as a model approach for E2 removal and metabolomics evaluation, and low levels of estrogen spiking concentration should be more addressed in future studies.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The research was financially supported by the National Natural Science Foundation of China (No. 31672468); National Key R&D Program of China (Project No. 2017YFD0701700); and Thousand Talents Program (Youth Project 2016).References
- C. P. Silva, M. Otero and V. Esteves, Processes for the elimination of estrogenic steroid hormones from water: a review, Environ. Pollut., 2012, 165, 38–58 CrossRef CAS PubMed.
- T. Hashimoto and T. Murakami, Removal and degradation characteristics of natural and synthetic estrogens by activated sludge in batch experiments, Water Res., 2009, 43, 573–582 CrossRef CAS PubMed.
- J. A. Herrera-Melian, R. Guedes-Alonso, A. Borreguero-Fabelo, J. J. Santana-Rodriguez and Z. Sosa-Ferrera, Study on the removal of hormones from domestic wastewaters with lab-scale constructed wetlands with different substrates and flow directions, Environ. Sci. Pollut. Res., 2018, 25, 20374–20384 CrossRef CAS PubMed.
- H. Dai, S. Gao, C. Lai, H. He, F. Han and X. Pan, Biochar enhanced microbial degradation of 17β-estradiol, Environ. Sci.: Processes Impacts, 2019, 21(10), 1736–1744 RSC.
- N. Elnwishy and N. Sedky, E2, an aquatic hazard worldwide, J. Agric. Sci., 2016, 8, 147 Search PubMed.
- B. Petrie, R. Barden and B. Kasprzyk-Hordern, A review on emerging contaminants in wastewaters and the environment: current knowledge, understudied areas and recommendations for future monitoring, Water Res., 2015, 72, 3–27 CrossRef CAS PubMed.
- C. P. Yu, R. A. Deeb and K. H. Chu, Microbial degradation of steroidal estrogens, Chemosphere, 2013, 91, 1225–1235 CrossRef CAS PubMed.
- J. P. Bagnall, A. Ito, E. J. McAdam, A. Soares, J. N. Lester and E. Cartmell, Resource dependent biodegradation of estrogens and the role of ammonia oxidising and heterotrophic bacteria, J. Hazard. Mater., 2012, 239–240, 56–63 CrossRef CAS PubMed.
- E. J. McAdam, J. P. Bagnall, Y. K. Koh, T. Y. Chiu, S. Pollard, M. D. Scrimshaw, J. N. Lester and E. Cartmell, Removal of steroid estrogens in carbonaceous and nitrifying activated sludge processes, Chemosphere, 2010, 81, 1–6 CrossRef CAS PubMed.
- M. R. Abargues, J. Ferrer, A. Bouzas and A. Seco, Fate of endocrine disruptor compounds in an anaerobic membrane bioreactor (AnMBR) coupled to an activated sludge reactor, Environ. Sci.: Water Res. Technol., 2018, 4, 226–233 RSC.
- C. P. Yu, H. Roh and K. H. Chu, 17beta-estradiol-degrading bacteria isolated from activated sludge, Environ. Sci. Technol., 2007, 41, 486–492 CrossRef CAS PubMed.
- S. M. Blunt, M. J. Benotti, M. R. Rosen, B. P. Hedlund and D. P. Moser, Reversible reduction of estrone to 17beta-estradiol by Rhizobium, Sphingopyxis, and Pseudomanas isolates from the Las Vegas Wash, J. Environ. Qual., 2017, 46, 281–287 CrossRef CAS PubMed.
- S. Sathyamoorthy, K. Chandran and C. A. Ramsburg, Biodegradation and cometabolic modeling of selected beta blockers during ammonia oxidation, Environ. Sci. Technol., 2013, 47, 12835–12843 CrossRef CAS PubMed.
- W. Ismail and Y.-R. Chiang, Oxic and anoxic metabolism of steroids by bacteria, J. Biorem. Biodegrad., 2011, S1, 001 Search PubMed.
- H. Roh and K. H. Chu, A 17beta-estradiol-utilizing bacterium, Sphingomonas strain KC8: part I – characterization and abundance in wastewater treatment plants, Environ. Sci. Technol., 2010, 44, 4943–4950 CrossRef CAS PubMed.
