Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ciprofloxacin conjugated to diphenyltin(IV): a novel formulation with enhanced antimicrobial activity†
M. P.
Chrysouli
a,
C. N.
Banti
 *a,
N.
Kourkoumelis
b,
E. E.
Moushi
c,
A. J.
Tasiopoulos
*a,
N.
Kourkoumelis
b,
E. E.
Moushi
c,
A. J.
Tasiopoulos
 d,
A.
Douvalis
e,
C.
Papachristodoulou
f,
A. G.
Hatzidimitriou
g,
T.
Bakas
e and
S. K.
Hadjikakou
d,
A.
Douvalis
e,
C.
Papachristodoulou
f,
A. G.
Hatzidimitriou
g,
T.
Bakas
e and
S. K.
Hadjikakou
 *ah
*ah
aInorganic and Analytical Chemistry, Department of Chemistry, University of Ioannina, 45110 Ioannina, Greece. E-mail: cbanti@uoi.gr; shadjika@uoi.gr; Tel: x30-26510-08374 Tel: x30-26510-08362
bMedical Physics Laboratory, Medical School, University of Ioannina, Ioannina, Greece
cDepartment of Life Sciences, The School of Sciences, European University Cyprus, Nicosia, Cyprus
dDepartment of Chemistry, University of Cyprus, 1678 Nicosia, Cyprus
eMössbauer Spectroscopy and Physics of Material Laboratory, Department of Physics, University of Ioannina, Ioannina, Greece
fDepartment of Physics, University of Ioannina, Ioannina, Greece
gDepartment of Chemistry, Aristotle University of Thessaloniki, Thessaloniki, Greece
hUniversity Research Center of Ioannina (URCI), Institute of Materials Science and Computing, Ioannina, Greece
First published on 30th June 2020
Abstract
The metalloantibiotic of formula Ph2Sn(CIP)2 (CIPTIN) (HCIP = ciprofloxacin) was synthesized by reacting ciprofloxacin hydrochloride (HCIP·HCl) (an antibiotic in clinical use) with diphenyltin dichloride (Ph2SnCl2DPTD). The complex was characterized in the solid state by melting point, FT-IR, X-ray Powder Diffraction (XRPD) analysis, 119Sn Mössbauer spectroscopy, X-ray Fluorescence (XRF) spectroscopy, and Thermogravimetry/Differential Thermal Analysis (TG-DTA) and in solution by UV-Vis, 1H NMR spectroscopic techniques and Electrospray Ionisation Mass Spectrometry (ESI-MS). The crystal structure of CIPTIN and its processor HCIP was also determined by X-ray crystallography.
The antibacterial activity of CIPTIN, HCIP·HCl, HCIP and DPTD was evaluated against the bacterial species Pseudomonas aeruginosa (P. aeruginosa), Escherichia coli (E. coli), Staphylococcus aureus (S. aureus) and Staphylococcus epidermidis (S. epidermidis), by the means of Minimum Inhibitory Concentration (MIC), Minimum Bactericidal Concentration (MBC) and Inhibition Zones (IZs). CIPTIN shows lower MIC values than those of HCIP·HCl (up to 4.2-fold), HCIP (up to 2.7-fold) or DPTD (>135-fold), towards the tested microbes. CIPTIN is classified into bactericidal agents according to MBC/MIC values. The developing IZs are 40.8 ± 1.5, 34.0 ± 0.8, 36.0 ± 1.1 and 42.7 ± 0.8 mm, respectively which classify the microbes P. aeruginosa, E. coli, S. aureus and S. epidermidis to susceptible ones to CIPTIN. These IZs are greater than the corresponding ones of HCIP·HCl by 1.1 to 1.5-fold against both the tested Gram negative and Gram positive bacteria. CIPTIN eradicates the biofilm of P. aeruginosa and S. aureus more efficiently than HCIP·HCl and HCIP. The in vitro toxicity and genotoxicity of CIPTIN were tested against human skin keratinocyte cells (HaCaT) (IC50 = 2.33 μM). CIPTIN exhibits 2 to 9-fold lower MIC values than its IC50 against HaCaT, while its genotoxic effect determined by micronucleus assay is equivalent to the corresponding ones of HCIP·HCl or HCIP.
