Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Low carbon strategies for sustainable bio-alkane gas production and renewable energy†
Mohamed
Amer‡
 a,
Emilia Z.
Wojcik‡
a,
Emilia Z.
Wojcik‡
 a,
Chenhao
Sun‡
a,
Robin
Hoeven‡
a,
Chenhao
Sun‡
a,
Robin
Hoeven‡
 ab,
John M. X.
Hughes‡
a,
Matthew
Faulkner‡
ab,
John M. X.
Hughes‡
a,
Matthew
Faulkner‡
 a,
Ian Sofian
Yunus
c,
Shirley
Tait
a,
Linus O.
Johannissen
a,
Ian Sofian
Yunus
c,
Shirley
Tait
a,
Linus O.
Johannissen
 a,
Samantha J. O.
Hardman
a,
Derren J.
Heyes
a,
Guo-Qiang
Chen
ad,
Michael H.
Smith
b,
Patrik R.
Jones
c,
Helen S.
Toogood
a,
Samantha J. O.
Hardman
a,
Derren J.
Heyes
a,
Guo-Qiang
Chen
ad,
Michael H.
Smith
b,
Patrik R.
Jones
c,
Helen S.
Toogood
 a and
Nigel S.
Scrutton
a and
Nigel S.
Scrutton
 *abd
*abd
aEPSRC/BBSRC Future Biomanufacturing Research Hub, BBSRC/EPSRC Synthetic Biology Research Centre, Manchester Institute of Biotechnology and School of Chemistry, The University of Manchester, Manchester, M1 7DN, UK. E-mail: nigel.scrutton@manchester.ac.uk
bC3 Biotechnologies Ltd, The Railway Goods Yard, Middleton-in-Lonsdale, Lancashire LA6 2NF, UK
cDepartment of Life Sciences, Imperial College London, Sir Alexander Fleming Building, London SW7 2AZ, UK
dSchool of Life Sciences, Tsinghua University, 100084 Beijing, China
First published on 19th May 2020
Abstract
Propane and butane are the main constituents of liquefied petroleum gas and are used extensively for transport and domestic use. They are clean burning fuels, suitable for the development of low carbon footprint fuel and energy policies. Here, we present blueprints for the production of bio-alkane gas (propane and butane) through the conversion of waste volatile fatty acids by bacterial culture. We show that bio-propane and bio-butane can be produced photo-catalytically by bioengineered strains of E. coli and Halomonas (in non-sterile seawater) using fatty acids derived from biomass or industrial waste, and by Synechocystis (using carbon dioxide as feedstock). Scaled production using available infrastructure is calculated to be economically feasible using Halomonas. These fuel generation routes could be deployed rapidly, in both advanced and developing countries, and contribute to energy security to meet global carbon management targets and clean air directives.
Broader contextThere is an urgent need to develop sustainable and renewable biofuels to address the depletion of fossil fuels and the consequences of their combustion on climate change. Commercially viable bio-LPG production (propane and butane blends) would answer both concerns by reducing the demand on petroleum and natural gas usage, and improving air quality by utilising a cleaner-burning fuel. A secondary global concern is the disposal and/or recycling of organic waste, enabling sustainable energy capture and utilisation, and improvement in the environment and living conditions. Both of these concerns can be met by generating biologically-sourced alkane gases through cultivation of engineered microbial hosts fed on waste carbon sources. The microbial ‘chassis’ could be engineered to utilise specific waste types (e.g. biodiesel waste or salted milk whey), and low cost bioprocess ‘hubs’ could be localised at existing waste generating industries. This would increase the recycling of industrial waste, thereby reducing the industries carbon footprint, improving waste management strategies and generating further income. |
Introduction
The race to develop economically viable microbial biofuels is a consequence of the pressing need to reduce carbon emissions, improve air quality and implement renewable and sustainable fuel strategies.1 Current over reliance on fossil fuels has led to concerns over energy security and climate change. This has driven new policies to restrict greenhouse gas emissions, increase the recycling of waste biomaterials and accelerate the delivery of the bioeconomy.2 Effective sustainable biofuels strategies would comprise scalable production of transportable and clean-burning fuels derived from a robust microbial host, cultivated on renewable waste biomass or industrial waste streams, with minimal downstream processing, and (limited) use of fresh water. Embedding production techniques within existing infrastructures for waste processing and fuel distribution would minimise expenditure. Tailoring to specific waste streams would support local economies, waste management, energy self-sufficiency, and carbon reduction in both advanced and developing countries.Propane is an ideal biofuel. This simple hydrocarbon gas is a highly efficient, clean-burning fuel requiring little energy to store in a liquefied state.2 It is currently obtained from natural gas and petroleum refining. Propane is the third most widely used transportation fuel (20 million tons per annum globally), with existing infrastructure and global markets well established. It is also used for domestic heating and cooking, non-greenhouse gas refrigerants and aerosol propellants.3 Its ‘drop-in’ nature boosts the calorific value of current methane/biogas supplies, with lower energy requirements for liquefaction and storage. The only existing alternative production method is the Nesté process, an energy intensive, catalytic chemical conversion of biodiesel waste (glycerol) reliant on natural gas derived hydrogen.4 No natural biosynthetic routes to propane are known. Engineered biological pathways to propane have been developed based on decarbonylation of butyraldehyde incorporating natural or engineered variants of the enzyme aldehyde deformylating oxygenase (ADO).5–9 The low turnover number of ADO (∼3–5 h−1), however, limits implementation of these pathways in scaled bio-propane production.5,6,8
Here we describe blueprints for the scaled and economic production of bio-alkane gas (propane and butane, or ‘Bio-LPG’) using engineered forms of a recently discovered, blue light-dependent, fatty acid photodecarboxylase (FAP) that catalyzes decarboxylation of fatty acids to n-alkanes or n-alkenes (Fig. 1).10,11 We have taken a systems engineering approach to convert waste VFAs to bio-alkane gas in live bacterial cultures. The strategies we describe could enable environment-friendly in situ gas generation (e.g. in rural and/or arid communities), dependent on the availability of abundant waste resources, and implemented with CO2 capture. These low carbon strategies could provide economic, sustainable, secure and clean alternatives to extant petrochemical LPG supplies.
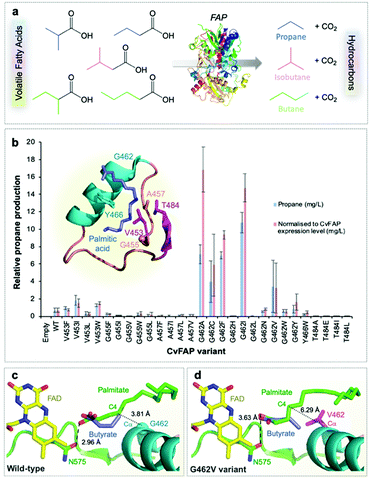 | ||
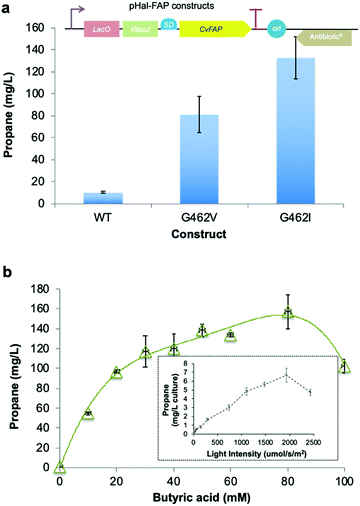
| Fig. 1 Structure-guided molecular engineering of CvFAP. (a) Photocatalytic gaseous hydrocarbon production from short chain volatile fatty acids. (b) Comparative propane production screen of variants of CvFAP in E. coli. Transformed E. coli cultures (three biological replicates) were grown at 37 °C in LB medium containing kanamycin (30 μg mL−1) to a density of OD600 ∼ 0.6–0.8. CvFAP expression was induced with IPTG (0.1 mM) and cultures were supplemented with 10 mM butyric acid. Triplicate aliquots (1 mL) of each culture were sealed into 5 mL glass vials and incubated at 30 °C for 16–18 h at 200 rpm, illuminated with a blue LED panel. Headspace gas was analysed for hydrocarbon content using a Micro GC. Data were normalized by dividing the propane titres (mg L−1 culture) by the relative protein concentration compared to the wild type (WT) enzyme (Fig. S2; ESI†). Error bars represent one standard deviation for triplicate biological repeats (n = 3). Inset: Structure of the palmitic acid binding region of CvFAP (PDB: 5NCC) shown as a cartoon with secondary structure colouring. Models of butyrate and palmitate in the active site of (c) wild-type and (d) G462V variant of CvFAP. The position of palmitate in the wild-type enzyme is crystallographically determined (PDB: 5NCC). The positions of the remaining ligands were determined by Autodock Vina, and mutagenesis to G462V was simulated using SwissPDBViewer 4.10. The protein is shown as a cartoon with secondary structure coloring, with selected residues shown as sticks. FAD, palmitate and butyrate are shown as atom-colored sticks with yellow, green and blue carbons, respectively. In panels c and d, the dashed line shows a hydrogen bond between palmitate and the wild-type enzyme, while the dotted lines indicate the distance between the C4 carbon of palmitate and the Cα atom of residue 462. All crystal structure images were generated in Pymol. | ||
Experimental
Materials, services and equipment
All chemicals and solvents were of analytical grade or better. Gene sequencing and oligonucleotide synthesis were performed by Eurofins MWG (Ebersberg, Germany). All oligonucleotide sequences can be found in Tables S2–S4 (ESI†). Gene synthesis was performed by Geneart (Thermo Fisher), with codon-optimization for E. coli or Synechocystis. The mounted high-power blue LEDs and LED drivers were from Thorlabs (Ely, U.K.), with spectra centered at 455 nm (bandwidth (FWHM) 18 nm, 1020 mW typical output) and 470 nm (FWHM 25 nm, 710 mW typical output). The custom-built LED blue light array had area of 396 cm2 of relatively consistent light intensity and a fixed average culture-to-LED distance of 8 cm (Fig. S1; ESI†). Light ‘intensity’ was measured with a Li-Cor light meter with a Quantum sensor in μmol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) photons
photons![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) m−2
m−2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s−1 (or μE), with background light value subtracted. The photobioreactor was a thermostatic flat panel FMT 150 (500 mL; Photon Systems Instruments, Czech Republic) with integral culture monitoring (OD 680/720 nm), pH and feeding control and an LED blue light panel (465 nm; maximum PPFD = 1648 μE photons).
s−1 (or μE), with background light value subtracted. The photobioreactor was a thermostatic flat panel FMT 150 (500 mL; Photon Systems Instruments, Czech Republic) with integral culture monitoring (OD 680/720 nm), pH and feeding control and an LED blue light panel (465 nm; maximum PPFD = 1648 μE photons).