- A. Hu, J. He, K. H. Chu and C. P. Yu, Genome sequence of the 17beta-estradiol-utilizing bacterium Sphingomonas strain KC8, J. Bacteriol., 2011, 193, 4266–4267 CrossRef CAS PubMed.
- A. R. Johnsen, M. Hausner, A. Schnell and S. Wuertz, Evaluation of fluorescently labeled lectins for noninvasive localization of extracellular polymeric substances in Sphingomonas biofilms, Appl. Environ. Microbiol., 2000, 66, 3487–3491 CrossRef CAS PubMed.
- N. H. Tran, T. Urase, H. H. Ngo, J. Hu and S. L. Ong, Insight into metabolic and cometabolic activities of autotrophic and heterotrophic microorganisms in the biodegradation of emerging trace organic contaminants, Bioresour. Technol., 2013, 146, 721–731 CrossRef CAS.
- A. R. Ribeiro, M. F. Carvalho, C. M. M. Afonso, M. E. Tiritan and P. M. L. Castro, Microbial degradation of 17-estradiol and 17-ethinylestradiol followed by a validated HPLC-DAD method, J. Environ. Sci. Health, Part B, 2010, 45, 265–273 CrossRef CAS PubMed.
- D. T. Tan, W. A. Arnold and P. J. Novak, Impact of organic carbon on the biodegradation of estrone in mixed culture systems, Environ. Sci. Technol., 2013, 47, 12359–12365 CrossRef CAS PubMed.
- P. Arulazhagan and N. Vasudevan, Role of nutrients in the utilization of polycyclic aromatic hydrocarbons by halotolerant bacterial strain, J. Environ. Sci., 2011, 23, 282–287 CrossRef CAS.
- H. J. Zhang, L. Wang, Y. Li, P. F. Wang and C. Wang, Background nutrients and bacterial community evolution determine C-13-17 beta-estradiol mineralization in lake sediment microcosms, Sci. Total Environ., 2019, 651, 2304–2311 CrossRef CAS PubMed.
- A. Joss, H. Andersen, T. Ternes, P. R. Richle and H. Siegrist, Removal of estrogens in municipal wastewater treatment under aerobic and anaerobic conditions: consequences for plant optimization, Environ. Sci. Technol., 2004, 38, 3047–3055 CrossRef CAS PubMed.
- Y. X. Ren, K. Nakano, M. Nomura, N. Chiba and O. Nishimura, Effects of bacterial activity on estrogen removal in nitrifying activated sludge, Water Res., 2007, 41, 3089–3096 CrossRef CAS PubMed.
- K. Wu, T. H. Lee, Y. L. Chen, Y. S. Wang, P. H. Wang, C. P. Yu, K. H. Chu and Y. R. Chiang, Metabolites Involved in Aerobic Degradation of the A and B Rings of Estrogen, Appl. Environ. Microbiol., 2019, 85(3), e02223-18 CrossRef PubMed.
- Z. Du, Y. Chen and X. Li, Quantitative proteomic analyses of the microbial degradation of estrone under various background nitrogen and carbon conditions, Water Res., 2017, 123, 361–368 CrossRef CAS PubMed.
- Z. Li, R. Nandakumar, N. Madayiputhiya and X. Li, Proteomic analysis of 17beta-estradiol degradation by Stenotrophomonas maltophilia, Environ. Sci. Technol., 2012, 46, 5947–5955 CrossRef CAS PubMed.
- J. Xu, L. Zhang, J. L. Hou, X. L. Wang, H. Liu, D. N. Zheng and R. B. Liang, iTRAQ-based quantitative proteomic analysis of the global response to 17beta-estradiol in estrogen-degradation strain Pseudomonas putida SJTE-1, Sci. Rep., 2017, 7, 41682 CrossRef CAS.
- J. K. Nicholson and J. C. Lindon, Systems biology - metabonomics, Nature, 2008, 455, 1054–1056 CrossRef CAS PubMed.
- Y. S. Keum, J. S. Seo, Q. X. Li and J. H. Kim, Comparative metabolomic analysis of Sinorhizobium sp C4 during the degradation of phenanthrene, Appl. Microbiol. Biotechnol., 2008, 80, 863–872 CrossRef CAS PubMed.