Introduction
Ciprofloxacin, a second-generation fluoroquinolone, is effective against both Gram-positive and Gram-negative pathogenic bacteria.1 It exhibits good bioavailability or tissue permeability, low incidence of toxic effects and bacteriostatic activity.2 Generally, bacteriocidal agents are preferred than bacteriostatic ones, against infections, since they limit the development of resistance towards bacteria.3 However, microbes such as P. aeruginosa, E. coli, S. aureus and S. epidermidis are reported to have developed resistance towards ciprofloxacin.4 Hence, efforts were made in recent years to modify ciprofloxacin with metal ions, in order to increase its antibacterial activity against resistant strains.5–8The conjugation of metals with drugs (CoMeDs) is a new research area being investigated for discovery of new synergistic therapeutic modalities. The term “conjugation” is used for polymeric drug formulations, to emphasize the synergistic effect of a metal with a drug.9,10 CoMeDs combines metals with specific classes of drugs aiming to create new agents possessing altogether new properties than with the drugs from which they were prepared.9,10 From this point of view, organotin(IV) compounds which contain active drugs as ligands are expected to exhibit enhanced effectiveness due to the synergy which is developing between the RnSn(IV) (R = alkyl or aryl group; n = 1 to 3) moiety and the drug. Besides, organotin compounds are commercially used as agricultural biocides.11,12 Recently, Pokharia et al. have shown that triorganotin(IV) complexes of ciprofloxacin (R3Sn(CIP) (R = Me-, Cy-)) exhibit better antibacterial effect than free ciprofloxacin against E. faecalis, S. aureus, K. pneumoniae, E. coli, P. aeruginosa, and P. mirabilis.13
With the aim to develop new antimicrobial agents with enhanced activity capable of overcoming bacterial resistance,9,10,14–17 the metalloantibiotic of ciprofloxacin (HCIP, Scheme 1) having the formula Ph2Sn(CIP)2 (CIPTIN) was synthesized, characterized and evaluated against bacterial strains P. aeruginosa, E. coli, S. aureus and S. epidermidis. Moreover, the combination of an antibiotic, ciprofloxacin, which possesses pharmacological significance, and a metal with known biological activity, tin(IV), is expected to throw light on principles that govern the synergy between metals and drugs in these metallodrugs.
Results and discussion
General aspects
A solution of HCIP·HCl in water was initially treated with an excess amount of KOH in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 molar ratio to form KCIP. A solution of DPTD (Ph2SnCl2, diphenyltin dichloride) in MeOH (1
1.5 molar ratio to form KCIP. A solution of DPTD (Ph2SnCl2, diphenyltin dichloride) in MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 metal to KCIP) was added to the previous one under continuous stirring for 30 min, (Scheme 2) and the suspension was filtered off.
2 metal to KCIP) was added to the previous one under continuous stirring for 30 min, (Scheme 2) and the suspension was filtered off.
Crystals of CIPTIN were grown by slow evaporation of ddH2O/MeOH solution. Besides, crystals of HCIP were isolated from the filtrate of the intermediate solution (Scheme 2). CIPTIN is stable when it is stored under ambient conditions and it is soluble in MeOH, CH2Cl2, DMSO and acetone.