E. coli strain BL21(DE3) was modified by chromosomal deletion of two aldehyde reductase genes yqhD and yjgB (BL21(DE3)ΔyqhD/ΔyjgB/KanR; GenBank: ACT44688.1 and AAA97166.1, respectively) as described previously.5 The kanamycin selection gene was removed using the Flp-mediated excision methodology (BL21(DE3)ΔyqhD/ΔyjgB).12Synechocystis sp. PCC 6803 was modified by chromosomal deletion of the acyl-ACP synthetase (Δaas) gene as described previously.13,14Halomonas strains TD0115 and TQ10, and modified pSEVA plasmids have been described previously.16Halomonas strain TQ10-MmP1 is a modified version of the TQ10 strain, which had been cured of a recombinant plasmid.
Gene synthesis, sub cloning and mutagenesis
The following N-terminally truncated (ΔN) FAP enzymes were synthesized (Table S1, ESI†): CvFAPWT from Chlorella variabilis NC64A10 (Genbank: A0A248QE08; ΔN-61 amino acids truncated); CcFAP from Chondrus crispus (UniProt: R7Q9C0; ΔN-50), ChFAP from Chrysochromulina sp. (UniProt: A0A0M0JFC3), CmFAP from Cyanidioschyzon merolae (UniProt: M1VK13; ΔN-64), CrFAP from Chlamydomonas reinhardtii (UniProt: A8JHB7; ΔN-31), CsFAP from Coccomyxa subellipsoidea (UniProt: I0YJ13; ΔN-43), GpFAP from Gonium pectorale (UniProt: A0A150GC51; ΔN-38) and PtFAP from Phaeodactylum tricornutum (UniProt: B7FSU6).10 Each gene was sub cloned into pETM11 with a N-His6-tag for rapid protein purification. The gene encoding thioesterase Tes4 from Bacteroides fragilis (UniProt: P0ADA1) was obtained from plasmid pET-TPC4, as described previously.5 For the valine to propane pathway, leucine 2-oxoglutarate transaminase from E. coli (ilvE; P0AB80); 3-hydroxypropionaldehyde dehydrogenase from E. coli (Hpad; P23883) and branched-chain keto acid decarboxylase from Lactococcus lactis (KdcA; Q6QBS4) were synthesised and sub-cloned into pET21b (C-His6-tag), pETM11 (N-His6-tag) and pET28b (N-His6-tag), respectively.Variant CvFAPG462V was generated by site-directed mutagenesis of the wild-type using the QuikChange whole plasmid synthesis protocol (Stratagene) with CloneAmp HiFi PCR premix (Clontech). Additional variants (e.g. G462N/W/L/C/I/F/A/H/Y and those at neighbouring positions; see Fig. 1) were generated using the Q5 and QuikChange site directed mutagenesis kits (New England Biolabs and Novagen, respectively). PCR products were analysed by agarose gel electrophoresis, followed by gel purification (NucleoSpin Gel), or purified using the PCR clean-up kit (Macherey-Nagel). Constructs were transformed into E. coli strain NEB5α (New England Biolabs) for plasmid recircularization and production. The presence of the mutations was confirmed by DNA sequencing followed by transformation into E. coli strains BL21(DE3) and BL21(DE3)ΔyqhD/ΔyjgB5 for functional expression studies.
Molecular modelling
Substrates palmitic and butyric acid were docked into chain A of the crystal structure of the palmitic acid bound CvFAP structure 5NCC using Autodock vina.17 Non-polar hydrogen assignment was performed using AutoDock Tools 1.5.6. A cubic search volume with 15 Å sides was defined with the coordinates of C6 of palmitic acid as the centre, and an exhaustiveness of fifty. Twenty conformations were analysed and the lowest-energy conformation with the substrate in the correct orientation (carboxylate pointing towards the FAD) was selected. Mutations were performed in SwissPDBViewer 4.10,18 using the exhaustive search function to identify the best rotamer for the mutated residue.Multi-enzyme construct generation
N-His6-CvFAPG462V was sub-cloned into plasmids pET21b and pBbA1c19 by PCR-mediated In-Fusion cloning. Plasmids were transformed into E. coli strain NEB5α, BL21(DE3) and BL21(DE3)ΔyqhD/ΔyjgB5 for functional expression studies. The multi-gene valine to propane construct was assembled with CvFAP variant G462I in pBbE1k with a single pTrc promoter (pTrc-ilvE-Hpad-KcdA-CvFAPG462I) by overlap extension PCR, with vector linearisation and insert(s) amplifications performed by PCR.Halomonas construct generation
CvFAPwt, CvFAPG462V and CvFAPG462I coding sequences were amplified from pETM11 (lacking His6-tag) and inserted (NcoI-XhoI) into Halomonas-compatible plasmid pHal2 downstream of the MmP1 IPTG-inducible phage T7-like RNA polymerase promoter. This promoter is composed of an optimized MmP1-lacO-RiboJ-RBS sequence16,20 and the CvFAP translation initiation site (bold) comprises part of an NdeI restriction site (underlined): TTTGTTTAACTTTAAGAAGGAGATATA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) . Promoter induction only occurs in Halomonas strain TQ10-MmP1, which contains the cognate chromosome-integrated MmP1 phage RNA polymerase gene.16,20 pHal2 is derived from pSEVA44121 and contains the pRO1600 broad host range replication origin; the pRP4 origin of conjugative transfer (oriT); and genes conferring spectinomycin/streptomycin and kanamycin resistance, for selection in Halomonas TQ10-MmP1 and in the E. coli conjugative donor strain S17-1,22 respectively. pHal2-CvFAP variants were introduced into Halomonas TQ10-MmP1 by conjugation as follows. Kanamycin-resistant transformed colonies of donor E. coli S17-7 were mixed with TQ10-MmP1 on YTS agar plates (yeast extract 5 g L−1, tryptone 10 g L−1, NaCl 30 g L−1, agar 15 g L−1), incubated overnight at 37 °C, then streaked onto YTN6 agar (5 g L−1 yeast extract; 10 g L−1 tryptone, 60 g L−1 NaCl, pH 9, 15 g L−1 agar) containing spectinomycin (50 μg mL−1) to select for Halomonas transconjugants. Plasmid content of the transconjugants was confirmed by DNA isolation, restriction mapping and sequencing.
. Promoter induction only occurs in Halomonas strain TQ10-MmP1, which contains the cognate chromosome-integrated MmP1 phage RNA polymerase gene.16,20 pHal2 is derived from pSEVA44121 and contains the pRO1600 broad host range replication origin; the pRP4 origin of conjugative transfer (oriT); and genes conferring spectinomycin/streptomycin and kanamycin resistance, for selection in Halomonas TQ10-MmP1 and in the E. coli conjugative donor strain S17-1,22 respectively. pHal2-CvFAP variants were introduced into Halomonas TQ10-MmP1 by conjugation as follows. Kanamycin-resistant transformed colonies of donor E. coli S17-7 were mixed with TQ10-MmP1 on YTS agar plates (yeast extract 5 g L−1, tryptone 10 g L−1, NaCl 30 g L−1, agar 15 g L−1), incubated overnight at 37 °C, then streaked onto YTN6 agar (5 g L−1 yeast extract; 10 g L−1 tryptone, 60 g L−1 NaCl, pH 9, 15 g L−1 agar) containing spectinomycin (50 μg mL−1) to select for Halomonas transconjugants. Plasmid content of the transconjugants was confirmed by DNA isolation, restriction mapping and sequencing.
Chromosomal insertion of the CvFAPG462I gene with the MmP1 promoter (pHal2-derived) and valine to propane pathways (IPTG-inducible and pPorin 69) were performed using a novel suicide vector (pSH) protocol based on previously published methods.23,24 The insertion plasmids contained the biocatalytic and chloramphenicol resistance (CamR) genes surrounded by homology arms, an I-SceI restriction site and a colE1 ori that is not compatible with replication in Halomonas. This plasmid was co-conjugated into Halomonas TQ10-MmP1 with a second spectinomycin-resistant plasmid (pSceI) expressing the gene for the restriction enzyme I-SceI. Expression of I-SceI enabled the linearization of pSH plasmids, facilitating chromosomal integration.23,24 The sites for integration were chosen based on the intergenic regions in Halomonas showing prior high recombinant protein expression.23 Successful integration was seen as growth of Halomonas on chloramphenicol-selective medium, as the pSH plasmid is not replicated. Integration was confirmed by colony PCR, genomic sequencing and in vivo propane production after pSceI plasmid curing.23,24
Synechocystis construct generation
Two versions of the non-His6-tagged C. variabilis CvFAPG462V gene with identical amino acid sequences were constructed in Synechocystis sp. PCC 6803 (fapG462V_Ecoli and fapG462V_cyano), differing by applying codon optimisation for E. coli and Synechocystis, respectively. For fapG462V_cyano, plasmid pIY505 (pJET-‘FAP’)14 variant G462V was generated by site-directed mutagenesis using the QuikChange whole plasmid synthesis protocol. To construct pJET-fapG462V_Ecoli, the non-His6-tagged gene in pETM11 was amplified by PCR and cloned into the blunt-ended pJET1.2 plasmid. The gene encoding Tes4 was amplified from construct pET-TPC414 and ligated into blunt pJET1.2 plasmid. To clone the mutated fap genes and/or tes4 genes into the erythromycin resistant RSF1010 plasmid, the Biopart Assembly Standard for Idempotent Cloning (BASIC) method was used as described previously.13,14,25 Gene expression was controlled using either the cobalt-inducible Pcoa or constitutive Ptrc (no lacI) promoters. Prefix and suffix linkers used to create the plasmids are listed in Tables S3 and S4 (ESI†). The following constructs were generated: (i) pIY894: Ptrc-fapG462V_cyano; (ii) pIY918: Ptrc-tes4, fapG462V_Ecoli; (iii) pIY906: Pcoa-tes4, fapG462V_cyano; and (iv) pIY845: Pcoa-tes4. Plasmid assembly was validated by DNA sequencing.Plasmids were transformed into the E. coli helper/cargo strain (100 μL; E. coli HB101 strain carrying the pRL623 and RSF1010 plasmids), conjugal strain (E. coli ED8654 strain carrying pRL443 plasmid)26 and Synechocystis sp. PCC 6803 lacking acyl-ACP synthetase (encoded by slr1609; Δaas strain; OD730 ∼ 1) using the tri-parental conjugation method described previously.13,14 Each strain had been pre-treated by washing with LB and BG11-Co medium for E. coli and Synechocystis, respectively, to remove antibiotics. The mixture was incubated for 2 h (30 °C, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μE), then spread onto BG11 agar plates without antibiotic, and incubated for 2 d (30 °C, 60 μE). Cells were scraped from the agar plate, resuspended in 500 μL of BG11-Co medium, and transferred onto a new agar plate containing 20 μg mL−1 erythromycin. Cells were allowed to grow for one week until colonies appeared.