- N. Pfenning, Rhodocyclus purpureus gen. nov. and sp. nov., a ringshaped, vitamin B12-requiring member of the Family Rhodospirillaceae, Int. J. Syst. Bacteriol., 1978, 28, 283–288 CrossRef.
- A. Guidhe, F. A. Ansari, P. Singh and F. Bux, Heterotrophic cultivation of microalgae using aquaculture wastewater: a biorefinery concept for biomass production and nutrient remediation, Ecological Engineering, 2017, 99, 47–53 CrossRef.
- Y. J. Feng, W. H. He, J. Liu, X. Wang, Y. P. Qu and N. Q. Ren, A horizontal plug flow and stackable pilot microbial fuel cell for municipal wastewater treatment, Bioresour. Technol., 2014, 156, 132–138 CrossRef CAS.
- I. Othman, A. N. Anuar, Z. Ujang, N. H. Rosman, H. Harun and S. Chelliapan, Livestock wastewater treatment using aerobic granular sludge, Bioresour. Technol., 2013, 133, 630–634 CrossRef CAS PubMed.
- L. Fernandez, A. Louvado, V. I. Esteves, N. C. Gomes, A. Almeida and A. Cunha, Biodegradation of 17beta-estradiol by bacteria isolated from deep sea sediments in aerobic and anaerobic media, J. Hazard. Mater., 2017, 323, 359–366 CrossRef CAS.
- J. Ivanisevic, D. Elias, H. Deguchi, P. M. Averell, M. Kurczy, C. H. Johnson, R. Tautenhahn, Z. J. Zhu, J. Watrous, M. Jain, J. Griffin, G. J. Patti and G. Siuzdak, Arteriovenous blood metabolomics: a readout of intra-tissue metabostasis, Sci. Rep., 2015, 5, 12757 CrossRef CAS PubMed.
- E. J. Want, I. D. Wilson, H. Gika, G. Theodoridis, R. S. Plumb, J. Shockcor, E. Holmes and J. K. Nicholson, Global metabolic profiling procedures for urine using UPLC-MS, Nat. Protoc., 2010, 5, 1005–1018 CrossRef CAS PubMed.
- W. B. Dunn, D. Broadhurst, P. Begley, E. Zelena, S. Francis-McIntyre, N. Anderson, M. Brown, J. D. Knowles, A. Halsall, J. N. Haselden, A. W. Nicholls, I. D. Wilson, D. B. Kell, R. Goodacre and The Human Serum Metabolome (HUSERMET) Consortium, Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry, Nat. Protoc., 2011, 6, 1060–1083 CrossRef CAS PubMed.
- S. Wiklund, E. Johansson, L. Sjostrom, E. J. Mellerowicz, U. Edlund, J. P. Shockcor, J. Gottfries, T. Moritz and J. Trygg, Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models, Anal. Chem., 2008, 80, 115–122 CrossRef CAS PubMed.
- J. Shi, S. Fujisawa, S. Nakai and M. Hosomi, Biodegradation of natural and synthetic estrogens by nitrifying activated sludge and ammonia-oxidizing bacterium Nitrosomonas europaea, Water Res., 2004, 38, 2323–2330 CrossRef CAS PubMed.
- H. B. Lee and D. Liu, Degradation of 17 beta-estradiol and its metabolites by sewage bacteria, Water, Air, Soil Pollut., 2002, 134, 353–368 CrossRef CAS.
- M. Isabelle, R. Villemur, P. Juteau and F. Lepine, Isolation of estrogen-degrading bacteria from an activated sludge bioreactor treating swine waste, including a strain that converts estrone to beta-estradiol, Can. J. Microbiol., 2011, 57, 559–568 CrossRef PubMed.
- J. X. Ke, W. Q. Zhuang, K. Y. H. Gin, M. Reinhard, L. T. Hoon and J. H. Tay, Characterization of estrogen-degrading bacteria isolated from an artificial sandy aquifer with ultrafiltered secondary effluent as the medium, Appl. Microbiol. Biotechnol., 2007, 75, 1163–1171 CrossRef CAS PubMed.
- K. J. Boor, Bacterial stress responses: what doesn't kill them can make them stronger, PLoS Biol., 2006, 4, 18–20 CrossRef CAS PubMed.