Solid state studies
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group with a = 7.9606(2), b = 8.5798(2) and c = 10.7739(3) Å, α = 87.868(4), β = 87.868(4), γ = 88.212(4)°, V = 732.43(4) Å3.18 In our study, we found that HCIP (anhydrous) crystallizes in the P
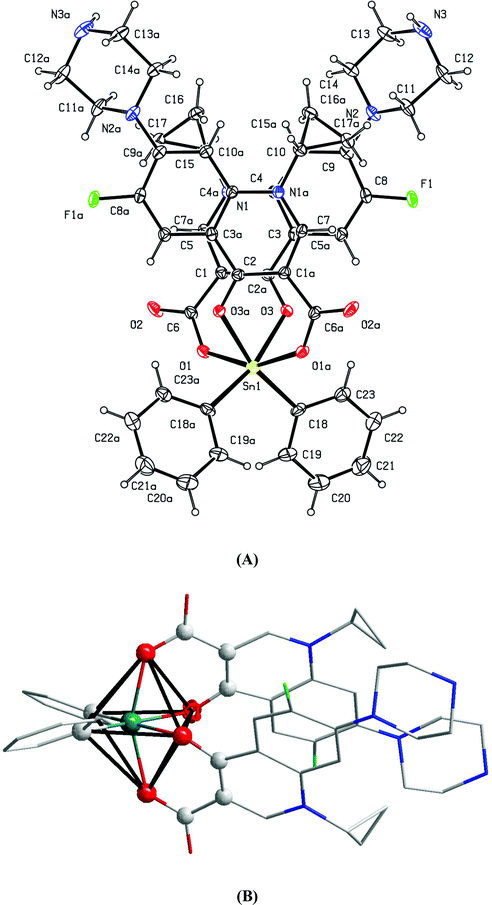
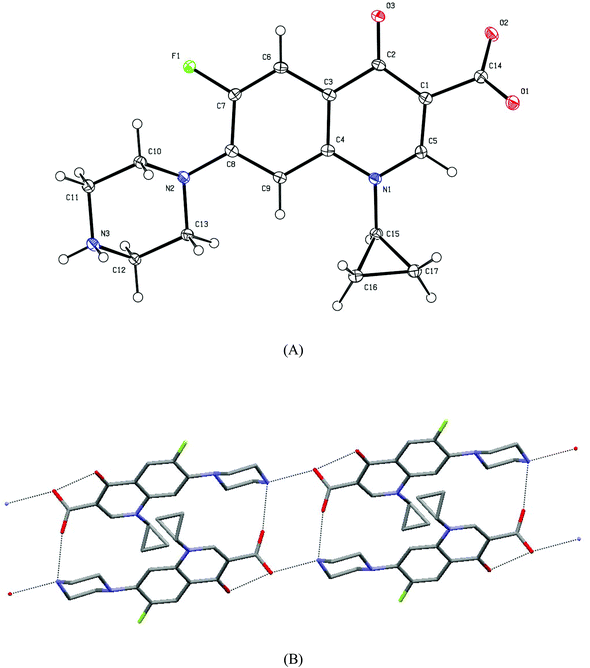
space group with a = 7.9606(2), b = 8.5798(2) and c = 10.7739(3) Å, α = 87.868(4), β = 87.868(4), γ = 88.212(4)°, V = 732.43(4) Å3.18 In our study, we found that HCIP (anhydrous) crystallizes in the P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group with almost identical unit cell parameters: a = 7.8945(7), b = 8.5010(8) and c = 10.7522(9) Å and α = 87.440(7), β = 84.496(7), γ = 88.912(7)°, V = 717.47 Å3. Since the synthesis of CIPTIN requires HCIP HCl as the starting material, HCIP is an intermediate of the methodology for the preparation. For this reason, a complete refinement of the crystal structure of HCIP was attempted independently to clarify the route followed. ORTEP diagrams of CIPTIN and HCIP along with their selected bond distances and angles are shown in Fig. 1 and 2.
space group with almost identical unit cell parameters: a = 7.8945(7), b = 8.5010(8) and c = 10.7522(9) Å and α = 87.440(7), β = 84.496(7), γ = 88.912(7)°, V = 717.47 Å3. Since the synthesis of CIPTIN requires HCIP HCl as the starting material, HCIP is an intermediate of the methodology for the preparation. For this reason, a complete refinement of the crystal structure of HCIP was attempted independently to clarify the route followed. ORTEP diagrams of CIPTIN and HCIP along with their selected bond distances and angles are shown in Fig. 1 and 2.
CIPTIN is a covalent organotin compound. It consists of two ciprofloxacin ligands and one Ph2Sn(IV) moiety. Ciprofloxacin coordinates to tin(IV) through the anionic oxygen atom of its de-protonated carboxylic group and the oxygen atom of its keto group. Thus, two oxygen atoms from each ciprofloxacin and two carbon atoms from the phenyl groups form a distorted octahedral (Oh) arrangement around the Sn(IV) ion. The equatorial plane of the octahedron around the Sn(IV) ion is constituted by two keto oxygen atoms (O3 and O3a) and two carbon atoms of phenyl groups (O3–Sn1–O3a = 80.96(11)° and O3–Sn1–C18a = 166.12(15)°). The axial positions are occupied by the two carboxylic oxygen atoms (O1 and O1a) (O1–Sn1–O1a = 155.64(11)°). The final arrangement of the ligands in CIPTIN lead to the Δ-cis-[Ph2Sn(CIP)2] stereoisomer (Fig. 1B). Only the Δ-cis-[Ph2Sn(CIP)2] isomer has been isolated in the crystal lattice indicating stereo-selectivity of the preparation reaction (Fig. 1B).