μE), then spread onto BG11 agar plates without antibiotic, and incubated for 2 d (30 °C, 60 μE). Cells were scraped from the agar plate, resuspended in 500 μL of BG11-Co medium, and transferred onto a new agar plate containing 20 μg mL−1 erythromycin. Cells were allowed to grow for one week until colonies appeared.
Protein expression and lysate production
Wild type CvFAP-pETM11 homologues in E. coli BL21(DE3) were cultured in LB Broth Miller (500 mL; Formedium) containing 30 μg mL−1 kanamycin at 37 °C with 180 rpm shaking until OD600nm = 0.2. The temperature was maintained at 25 °C until OD600nm = 0.6–0.8. Recombinant protein production was induced with 50 μM IPTG, and maintained at 17 °C overnight. Cells were harvested by centrifugation (8950 × g, 4 °C, 10 min), and analysed for protein content using 12% SDS-PAGE gels (Mini-PROTEAN TGX Stain-Free Precast Gels, Bio-Rad). Protein gels were imaged using a BioRad Gel Doc EZ Imager and the relative protein band intensity was determined using the BioRad ImageLab software.Cell pellets were resuspended in lysis buffer (1.2–1.7 mL g−1 pellet; 50 mM Tris pH 8 containing 300 mM NaCl, 10 mM imidazole, 10% glycerol, 0.25 μg mL−1 lysozyme, 10 μg mL−1 DNase I and 1 × protease inhibitors) and sonicated for 20 minutes (20 s on, 60 s off; 30% amplitude). Cell-free lysate was prepared by centrifugation at 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 minutes at 4 °C. Lysate samples were analysed for recombinant protein expression by SDS PAGE.
000 × g for 30 minutes at 4 °C. Lysate samples were analysed for recombinant protein expression by SDS PAGE.
Hydrocarbon production
In vitro propane production reactions (1 mL) of FAP homologues were composed of cell-free lysate and butyric acid (0.4 mM) in sealed 4 mL vials. Reactions were incubated at 30 °C for 24 h at 180 rpm under illumination (blue LED; 455 nm). In vivo propane production of CvFAPWT and variants in E. coli was performed by the following general protocol: Cultures (20–100 mL) in LB medium containing kanamycin (30 μg mL−1; pETM11) or ampicillin (50 μg mL−1; pET21b) were incubated for 4–6 h (OD600 ∼ 1) at 37 °C and 180 rpm, followed by induction with IPTG (100 μM) and butyric acid supplementation (1–1000 mM; pH 6.8). Triplicate aliquots (1–5 mL) each of 3 biological replicate cultures were sealed into vials (4–20 mL) and incubated at 30 °C for 16–18 h at 200 rpm, illuminated continuously with an LED (455 nm or 470 nm). Comparative in vivo studies with 10 mM butyric, isobutyric, valeric, 2-methylbutyric and isovaleric acids were performed as above, with culture induction at OD600 of 0.6–0.8. For all in vitro and in vivo alkane gas production studies, the headspace gas was analysed for propane content using a Micro GC. Data is expressed as mg hydrocarbon production per litre of fermenting culture.Propane production in Halomonas was performed by a modified E. coli protocol as follows: Cultures were grown in phosphate buffered (50 mM K2HPO4 pH 6.6) YTN6 medium containing spectinomycin (50 μg mL−1) for 5 h at 37 °C and 180 rpm, followed by IPTG induction at OD ∼ 1.6. The remainder of the in vivo propane production process was performed as above, with butyric acid concentrations of 10–80 mM. For studies with the valine pathway, amino acids (up to 30 mg L−1) were added after induction in place of VFAs. Autolysed brewery yeast extract (waste amino acid source) was produced by culturing waste brewery barley grains from a North of England supplier in YPD medium (10 g L−1 yeast extract, 20 g L−1 peptone and 20 g L−1 glucose), followed by autolysis (2 h at 50 °C) and autoclaving. Waste milk medium was composed of milk whey containing 60 g L−1 NaCl and pH adjusted to 9.0.
Propane production in Synechocystis was performed in BG11 medium13,14 using a modified protocol as follows: Starter cultures in BG11 medium were incubated at 30 °C under 30 μE white LED until OD720nm reached 1.0 (∼4 days). Replicate culture aliquots (2 mL) were harvested by centrifugation and re-suspended in 1 mL BG11 medium supplemented with sodium bicarbonate (150 mM), cobalt(II) nitrate hexahydrate (100 μM; for Pcoa cultures only), 50 μg mL−1 kanamycin, and 20 μg mL−1 erythromycin at 30 °C ± butyric acid (10 mM). Cultures were sealed within 4 mL gas tight vials and incubated at 30 °C for 24–48 h under blue light (average 63 μE).
Halomonas cultivation
Cultures were grown in phosphate buffered YTN6 medium containing spectinomycin (50 μg mL−1) for 5 h at 37 °C and 180 rpm. Recombinant protein expression was induced with IPTG (0.1 mM; OD600 ∼ 1.6), and cultures were supplemented with butyric acid (0–100 mM, buffered at pH 6.6). Triplicate aliquots (1 mL) of cultures were sealed into 4 mL glass vials and incubated at 30 °C for 16–18 h at 200 rpm, illuminated with a blue LED panel. For studies with the Synechocystis extract, cultures (1 mL) were incubated post induction with lysed Synechocystis extract in place of butyric acid. Headspace gas was analysed for hydrocarbon content using a Micro GC.Photobioreactor cultivation was performed with high salt glycerol medium at pH 6.8 (5 g L−1 yeast extract, 1 g L−1 glycerol, 60 g L−1 NaCl, 50 μg mL−1 spectinomycin and 0.5 mL L−1 antifoam; 400 mL) in batch mode, pre-equilibrated at 30 °C with 60–100% stirring output. An overnight starter culture (10 mL) of Halomonas TQ10-MmP1 containing pHal2-CvFAPG462V was added, and the culture was maintained at 30 °C with an airflow rate of 1.21 L min−1, automated pH maintenance, culture optical density monitoring and ambient room lighting until mid-log phase (4–5 hours). Protein induction by IPTG (0.1 mM) was followed by sodium butyrate addition (60–80 mM pH ∼ 6.8) with continual blue light exposure (1656 μE) for ∼48 h. Propane production was monitored at 15 min intervals by automated headspace sampling using a Micro GC, while aqueous butyrate and glycerol depletion were quantified by HPLC.
Synechocystis cultivation
The photobioreactor (400 mL) was set up in batch mode with starter culture diluted 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in fresh BG11+ medium (BG11 pH 8.013,14 containing TES buffer and 1 g L−1 sodium thiosulphate) with 150 mM NaHCO3. The culture was maintained at 30 °C with maximal stirring, airflow of 1.21 L min−1, illumination by a white LED (30 μE), automated pH maintenance (1 M acetic acid in 2 × BG11+) and optical density monitoring (680 nm and 720 nm). After reaching OD720nm of ∼0.5, cobalt(II) nitrate hexahydrate (100 μM) was added as required, the warm white illumination was increased to 60 μE and the integral actinic blue LED light panel provided 500–750 μE blue light. The culture was maintained at 30 °C for 18–48 hours, fed and not fed respectively. Manual headspace sampling for propane content was performed by Micro GC, and butyrate depletion was quantified by HPLC.
1 in fresh BG11+ medium (BG11 pH 8.013,14 containing TES buffer and 1 g L−1 sodium thiosulphate) with 150 mM NaHCO3. The culture was maintained at 30 °C with maximal stirring, airflow of 1.21 L min−1, illumination by a white LED (30 μE), automated pH maintenance (1 M acetic acid in 2 × BG11+) and optical density monitoring (680 nm and 720 nm). After reaching OD720nm of ∼0.5, cobalt(II) nitrate hexahydrate (100 μM) was added as required, the warm white illumination was increased to 60 μE and the integral actinic blue LED light panel provided 500–750 μE blue light. The culture was maintained at 30 °C for 18–48 hours, fed and not fed respectively. Manual headspace sampling for propane content was performed by Micro GC, and butyrate depletion was quantified by HPLC.
Analytical techniques
Propane levels were determined by headspace injection using an Agilent 490 Micro GC, containing an Al2O3/KCl column, a thermal conductivity detector (TCD) and a heated injector (110 °C; 100 ms injection) using helium as the carrier gas (10.2 psi). During continuous monitoring mode, fermenter exhaust gases were dried by passage through an ice-cooled condenser prior to entering the Micro GC. Compounds were separated isothermally (100 °C) over 120 s under static pressure conditions, with a sampling frequency of 100 Hz.Aqueous culture metabolites (glycerol and butyric acid) were analysed by HPLC using an Agilent 1260 Infinity HPLC with a 1260 ALS autosampler, TCC SL column heater and a 1260 refractive index detector (RID). Cell-free culture supernatant samples (10 μL injection) were analysed isocratically on an Agilent Hi-Plex H column (300 × 7.7 mm; 5 mM H2SO4) at 60 °C with a flow rate of 0.7 mL min−1 for 40 minutes. Analyte concentrations determined by Micro GC or HPLC were calculated by comparing the peak areas to a standard curve generated under the same running conditions.
Techno-economic analysis (TEA)
In this analysis,27–30 a number of assumptions were made to provide projected economics and establish benchmarks to assess the state of technology based on current research performance. Design basis and costing data for non-fermentation unit operations were obtained from earlier studies and publications. Rigorous structural and parametric optimisation, heat integration and site analysis were not included at this stage.The main tasks utilized were: (a) Conceptual design of a pilot-scale continuous process as the base case. Each reactor has a 1 m3 working volume, with an inside battery limit (ISBL) plant (fermentation and propane purification) cost of a process ∼£500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. (b) Construction of a plant model in MATLAB, to calculate the mass & energy balance for the main process streams. (c) Creation of case studies from the base case. A total of 11 case studies (including the base case) were created by introducing additional assumptions with positive impacts on process economy (Note S1; ESI†). (d) Estimation of propane production costs, based on known process parameters at laboratory scale with financial assumptions, carbon footprint and minimum propane selling prices (MPSP). The design basis, specifications and assumptions regarding the unit operations are summarized in Note S1 (ESI†).