- B. Stumpe and B. Marschner, Factors controlling the biodegradation of 17 beta-estradiol, estrone and 17 alpha-ethinylestradiol in different natural soils, Chemosphere, 2009, 74, 556–562 CrossRef CAS PubMed.
- Y. L. Chen, C. P. Yu, T. H. Lee, K. S. Goh, K. H. Chu, P. H. Wang, W. Ismail, C. J. Shih and Y. R. Chiang, Biochemical mechanisms and catabolic enzymes involved in bacterial estrogen degradation pathways, Cell Chem. Biol., 2017, 24, 1–13 CrossRef PubMed.
- C. Bereketoglu, K. Y. Arga, S. Eraslan and B. Mertoglu, Genome reprogramming in Saccharomyces cerevisiae upon nonylphenol exposure, Physiol. Genomics, 2017, 49, 549–566 CrossRef CAS PubMed.
- P. I. S. Pinto, M. D. Estevao and D. M. Power, Effects of estrogens and estrogenic disrupting compounds on fish mineralized tissues, Mar. Drugs, 2014, 12, 4474–4494 CrossRef PubMed.
- J. M. Hall, J. F. Couse and K. S. Korach, The multifaceted mechanisms of estradiol and estrogen receptor signaling, J. Biol. Chem., 2001, 276, 36869–36872 CrossRef CAS PubMed.
- M. B. Gu, J. Min and E. J. Kim, Toxicity monitoring and classification of endocrine disrupting chemicals (EDCs) using recombinant bioluminescent bacteria, Chemosphere, 2002, 46, 289–294 CrossRef CAS PubMed.
- H. Roh, N. Subramanya, F. M. Zhao, C. P. Yu, J. Sandt and K. H. Chu, Biodegradation potential of wastewater micropollutants by ammonia-oxidizing bacteria, Chemosphere, 2009, 77, 1084–1089 CrossRef CAS PubMed.
- S. Ji, Z. Liu, Z. Liu and H. Ren, Isolation and characterization of a bacterial strain that efficiently degrades sex steroid hormones, Front. Environ. Sci. Eng. China, 2007, 1, 325–328 CrossRef.
- M. Jia, T. Andreassen, L. Jensen, T. F. Bathen, I. Sinha, H. Gao, C. Y. Zhao, L. A. Haldosen, Y. H. Cao, L. Girnita, S. A. Moestue and K. Dahlman-Wright, Estrogen receptor a promotes breast cancer by reprogramming choline metabolism, Cancer Res., 2016, 76, 5634–5646 CrossRef CAS PubMed.
- F. Mercure, A. C. Holloway, D. R. Tocher, M. A. Sheridan, G. Van der Kraak and J. F. Leatherland, Influence of plasma lipid changes in response to 17beta-oestradiol stimulation on plasma growth hormone, somatostatin, and thyroid hormone levels in immature rainbow trout, J. Fish Biol., 2001, 59, 605–615 CAS.
- H. Havukainen, D. Munch, A. Baumann, S. Zhong, O. Halskau, M. Krogsgaard and G. V. Amdam, Vitellogenin recognizes cell damage through membrane binding and shields living cells from reactive oxygen species, J. Biol. Chem., 2013, 288, 28369–28381 CrossRef CAS.
- J. P. Sumpter and S. Jobling, Vitellogenesis as a biomarker for estrogenic contamination of the aquatic environment, Environ. Health Perspect., 1995, 103, 173–178 CAS.
- C. P. Czajka and K. L. Londry, Anaerobic biotransformation of estrogens, Sci. Total Environ., 2006, 367, 932–941 CrossRef CAS PubMed.
- T. Yi and W. F. Harper, The link between nitrification and biotransformation of 17 alpha-ethinylestradiol, Environ. Sci. Technol., 2007, 41, 4311–4316 CrossRef CAS.
- N. Xu, A. C. Johnson, M. D. Jurgens, N. R. Llewellyn, N. P. Hankins and R. C. Darton, Estrogen concentration affects its biodegradation rate in activated sludge, Environ. Toxicol. Chem., 2009, 28, 2263–2270 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9em00438f |
| This journal is © The Royal Society of Chemistry 2020 |