000. (b) Construction of a plant model in MATLAB, to calculate the mass & energy balance for the main process streams. (c) Creation of case studies from the base case. A total of 11 case studies (including the base case) were created by introducing additional assumptions with positive impacts on process economy (Note S1; ESI†). (d) Estimation of propane production costs, based on known process parameters at laboratory scale with financial assumptions, carbon footprint and minimum propane selling prices (MPSP). The design basis, specifications and assumptions regarding the unit operations are summarized in Note S1 (ESI†).
Results and discussion
Biocatalyst selection and redesign
We surmised that FAP could be engineered to increase the decarboxylation of butyric acid (and other short chain volatile fatty acids; VFAs)11 to form propane (and other hydrocarbon gases) to enable their production at scale (Fig. 1a). FAP from Chlorella variabilis NC64A has a reported reaction quantum yield of greater than 80% and it catalyzes a 1-step bioconversion of waste VFAs into alkanes.10 However, it has a reaction specificity that is strongly in favor of long chain fatty acids (C14–C18).10,11,31 We screened a range of previously identified potential FAP homologues10 for propane production with butyric acid. Direct kinetic comparison of each purified homologue was not possible as protein instability (aggregation) and flavin loss occurred to varying degrees during protein purification of each enzyme. Consequently, comparative propane production in live cells or cell-free lysates was the main approach taken in this study, with FAP concentrations estimated by quantification of the protein band density from SDS-PAGE (Fig. S2; ESI†). The presence of contaminating flavin and other chromophore (e.g. heme) -containing enzymes in cell lysates prevented active enzyme concentration determination by spectral analysis.The most suitable FAP enzymes for hydrocarbon gas production were identified as the Chlorella variabilis NC64A (CvFAP) and Chlamydomonas reinhardtii (CrFAP) homologues10,11,31 using biotransformation assays of cell-free extracts (Table S5; ESI†).10 We selected CvFAP as the target biocatalyst, given the availability of a three-dimensional crystal structure.10 Using structure-based engineering we identified regions in the natural substrate binding channel of CvFAP that when targeted by mutagenesis may be able to confer increased activity of the enzyme with short chain VFAs. We made a collection of twenty-eight CvFAP variants, targeting residues G462, G455, Y466, V453, T484 and A457 for substitution (Fig. 1b). A key substrate channel residue G462 was identified, and substituted for ten other residues (Val, Asn, Trp, Leu, Cys, Ile, Phe, Ala, His and Tyr; Fig. S2 and Table S6; ESI†). The side chains of these residues are in close proximity to bound palmitate in the co-crystal structure of CvFAPWT, and variants were designed to disfavor long chain fatty acid binding (Fig. 1b inset).10 Propane production in E. coli cultures exposed to blue light was measured and normalized according to each variant's relative expression level (Fig. S1 and S2; ESI†).
The most promising variants were found to be G462V and G462I (Fig. 1b; Table S6; ESI†), while variants at other positions produced less propane than CvFAPWT (Fig. 1b). Some variants appeared to show higher propane yields than G462V (e.g. G462A/F/C), but inconsistent expression levels of these variants within biological repeats made it difficult to accurately quantify this. Molecular docking simulations of CvFAPWT and variants G462V and G462I were performed using Autodock Vina17 to investigate the effects of the amino acid substitutions. This analysis predicted a 30–40-fold weaker binding of palmitate for the G462V/I variants (decrease in binding affinity of ∼2 kcal mol−1; Table S7; ESI†), with a small increase in binding of butyrate (∼30% tighter binding; Fig. 1c and d). This is consistent with the observation that purified CvFAPG462I exhibited an increase in propane production compared to wild-type (9-fold after 1.5 hours; Fig. S3; ESI†). Conversely, CvFAPWT showed a 2.6-fold higher rate of pentadecane production from palmitic acid after 45 minutes than CvFAPG462I.
Given the high variability in activity detected with some variants using cell lysates (Table S6; ESI†), more detailed activity assays were performed with purified CvFAP wild-type and G462I variant (normalised against FAD content; Fig. S3; ESI†). CvFAPG462I showed increased propane production (6.48 ± 1.04 μM propane) compared to wild-type enzyme (0.76 ± 0.31 μM propane), confirming the trend seen with the cell lysate screening data. We also determined that purified wild-type and variant CvFAPG462I are susceptible to rapid photoinactivation (Fig. S3; ESI†), so process scale production will require regeneration of the biocatalyst through continuous replenishment with live cell culture. This is possible by continuous biomass cultivation – an approach we have adopted in developing the design of bio-LPG production platforms described below.
Next, we investigated the effect of butyric acid concentration on propane production using live E. coli cultures. As butyric acid lowers the pH of LB medium (Fig. S4a; ESI†), we performed studies with the CvFAPG462I variant with pH control (buffered at pH 6.8; Table S8, ESI†). This showed propane production was maximal at around 50 mM butyric acid (Fig. S4b; ESI†). Under these conditions, the molar ratio of propane production to butyrate consumption was 1.9 to 1. Propane titres were also affected by the plasmid backbone used, and the position of fused purification tags to CvFAP. We observed a 6.4-fold increase in propane production using CvFAPG462V in plasmid pET21b compared to pETM11 (48.31 ± 2.66 mg L−1 culture). These levels are higher than maximal propane production observed previously with ADO (32 mg L−1 culture) using an E. coli strain upregulated for butyric acid production.5 This highlights the need to explore multiple plasmid backbones, and the effect of location and size of protein tags on gas production in vivo.
Tuneable bio-LPG blends
Photodecarboxylation of other short chain fatty acids (butyric, isobutyric, valeric, 2-methylbutyric and isovaleric acid) was investigated in vivo with wild-type and four CvFAP variants (G462V/A/I/F). The major gas produced, whether propane, butane or isobutane, was dependent on which VFA was supplied (Fig. 2a). Additional gases produced arose from naturally arising VFAs in cell extracts. Variant G462I showed the highest levels of gas production, especially with the branched chain substrates isovaleric and 2-methylbutyric acids (5-8-fold higher than with G462V; Table S9; ESI†). With CvFAPG462I, propane and butane production from butyric and valeric acid were less than 2-fold higher than with CvFAPG462V. Variants G462V and G462A generated similar levels of propane and butane, but G462A showed a greater variation in hydrocarbon titre (Fig. 2a; Table S9; ESI†). These data consolidate the finding that residue 462 is important in conferring activity with short chain carboxylic acids. The photodecarboxylation products of these VFAs can be used to make biological LPG-blends. As CvFAPG462V generated similar titres of propane and butane, this was chosen to explore the production of tuneable bio-LPG blends of varied composition.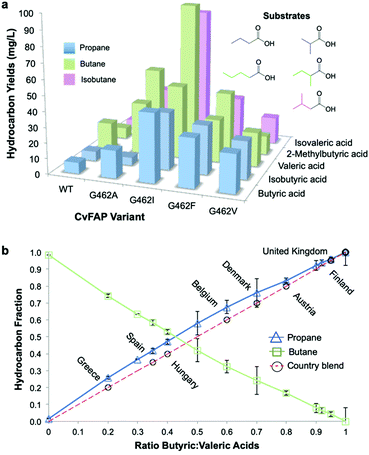 | ||
| Fig. 2 Tuneable bio-LPG blends. In vivo gaseous hydrocarbon production by wild type and variant CvFAP in E. coli BL21(DE3) ΔyqhD/ΔyjgB. Effect of (a) CvFAP-pETM11 variant and (b) butyric:valeric acid blends with CvFAPG462V-pBbA1c on hydrocarbon production. Cultures (20 mL; 3 biological replicates) were grown in LB medium containing kanamycin (50 μg mL−1) at 37 °C to OD600 ∼0.6–0.8. Recombinant protein expression was induced with IPTG (0.1 mM) followed by culture supplementation with fatty acid substrates (10 mM) after 1 h at 30 °C. Triplicate aliquots (1 mL) from each culture were sealed into 4 mL glass vials and incubated at 30 °C for 16–18 h at 200 rpm, illuminated with a blue LED panel. Headspace gas was analysed for hydrocarbon content using a Micro GC. aAll reactions designed to generate butane and isobutane also produced ∼2% propane. Errors for panel (a) are found in Table S9 (ESI†). Error bars represent one standard deviation for duplicate (n = 2) biological repeats for panel (b). | ||
The most common gases found in LPG blends are propane and n-butane. Blends may also contain isobutane, ethane, ethylene, propylene, butylene and isobutylene. The exact composition of LPG is country-specific, and can be varied between seasons as required, for example, in order to assure proper vaporisation in winter.32 In the UK, LPG is 100% propane, while in Italy the propane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) butane ratio varies from 90
butane ratio varies from 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 to 20
10 to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 (Fig. 2b). As CvFAPG462V can generate both propane and butane at similar titres, the possibility of producing country-specific bio-LPG blends by varying the ratio of externally supplied butyric:valeric acids was investigated. The butyric
80 (Fig. 2b). As CvFAPG462V can generate both propane and butane at similar titres, the possibility of producing country-specific bio-LPG blends by varying the ratio of externally supplied butyric:valeric acids was investigated. The butyric![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) valeric acid ratios fed to live cultures were closely correlated with propane
valeric acid ratios fed to live cultures were closely correlated with propane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) butane ratios measured in the culture headspace (Fig. 2b; Table S10; ESI†). This indicates the ease with which tuneable bio-LPG blends can be generated. Manipulation of the externally supplied VFA feed ratios, or modulation of VFA concentrations generated in vivo, could therefore offer simple routes to generate LPG blends in scaled production.
butane ratios measured in the culture headspace (Fig. 2b; Table S10; ESI†). This indicates the ease with which tuneable bio-LPG blends can be generated. Manipulation of the externally supplied VFA feed ratios, or modulation of VFA concentrations generated in vivo, could therefore offer simple routes to generate LPG blends in scaled production.
Engineered metabolic pathways for bio-alkane gas production
Upregulation of cellular VFA biosynthesis is an alternative means of biosynthesising alkane gas with engineered CvFAP biocatalysts. Ideally, the chemical precursor for a VFA biosynthetic pathway should be a major component of available waste feedstocks. Amino acids derived from protein-rich waste products are simple, cheap and readily available carbon sources. They are prevalent in salted milk whey from cheese manufacture,33 brewery waste yeast34 and proteinaceous food waste.35A pathway was constructed from valine to propane beginning with the deamination of valine to α-ketovalerate, catalysed by leucine 2-oxoglutarate transaminase (ilvE) from E. coli36 (Fig. 3a). Irreversible decarboxylation of α-ketovalerate leads to isobutyraldehyde catalysed by branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis.37 Isobutyric acid is then formed by the action of 3-hydroxypropionaldehyde dehydrogenase (Hpad) from E. coli,38 which is subsequently decarboxylated by CvFAPG462I to form propane (Fig. 3a). An ADO/ferredoxin-dependent decarbonylation of isobutyraldehyde to propane9 can provide a further ‘dark’ pathway to operate alongside the light-dependent pathway should this be required.
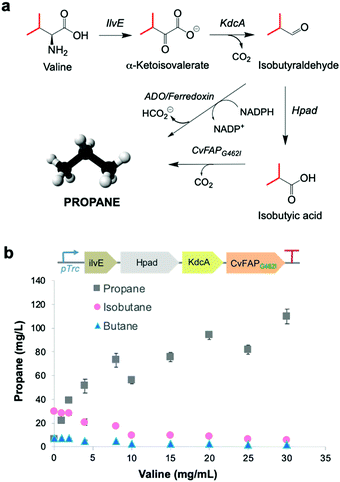 | ||
| Fig. 3 Metabolic pathway to bio-LPG. (a) Enzymatic CvFAP- and ADO-dependent pathways from valine to propane. Enzymes: ilvE = leucine 2-oxoglutarate transaminase; KdcA = thiamine diphosphate-dependent branched-chain keto acid decarboxylase; Hpad = 3-hydroxypropionaldehyde dehydrogenase. (b) Effect of valine supplementation on propane production in E. coli expressing a recombinant CvFAP-dependent pathway from valine. Culture conditions and gas measurement were performed as described in the legend of Fig. 2, except the cultures were supplemented with valine (0–30 mg mL−1) 1 h after IPTG-induction, instead of VFAs. The numerical data for panel b can be found in Table S11 (ESI†). Error bars represent one standard deviation for triplicate biological repeats (n = 3). Differences in ratios between the difference gases likely reflects competitive binding of the three amino acids for a common CvFAP binding site. | ||
This pathway was engineered in E. coli (Fig. 3b inset) and cultures were supplemented with valine (0–30 mg L−1). In each case, propane, isobutane and butane were detected in the headspace. Propane levels increased on feeding valine by up to 17-fold higher with 30 mg L−1 valine (109.7 ± 6.3 mg L−1 propane; Table S11; ESI†). (Iso)butane titres decreased with increased valine supplementation. These observations are likely due to an increase in (valine-derived) butyrate levels, leading to the favouring of propane production over other alkane gases, similar to that seen in Fig. 2b. Endogenous valine and/or butyrate levels are likely high because propane yields were about three-fold higher than the externally fed valine concentration (Fig. 3b). The propane titres observed using this pathway are comparable to those obtained by external feeding of butyrate to CvFAPG462V alone (Table S8; ESI†). To operate with this pathway, waste feedstocks would need to be protein rich (e.g. food waste). Amino acid content of thirty-nine samples of vegetal and dairy product food waste from EU industrial agro-food systems has been shown to have a valine content that varies from 6.4–29.4 mg g−1 waste,35 and also can act as general carbon sources. Therefore, these abundant wastes are attractive feedstocks for alkane gas production.
A microbial chassis for scaled bio-alkane gas production
E. coli was chosen to demonstrate laboratory scale production of bio-alkane gases using CvFAP variants as proof-of-concept. It may not be suitable, however, for scaled production. Microbial fuels and chemicals production is a costly process, and places high demands on both capital and operational expenditures. Typically, steel-based bioreactors with complex monitoring systems are used, with high running costs (e.g. energy-intensive aeration, mixing and downstream processing), production rates and titres. Sterilisation is required to minimise microbial contamination, and growth under aseptic conditions is necessary. There are also environmental concerns over waste processing and disposal, and production methods use large quantities of clean water. These multi-faceted issues can increase production costs. At the outset, by selecting Halomonas st. TD01 as a production host, we tackled many of these issues.39Halomonas grows at high salinity (e.g. 20% w/v NaCl) and at pH values as high as 12. Continuous cultures have been grown for over three years in industrial-scale vessels for the biomanufacture of polyhydroxyalkanoates at greater than 1000 tonnes scale, with no decline in growth potential.15 Seawater, waste-water and recycled water can be used without sterilisation, conserving fresh water and reducing energy expenditure. Bioreactors can be constructed using low cost materials (e.g. plastics, ceramics and cement). Scaled production of polyhydroxyalkanoates using Halomonas is at a 65% cost saving compared to E. coli.21 This suggests that distributed bio-LPG biomanufacture could be more profitable using Halomonas.
We constructed a Halomonas-compatible plasmid pHal2 containing CvFAP using a broad host range pSEVA-derived plasmid40 with an IPTG-inducible promoter (Fig. 4a inset).16,41 Halophilic in vivo alkane gas production with both CvFAPG462V and CvFAPG462I variants was performed with butyric acid (Fig. 4a), with the highest titres shown for CvFAPG462V (157.1 ± 17.1 mg L−1 culture) with 80 mM butyrate in buffered medium (Fig. 4b and Table S12; ESI†). Variant CvFAPG462I produced more propane than CvFAPG462V, and both variants produced several-fold more propane than the wild-type (Fig. 4a). These titres are about nine-fold greater than those reported for E. coli containing engineered ADO-dependent pathways, and five-fold greater than E. coli containing CvFAPG462V and fed with butyrate.5,6,8 Propane production progressively increased with light ‘intensity’ (up to about 2000 μmol s−1 m−2; Fig. 4b inset) but declined at higher light intensities, most likely due to increased photoinactivation of CvFAP (Fig. S3; ESI†), or light-dependent effects on cell viability. Halomonas therefore proves to be well able to support the production of propane, showing titres comparable to valine supplemented E. coli cultures expressing the KdcA-dependent pathway (Fig. 3).
For scaled-up production, engineered strains require waste biomass feedstock mixed with seawater and recycled water grown aerobically at high salinity with minerals, vitamins and VFAs. Wastewater streams (with salt supplementation) are suitable for production at inland sites. Autolysed spent brewery yeast or similar (e.g. hydrolysed ‘spent’ Halomonas cells) can be used to provide essential vitamins. Biodiesel waste (60–70% glycerine) is a cost-effective carbon source for bacterial growth.42 VFAs can be sourced readily from anaerobic digestion (AD) (e.g. 50 g L−1 butyrate from fed-batch fermentation of brown algae;43 36 g L−1 with kitchen waste44).
Bio-LPG production using waste feedstocks was investigated at laboratory scale in a flatbed photobioreactor. Non-sterile fermentations were performed using ‘clean’ (laboratory grade reagents) and ‘crude’ (seawater and waste glycerin) media. Seawater and biodiesel waste impurities affected Halomonas growth and propane production to only a minor extent (Fig. 5). Maximal propane production occurred 4–6 hours after induction, with an overall yield of ∼90 mg g−1 cells (over 2 days). A steady decline in propane production after the early peak rate was observed (Fig. 5b; Fig. S5; ESI†), attributed to plasmid instability and/or loss45 or possibly CvFAP inactivation. In addition, the continuous exposure of Halomonas (or microorganisms in general) to blue light may impact on cell viability, compromising further replenishment of the CvFAP catalyst in vivo. In spite of this, we have shown that waste feedstocks and seawater are capable of supporting bio-alkane gas production.
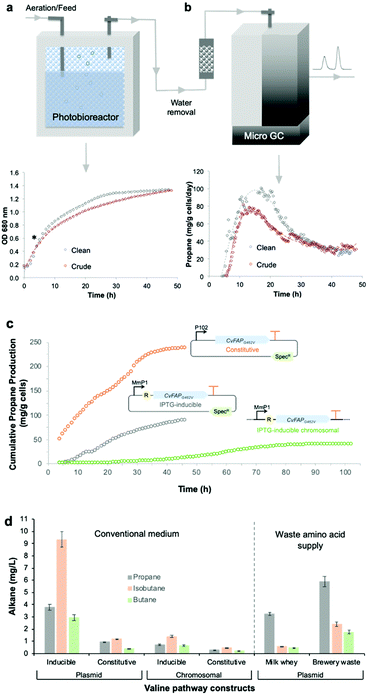 | ||
| Fig. 5 Propane production by the robust industrial chassis Halomonas. (a) Culture growth (OD 680 nm) and (b) propane production of Halomonas expressing IPTG-inducible pHal2-CvFAPG462V in the presence of analytical grade glycerol (‘clean’) or biodiesel waste (‘crude’). (c) Cumulative propane production by Halomonas expressing plasmid-borne or chromosomally integrated CvFAP variants. Cultures (400 mL) were grown in high salt glycerol medium at pH 6.8 containing 50 μg mL−1 spectinomycin (plasmid borne cultures only) and 0.5 mL L−1 antifoam. Conditions were maintained at 30 °C with 65–100% stirring, an airflow rate of 1.21 L min−1 in the dark until mid-log phase (4–5 hours). Recombinant protein expression was induced with IPTG (0.1 mM) where required, followed by the addition of sodium butyrate (60 mM pH ∼ 6.8) and blue light exposure (1656 μE). Cultures were maintained for 48–110 h and propane production was monitored at 15 to 20 minute intervals by automated headspace sampling using a Micro GC. In panel c, data for inducible and constitutively expressed plasmid-borne expression systems are indicated by grey and orange circles, respectively, while the chromosomally integrated expression system is indicated by green circles. R = lac repressor; SpecR = spectinomycin resistance gene. (d) Production of alkane gases in Halomonas using the valine-dependent pathway. The IPTG-inducible and constitutive promoters were pTrc and pPorin 69, respectively (Table S2; ESI†). General culture conditions (non-sterile) and gas measurements were performed in YTN6 media, as described in the legend of Fig. 4, containing autolysed brewery yeast without VFA addition. Milk whey medium (pH 9) was composed of cheesemaking residual salt whey from a North of England supplier supplemented with 60 g L−1 NaCl. Error bars represent one standard deviation for biological repeats (n = 3). | ||
Stable strains for bio-LPG production
Genome-integrated alkane gas producing Halomonas strains are required for scaled production to eliminate the need for antibiotic-mediated plasmid maintenance. We integrated an IPTG-inducible CvFAPG462I into the genome of Halomonas and tested its bio-alkane gas producing ability in a photobioreactor for 48 to up to ∼100 h.16 Cumulative propane yields were less than half those obtained with plasmid borne pHal2-CvFAPG462V (Fig. 5c), in spite of the G462I variant displaying higher activity than CvFAPG462V (Fig. 4a). This lower propane titre is not surprising as only one copy of CvFAPG462I was integrated into the genome. However, these titres compare favourably to plasmid-based expression (Fig. 5c). Propane production rates were maximal around 60 h (Fig. S6; ESI†). Beyond 60 h, cell viability of Halomonas was shown to decline, as evidenced by a reduction in viable cells (colonies) detected during plate counting assays. This is likely due to prolonged high intensity light exposure. Continuous culture replenishment of the photobioreactor with fresh ‘dark-grown’ culture would mitigate against this loss of cell viability.Next, we eliminated the need for IPTG induction of CvFAPG462V using a modified constitutive pPorin-like promoter.46 Constitutively-expressed CvFAPG462V cultures showed elevated (2.7-fold) propane yields (237 mg g−1 cells) compared with induced cultures (Fig. 5c; Fig. S6; ESI†), with a concomitant reduction in cell density attributed to slower cell growth. We also integrated a ‘valine-to-propane’ pathway (constitutive and inducible versions) into the Halomonas chromosome and demonstrated bio-alkane gas production. Higher overall alkane titres were obtained with the integrated IPTG-inducible pathway (1.19 ± 0.01 mg L−1 isobutane; Fig. 5d) compared to the constitutively expressed strain. Extensive industrial waste amino acid sources can be found in the dairy (salted milk whey) and brewery industries (autolysed yeast). We compared the bio-alkane gas production of the plasmid-borne KcdA-dependent pathway using these waste supplies and found in all cases, bio-alkane gases are produced (Fig. 5d). The titres and compositions of the produced gas were dependent on feedstocks, which reflects on the relative amino acid and/or overall nutritional compositions of each feedstock.
Bio-LPG from carbon dioxide
An ideal energy strategy would directly utilise CO2 as the carbon source for the production of biofuels. A microbial carbon capture solution could take advantage of the photosynthetic ability of cyanobacteria to fix CO2 into organic carbon. Synechcocystis PCC 6803 is an ideal host because it grows rapidly and is genetically tractable. It is tolerant to abiotic stress and growth requirements are well understood.47,48 Conversion of CO2 into medium chain-length fatty acids13 and long chain hydrocarbons14 has been described. We previously bioengineered Synechcocystis PCC 6803 by incorporating E. coli thioesterase A, which catalyses the conversion of fatty acyl-ACP to free fatty acids. We also knocked out the native fatty acyl ACP synthase gene (Δaas) to minimise the reverse reaction (Fig. 6a).14 These changes increased the availability of free fatty acid precursors in vivo, enabling hydrocarbon biosynthesis direct from CO2, instead of via an external carbon source.14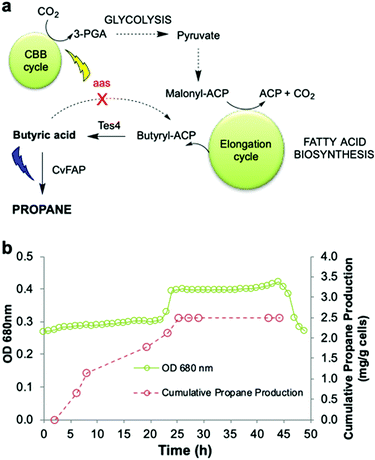 | ||
Fig. 6 Propane production employing natural photosynthetic CO2 capture. (a) Scheme for engineering Synechocystis to enable propane production by CO2 fixation. (b) Cumulative propane production of Synechocystis Δaas expressing CvFAPG462V and Tes4 (strain pIY819). The photobioreactor (400 mL) was set up in batch mode with starter culture diluted 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in fresh BG11+ medium (BG11 pH 8.013,14 containing TES buffer and 1 g L−1 sodium thiosulphate) in the presence of 150 mM NaHCO3. Both pH control and CO2 supply were maintained using 1 M NaHCO3 in 2 × BG11+. The culture was maintained at 30 °C with maximal stirring with an airflow rate of 1.21 L min−1, illumination of warm white light (30 μE), automated pH maintenance (1 M acetic acid in 2 × BG11+) and optical density monitoring (680 nm). After reaching an optical density of ∼0.3 (∼20 h), cobalt(II) nitrate hexahydrate (150 μM) was added, warm white illumination was increased to 60 μE and the integral actinic blue LED light panel was activated to provide 500–750 μE blue light (460–480 nm). The culture was maintained at 30 °C for 18–48 hours, with manual headspace sampling to quantify propane by Micro GC. 1 in fresh BG11+ medium (BG11 pH 8.013,14 containing TES buffer and 1 g L−1 sodium thiosulphate) in the presence of 150 mM NaHCO3. Both pH control and CO2 supply were maintained using 1 M NaHCO3 in 2 × BG11+. The culture was maintained at 30 °C with maximal stirring with an airflow rate of 1.21 L min−1, illumination of warm white light (30 μE), automated pH maintenance (1 M acetic acid in 2 × BG11+) and optical density monitoring (680 nm). After reaching an optical density of ∼0.3 (∼20 h), cobalt(II) nitrate hexahydrate (150 μM) was added, warm white illumination was increased to 60 μE and the integral actinic blue LED light panel was activated to provide 500–750 μE blue light (460–480 nm). The culture was maintained at 30 °C for 18–48 hours, with manual headspace sampling to quantify propane by Micro GC. | ||
In this work, we incorporated the gene for CvFAPG462V into wild-type Synechocystis and a Δaas gene knockout strain in the presence or absence of Tes4 (a butyryl-ACP thioesterase from Bacteroides fragilis)9 under the control of a cobalt-inducible promoter (Pcoa) or a constitutive promoter (Ptrc) (Fig. 6a and Fig. S7; ESI†). CvFAPG462V was chosen for the experiments in Synechocystis due to the reproducibility of its high expression levels (Fig. S2; ESI†). Under batch culture conditions, neither the wild-type nor the Tes4/Δaas strains produced detectable propane. Only low levels of propane (11–14 μg per L culture per d) were produced when using strains carrying the CvFAPG462V gene that were supplied externally with butyrate (Fig. S7; ESI†). Encouraged by these findings, we then cultivated the Synechocystis Tes4/Δaas strain carrying CvFAPG462V in the photobioreactor under photosynthetic conditions (see Experimental section) with supplementary blue light exposure. This strain showed moderate propane production (11.1 ± 2.4 mg propane per L culture per day), which is equivalent to ∼12.2 ± 2.6 mg propane per g cells per day (Fig. 6b and Fig. S7; ESI†). To the best of our knowledge, this is the first demonstration of the direct conversion of CO2 into propane.
A factor to consider when using Synechocystis for propane production is the photobleaching of photosynthetic pigments in the presence of high intensity blue light.49 This fixes a practical upper limit on blue light intensity to maximise CvFAP activity whilst minimizing the extent of photobleaching (Fig. S8 and S3, respectively; ESI†). Photobleaching was apparent when light exposure was maintained between 500 to 800 μE, but not during prolonged exposure (140 h) between 300 to 500 μE (Fig. S8; ESI†).
A complementary strategy is to feed bio-alkane-producing Halomonas with osmotically lysed Synechocystis; the latter acting both as a carbon (growth) and butyrate feedstocks.50 This bypasses photobleaching effects, but retains the ability to capture CO2. Synechocystis can also be degraded by AD to generate VFAs for Halomonas gas production. To test the feasibility of this approach, we fed batch cultures (1 mL aliquots) of Halomonas expressing CvFAP with lysed Synechocystis as supplementary carbon and butyrate sources, and produced propane gas (25.3 ± 5.8 mg L−1 culture; Fig. 6b). These titres were enhanced compared to control cultures reliant on only endogenous butyrate alone (YTN6 medium; 0.9 ± 0.1 mg L−1 culture). Use of Synechocystis as a feedstock could enable Halomonas to produce bio-LPG from industrial or atmospheric CO2 rather than being reliant on waste organic matter. The International Energy Agency has estimated that carbon capture and storage/utilisation (CCS) could potentially contribute to a 19% reduction in CO2 emissions by 2050 using existing technologies.51,52 A coupled Synechocystis–Halomonas process could further enhance management of industrial CO2 emissions as well as generate bio-LPG to meet energy demands.
Vision and economics of bio-LPG production
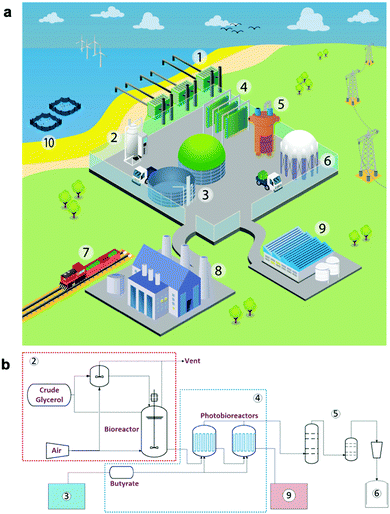
Multiple designs of scaled bio-LPG production hubs have been envisaged, which differ in waste feedstock supply, bioreactor design (∼1000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 L scale) and light requirements. A prototype design based on Halomonas cultivation, could be located in a coastal region with on-site seawater, an anaerobic digester (AD) for VFA production and optionally a cyanobacteria photobioreactor for CO2 fixation and VFA supply (Table S11; ESI†). The AD plant would be tuned to generate a specific VFA blend by modulating the waste composition (e.g. oil and salt concentrations), microbial consortium and running conditions (e.g. temperature). The site could also contain multiple photobioreactors for bio-LPG production (Fig. 7) fed from a dark bioreactor for Halomonas propagation prior to bio-alkane gas production. These photobioreactors could be classical flat-bed photobioreactors, or even low-cost pressurised polyethylene bags with external illumination.53 Propane could be harvested using gas-scrubbing methodologies,54 linked to existing desiccant moisture removal and liquefaction technology. On-site generated bio-LPG could be used to feed adjacent heavy industry, or be transported using local distribution infrastructures. For on-site usage of the alkane gas blends, the exact gas composition (e.g. propane vs. butane) does not need to conform to local government requirements if it is not sold under the ‘LPG’ label. This option allows the usage of local organic waste that may not generate a specific ratio of VFAs, or if the waste composition is likely to vary considerably. Also, small distributed plants can utilise local power generated by solar, wind or tidal technology to power LED illumination, considerably reducing operating costs.
000 L scale) and light requirements. A prototype design based on Halomonas cultivation, could be located in a coastal region with on-site seawater, an anaerobic digester (AD) for VFA production and optionally a cyanobacteria photobioreactor for CO2 fixation and VFA supply (Table S11; ESI†). The AD plant would be tuned to generate a specific VFA blend by modulating the waste composition (e.g. oil and salt concentrations), microbial consortium and running conditions (e.g. temperature). The site could also contain multiple photobioreactors for bio-LPG production (Fig. 7) fed from a dark bioreactor for Halomonas propagation prior to bio-alkane gas production. These photobioreactors could be classical flat-bed photobioreactors, or even low-cost pressurised polyethylene bags with external illumination.53 Propane could be harvested using gas-scrubbing methodologies,54 linked to existing desiccant moisture removal and liquefaction technology. On-site generated bio-LPG could be used to feed adjacent heavy industry, or be transported using local distribution infrastructures. For on-site usage of the alkane gas blends, the exact gas composition (e.g. propane vs. butane) does not need to conform to local government requirements if it is not sold under the ‘LPG’ label. This option allows the usage of local organic waste that may not generate a specific ratio of VFAs, or if the waste composition is likely to vary considerably. Also, small distributed plants can utilise local power generated by solar, wind or tidal technology to power LED illumination, considerably reducing operating costs.
The selection of suitable sites for bio-alkane gas production will be dictated by the need to use locally supplied waste feedstocks and seawater. The current global price for non-AD butyrate is around £2–3 per kg, which is not cost-effective for bio-alkane gas production. Local food waste could be used as both a carbon source and amino acid supply (KdcA pathway utilization) in place of AD-generated VFA blends. For example, the UK generates approximately 7 million tonnes of household food waste annually, of which 0.6–0.7 million tonnes can be collected by local authorities and treated through waste management systems.55 Based on laboratory scale studies, food waste has a great potential as a VFA fermentation substrate due to high VFA yield (up to 0.43 gVFA per g substrate).30
The feasibility of this approach was investigated by performing a preliminary Techno-Economic Analysis (TEA) and carbon footprint analysis, limited to the process itself, of a prototype design for Halomonas bio-alkane gas production (Note S1, including Fig. S9–S11 and Tables S13–S26; ESI†). This analysis is intended to understand the gap between the early-stage research and commercial realisation, and to illustrate potential strategies to bring further process improvements. The TEA contains a high degree of uncertainty because the process is still at a low technology readiness level. Nevertheless, the analysis results can be used to highlight bottlenecks and hotspots which have significant impact on the economic or environmental aspect of the process, so that future research and process design can be directed in the most effective direction. The base case was modelled on a conceptual design of a pilot-scale continuous process with one 1 m3 bioreactor for biomass synthesis, two 1 m3 photobioreactors in series for propane synthesis, and one 1.72 m3 anaerobic digester to generate butyrate feed. Ten further TEA cases were formulated by introducing additional measures and strategies to strengthen the economic potential of the process (Note S1; ESI†). Examples of cost reducing strategies include implementing non-sterile fermentation, valorization of side-streams to produce additional valuable chemicals, optimization of cell productivity and process scale-up. The design basis, process specifications and engineering as well as financial assumptions are summarized in Note S1 (ESI†). Based on this information, the TEA model generated estimates for plant performance, production costs, minimum propane selling prices (MPSP) and CO2 emissions for all the cases.
In comparison to chemical routes, CvFAP catalyst ‘poisoning’ by photoinactivation is overcome by continuous replenishment from a ‘dark’ bioreactor (biomass production in the dark). Reduction in energy costs associated with light supply (CvFAP photoactivation; cyanobacterial growth) is central to production costs savings. This could be managed by using solar energy or wind farm electricity. Substituting blue LEDs for concentrated wavelength-filtered sunlight (e.g. 425–475 nm) could reduce the energy burden and associated costs. The daily blue light intensity in the Northern United Kingdom56 averages around 29.0 W m−2, while the required photobioreactor intensity is up to 424 W m−2, dependent on microbial chassis. A 15-fold solar concentration is required to generate the required blue light intensities which could be met using existing solar concentration technologies, similar to the Australian National University parabolic trough or the Entech Incorporated Fresnel lens (each achieving 30-fold concentration).57 To allow diurnal propane production, light supplementation via LEDs could be supplied outside daylight hours. Alternatively, Halomonas could be engineered to include an alternative non-light requiring ADO-dependent pathway from valine (Fig. 3a), enabling propane production during the dark phase.
A cost-effective solution to bio-alkane production requires a significant reduction in illumination costs. This was modelled by the utilisation of natural sunlight with solar concentrators, cleaner wind power and the localisation of the bioreactors within developing countries with lower operating costs (Case 6, 10 and 11; Tables S16, S23–S24; ESI†) In addition, process scaling-up and the generation of secondary revenue are necessary to increase the cost-effectiveness of the process (Case 5 & 9). This could include the conversion of waste Halomonas and Synechocystis biomass to fertiliser, further processing through AD plants as a source of VFAs, or desiccated to produce fish food at larger scale.
As about 25% of the carbon content of supplied butyrate would be lost as CO2, the energy ratio of propane production (i.e. energy output/energy input) at maximal productivity is estimated to be 3.18. (Case 11; Tables S17, S23–S24; ESI†). The projected propane yields were estimated at 358 tonnes per year for a pilot production system scaled-up by 10-fold, with projected combined revenue of £3.1 M (primary and secondary products; Table S24; ESI†). This is based on developing a Halomonas strain with multi-copy insertion of CvFAPG462I to ensure stable propane titres similar to plasmid-borne systems. The TEA study also predicted a 300-fold decrease in propane production cost at scale (US $626.80 kg−1 to US $1.89 kg−1) in comparison to traditional, unoptimised biotechnological approaches.
The UK LPG market is ca. 82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970 barrels per day,55 equivalent to ∼0.086 tonnes of propane per barrel or 2
970 barrels per day,55 equivalent to ∼0.086 tonnes of propane per barrel or 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 540
540![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tonnes per year. If the future transport sector were to use ca. 10% of the current market for bio-LPG as a drop-in fuel this would require around 710 of the said pilot process operating at maximal productivity and consume 225 tonne of crude glycerol per year. After four decades of technology development, ethanol derived from starch or cellulosic biomass is currently the dominant biofuel for liquid transportation and power generation. Bioethanol can now be made economically and at large scales sufficient to contribute to a nation's fuel market; this is not yet feasible for other newly emerged biofuels e.g. butanol (non-ABE derived), biofene (farnesene), and bisabolene. Ethanol biorefineries have the capacity to utilise biomass feedstock, transforming most of the components into valuable products, which are integrated readily with existing industrial infrastructures. These features are desirable also for bio-alkane gas production processes, if they are to meet the ambitious goal of utilising propane/alkane blends as drop-in-fuels. Also, fuelling vehicles with Bio-LPG is one way to diversify the availability of clean fuels and to increase energy security so that economies are not over reliant on provision of ethanol. Bio-LPG has its own niche in the transportation fuel sector – for example, it is suitable for high-mileage vehicles by offering improved engine life and lower maintenance costs. As with ethanol production, Bio-LPG production will require scaled technology development and optimisation, and more detailed TEA evaluation as the technologies mature.
000 tonnes per year. If the future transport sector were to use ca. 10% of the current market for bio-LPG as a drop-in fuel this would require around 710 of the said pilot process operating at maximal productivity and consume 225 tonne of crude glycerol per year. After four decades of technology development, ethanol derived from starch or cellulosic biomass is currently the dominant biofuel for liquid transportation and power generation. Bioethanol can now be made economically and at large scales sufficient to contribute to a nation's fuel market; this is not yet feasible for other newly emerged biofuels e.g. butanol (non-ABE derived), biofene (farnesene), and bisabolene. Ethanol biorefineries have the capacity to utilise biomass feedstock, transforming most of the components into valuable products, which are integrated readily with existing industrial infrastructures. These features are desirable also for bio-alkane gas production processes, if they are to meet the ambitious goal of utilising propane/alkane blends as drop-in-fuels. Also, fuelling vehicles with Bio-LPG is one way to diversify the availability of clean fuels and to increase energy security so that economies are not over reliant on provision of ethanol. Bio-LPG has its own niche in the transportation fuel sector – for example, it is suitable for high-mileage vehicles by offering improved engine life and lower maintenance costs. As with ethanol production, Bio-LPG production will require scaled technology development and optimisation, and more detailed TEA evaluation as the technologies mature.
The above example illustrates how an integrated biorefinery strategy could in principle be used to supply local energy requirements and generate income, while recycling industrial CO2 and food waste. A second strategy utilizing the multi-step pathway from amino acids could be employed, using food, brewery or dairy waste to supply the necessary amino acids (in place of AD-generated butyrate). There are likely other configurations around these examples that could likewise be deployed at scale, enabling bio-LPG manufacture from waste biomass and atmospheric/industrial CO2, and provide renewable energy solutions for localised economies around the globe. That said, further exploration of the TEAs will be required coupled to further rounds of microbial strain optimization, as individual bio-production formats are scaled at higher technology readiness levels.
Conclusions
The development of any synthetic biological solution for chemicals production into a commercially viable process requires the consideration of both (bio)catalytic process optimisation and an understanding of the techno-economic challenges of developing scaled bioprocesses. We tackled both of these challenges when developing a series of biocatalytic solutions to alkane gas production. We investigated (i) single vs. multi-catalytic pathways from abundant waste feedstocks, (ii) multiple chassis screening and development, (iii) lab-scale production in vivo and (iv) finally proposed designs for multiple scaled bio-LPG production ‘hubs’, utilising local waste materials and taking advantage of the available infrastructure. This combinatorial approach is key in any commercial development as it focuses the research and development beyond simple proof-of-principle demonstration, and directs progression towards practical solutions to process/economic bottlenecks.We have shown that sustainable and renewable solutions to highly efficient and clean-burning bio-alkane fuels (tuneable bio-LPG) is possible. Further optimisation of the microbial chassis to achieve stable high titres, coupled to improvements in the bio-LPG bioprocess hub design will drive the concept towards the realisation of a commercially-viable process. Localisation of these hubs close to existing waste-generating heavy industry in both advanced and developing countries will assist with waste management, reduce the carbon footprint, and increase energy security. This could positively contribute towards global carbon management targets and clean air directives.
Conflicts of interest
A patent application (PCT/EP2019/060013) entitled ‘Hydrocarbon production’ is pending in relation to the production of hydrocarbon gases in engineered microbial strains. P. R. J. is a board member, and M. S. and N. S. S. are founding directors of C3 Biotechnologies Ltd.Acknowledgements
The work was supported by C3 Biotechnologies Ltd, the UK Engineering and Physical Sciences Research Council (EP/S01778X/1; EP/J020192/1), the European Union's Horizon 2020 research and innovation programme project PHOTOFUEL under grant agreement no. 640720 and the Biotechnology and Biological Sciences Research Council (BB/M017702/1; BB/L010798/1). MA was funded by a PhD scholarship from the Newton-Mosharafa fund. EZW was funded by a BBSRC Case PhD studentship. IY was funded by a PhD scholarship from Indonesia Endowment Fund for Education (LPDP). This is a contribution from the EPSRC/BBSRC Future Biomanufacturing Research Hub and the BBSRC/EPSRC Synthetic Biology Research Centre SYNBIOCHEM. Graphical design work for Fig. 7a was completed by Mr Matthew Dykes.References
- T. D. H. Bugg and M. G. Resch, Curr. Opin. Chem. Biol., 2015, 29, v–vii CrossRef CAS PubMed.
- E. Johnson, Biofuels, Bioprod. Biorefin., 2017, 11, 887–896 CrossRef CAS.
- E. Johnson, Biofuels, Bioprod. Biorefin., 2015, 9, 627–629 CrossRef CAS.
- Z. Y. Zakaria, N. F. Mohamad and N. A. S. Amin, J. Appl. Sci., 2010, 10, 1166–1170 CrossRef CAS.
- P. Kallio, A. Pásztor, K. Thiel, M. K. Akhtar and P. R. Jones, Nat. Commun., 2014, 5, 4731 CrossRef CAS PubMed.
- N. Menon, A. Pásztor, B. R. Menon, P. Kallio, K. Fisher, M. K. Akhtar, D. Leys, P. R. Jones and N. S. Scrutton, Biotechnol. Biofuels, 2015, 8, 61 CrossRef PubMed.
- A. Schirmer, M. Rude, X. Li, E. Popova and S. del Cardayre, Science, 2010, 329, 559–562 CrossRef CAS PubMed.
- M. J. Sheppard, A. M. Kunjapur and K. L. J. Prather, Metab. Eng., 2016, 33, 28–40 CrossRef CAS PubMed.
- L. Zhang, Y. Liang, W. Wu, X. Tan and X. Lu, Biotechnol. Biofuels, 2016, 9, 80 CrossRef PubMed.
- D. Sorigué, B. Légeret, S. Cuiné, S. Blangy, S. Moulin, E. Billon, P. Richaud, S. Brugière, Y. Couté, D. Nurizzo, P. Müller, K. Brettel, D. Pignol, P. Arnoux, Y. Li-Beisson, G. Peltier and F. Beisson, Science, 2017, 357, 903–907 CrossRef PubMed.
- W. Zhang, M. Ma, M. Huijbers, G. A. Filonenko, E. A. Pidko, M. van Schie, S. de Boer, B. O. Burek, J. Bloh, W. J. H. Van Berkel, P. W. A. Smith and F. Hollmann, J. Am. Chem. Soc., 2019, 141, 3116–3120 CrossRef CAS PubMed.
- K. A. Datsenko and B. L. Wanner, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6640–6645 CrossRef CAS PubMed.
- I. S. Yunus and P. R. Jones, Metab. Eng., 2018, 44, 81–88 Search PubMed.
- I. S. Yunus, J. Wichmann, R. Wördenweber, K. J. Lauersen, O. Kruse and P. R. Jones, Metab. Eng., 2018, 49, 201–211 CrossRef CAS PubMed.
- W. Tao, L. Lv and G.-Q. Chen, Microb. Cell Fact., 2017, 1–11, DOI:10.1186/s12934-017-0655-3.
- H. Zhao, H. M. Zhang, X. Chen, T. Li, Q. Wu, Q. Ouyang and G.-Q. Chen, Metab. Eng., 2017, 39, 128–140 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- N. Guex and M. C. Peitsch, Electrophoresis, 1997, 18, 2714–2723 CrossRef CAS PubMed.
- T. S. Lee, R. A. Krupa, F. Zhang, M. Hajimorad, W. J. Holtz, N. Prasad, S. K. Lee and J. D. Keasling, J. Biol. Eng., 2011, 5, 15–17 CrossRef PubMed.
- C. Lou, B. Stanton, Y.-J. Chen, B. Munsky and C. A. Voigt, Nat. Biotechnol., 2012, 30, 1137–1142 CrossRef CAS PubMed.
- J. Ye, W. Huang, D. Wang, F. Chen, J. Yin, T. Li, H. Zhang and G.-Q. Chen, Biotechnol. J., 2018, 13, 1800074 CrossRef PubMed.
- R. Simon, U. Priefer and A. Puhler, Biotechnology, 1983, 1, 784–791 CrossRef CAS.
- X.-Z. Fu, D. Tan, G. Aibaidula, Q. Wu, J.-C. Chen and G.-Q. Chen, Metab. Eng., 2014, 23, 78–91 CrossRef CAS PubMed.
- Q. Qina, C. Linga, Y. Zhaoa, T. Yange, J. Yinb, Y. Guoa and G.-Q. Chen, Metab. Eng., 2018, 47, 219–229 CrossRef PubMed.
- M. Storch, A. Casini, B. Mackrow, T. Fleming, H. Trewhitt, T. Ellis and G. S. Baldwin, ACS Synth. Biol., 2015, 4, 781–787 CrossRef CAS PubMed.
- J. Elhai, A. Vepritskiy, A. M. Muro-Pastor, E. Flores and C. P. Wolk, J. Bacteriol., 1997, 179, 1998–2005 CrossRef CAS PubMed.
- R. Anggit, M. T. Ho and D. E. Wiley, Ind. Eng. Chem. Res., 2013, 52, 16887–16901 CrossRef.
- G. L. Rorrer, in Springer Handbook of Marine Biotechnology, ed. S.-K. Kim, Springer, 2015, pp. 257–294 Search PubMed.
- C. Selim, G. Yilan, B. S. Akbulut, A. Poli and D. Kazan, J. Biosci. Bioeng., 2012, 114, 45–52 CrossRef PubMed.
- E. A. Tampio, L. Blasco, M. M. Vainio, M. M. Kahala and S. E. Rasi, GCB Bioenergy, 2019, 11, 72–84 CrossRef CAS.
- M. M. E. Huijbers, W. Zhang, F. Tonin and F. Hollmann, Angew. Chem., Int. Ed., 2018, 57, 13648–13651 CrossRef CAS PubMed.
- https://www.elgas.com.au/blog/1972-lpg-contains-which-gases-gases-present-in-lpg-gases-used .
- A. R. Prazeres, J. Rivas, Ú. Paulo, F. Ruas and F. Carvalho, Environ. Sci. Pollut. Res. Int., 2016, 23, 13062–13075 CrossRef CAS PubMed.
- G. D. Zupančič, M. Panjičko and B. Zelić, Food Technol. Biotechnol., 2017, 55, 187–196 Search PubMed.
- B. Prandi, A. Faccini, F. Lambertini, M. Bencivenni, M. Jorba, B. van Droogenbroek, G. Bruggeman, J. Schöber, J. Petrusan, K. Elst and S. Sforza, Food Chem., 2019, 286, 567–575 CrossRef CAS PubMed.
- X. Yu, X. Wang and P. C. Engel, FEBS J., 2014, 28, 391–400 CrossRef PubMed.
- D. Gocke, T. Graf, H. Brosi, I. Frindi-Wosch, L. Walter, M. Müller and M. Pohl, J. Mol. Catal. B: Enzym., 2009, 61, 30–35 CrossRef CAS.
- J.-E. E. Jo, R. S. Mohan, C. Rathnasingh, E. Selvakumar, W.-C. C. Jung and S. Park, Appl. Microbiol. Biotechnol., 2008, 81, 51–60 CrossRef CAS PubMed.
- D. Tan, Y.-S. Xue, G. Aibaidula and G.-Q. Chen, Bioresour. Technol., 2011, 102, 8130–8136 CrossRef CAS PubMed.
- R. Silva-Rocha, E. Martinez-Garcia, B. Calles, M. Chavarria, A. Arce-Rodriguez, A. de las Heras, A. D. Paez-Espino, G. Durante-Rodriguez, J. Kim, P. I. Nikel, R. Platero and V. de Lorenzo, Nucleic Acids Res., 2012, 41, D666–D675 CrossRef PubMed.
- D. Tan, Q. Wu, J.-C. Chen and G.-Q. Chen, Metab. Eng., 2014, 26, 34–47 CrossRef CAS PubMed.
- S. Chozhavendhan, R. Praveen Kumar, S. Elavazhagan, B. Barathiraja, M. Jayakumar and S. J. Varjani, in Waste to Wealth. Energy, Environment, and Sustainability, ed. R. Singhania, R. Agarwal, R. Kumar and R. Sukumaran, Springer Singapore, Singapore, 2017, vol. 41, pp. 65–82 Search PubMed.
- J.-H. Song, J.-R. S. Ventura, C.-H. Lee and D. Jahng, Biotechnol. Bioprocess Eng., 2011, 16, 42–49 CrossRef CAS.
- B. Zhang, L. L. Zhang, S. C. Zhang, H. Z. Shi and W. M. Cai, Environ. Technol., 2005, 26, 329–339 CrossRef CAS PubMed.
- K. Friehs, Adv. Biochem. Eng./Biotechnol., 2004, 86, 47–82 CrossRef CAS PubMed.
- T. Li, T. Li, W. Ji, Q. Wang, H. Zhang, G.-Q. Chen, C. Lou and Q. Ouyang, Biotechnol. J., 2016, 11, 219–227 CrossRef CAS PubMed.
- P. van Alphen, H. Abedini Najafabadi, F. Branco Dos Santos and K. J. Hellingwerf, Biotechnol. J., 2018, 13, e1700764.
- X. Zang, B. Liu, S. Liu, K. K. I. U. Arunakumara and X. Zhang, J. Microbiol., 2007, 45, 241–245 CAS.
- R. P. Sinha, P. Richter, J. Faddoul, M. Braun and D. P. Häder, Photochem. Photobiol. Sci., 2002, 8, 553–559 RSC.
- M. Lai and E. I. Lan, Biotechnol. Bioeng., 2019, 116, 893–903 CAS.
- I. E. Agency, Energy Technology Perspectives, OECD, 2006 Search PubMed.
- R. Baciocchi, A. Corti, G. Costa, L. Lombardi and D. Zingaretti, Energy Procedia, 2011, 4, 4985–4992 CrossRef CAS.
- Q. Huang, F. Jiang, L. Wang and C. Yang, Engineering, 2017, 3, 318–329 CrossRef.
- A. Raksajati, M. T. Ho and D. E. Wiley, Ind. Eng. Chem. Res., 2013, 52, 16887–16901 CrossRef CAS.
- https://www.eia.gov/dnav/pet/pet_move_expc_a_EPLL_EEX_mbbl_m.htm, accessed 13 March, 2020.
- M. S. Islim, S. Videv, M. Safari, E. Xie, J. J. D. McKendry, E. Gu, M. D. Dawson and H. Haas, J. Lightwave Technol., 2018, 36, 2376–2386 Search PubMed.
- F. Muhammad-Sukki, R. Ramirez-Iniguez, S. G. McMeekin, B. G. Stewart and B. Clive, Int. J. Appl. Sci., 2010, 1, 1–15 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional results are provided. See DOI: 10.1039/d0ee00095g |
| ‡ Each author contributed equally to this manuscript. |
| This journal is © The Royal Society of Chemistry 2020 |