The two C–O bond distances of the carboxylic group (O1–C6 = 1.298(5) and O2–C6 = 1.216(5) Å) are almost equivalent, in CIPTIN, confirming deprotonation which it undergoes. The O1–C6 bond is slightly longer than the corresponding O2–C6 bond due to the donation of the O1 atom towards Sn(IV). The C–O bond length of the keto group (O3–C2 = 1.284(5) Å) suggests the retention of the double nature of this bond.
The Sn–O bond distances (Sn1–O1 = 2.135(3) and Sn1–O3 = 2.158(3) Å) are in accordance with the corresponding distances found previously in [(n-Bu-)2Sn(nap)2] (napH = naproxen) (Sn1–O1 = 2.136(7) and Sn1–O2 = 2.506(6) Å),19 in [Me2Sn(salH)2] (salH2 = salicylic acid) (Sn1–O171 = 2.1069(18), Sn1–O271 = 2.1087(15), Sn1–O172 = 2.5145(15) and Sn1–O272 = 2.5773(15) Å)20 and in [(n-Bu)2Sn(salH)2] (Sn1–O172 = 2.121(3), Sn1–O272 = 2.105(4), Sn1–O171 = 2.565(4) and Sn1–O271 = 2.632(4) Å).20 However, in the cases of [(n-Bu-)2Sn(nap)2], [Me2Sn(salH)2] and [(n-Bu)2Sn(salH)2], the ligands chelate to Sn(IV), through their carboxylic groups.
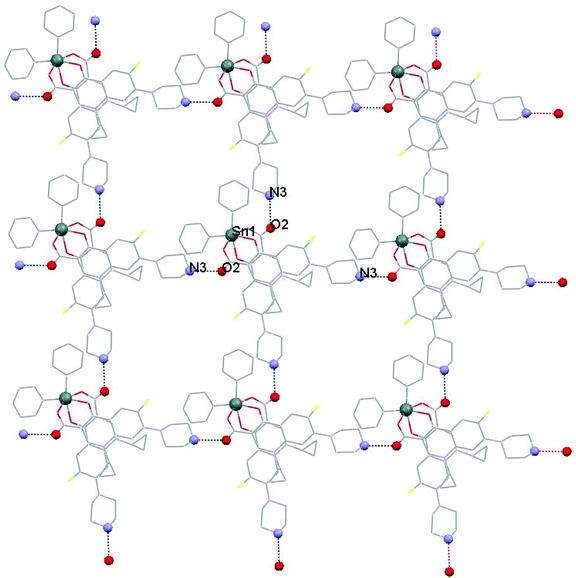
Strong hydrogen bonding interactions N3[H3]⋯O2 = 2.733(6) Å lead to 2D layers (Fig. 3). The layers consist of squares which are formed by four CIPTIN molecules (Sn⋯Sn = 13.969 Å and Sn–Sn–Sn = 87.03°).
The two C–O bond distances of the carboxylic group in anhydrous HCIP (O1–C14 = 1.247(3) and O2–C14 (1.266(3) Å) are equivalent suggesting its de-protonation (Fig. 2). The corresponding C1–O1 and C1–O2 bond lengths of the HCIP hexahydrate (HCIP·6H2O) are 1.259(2) and 1.255(2) Å respectively.8 Moreover, the O1–C14–O2 angle is 125.3(2)°. This is in accordance with the corresponding bond angle found in HCIP·6H2O (125.11(12)°).8 The N3–C11 and N3–C12 bond distances, on the other hand, are 1.481(3) and 1.485(3) Å, respectively, suggesting single N–C bonds (Fig. 2). This is further supported by the C11–N3–C12 bond angle of 109.0(2)°, which is close to the ideal 108°28′ of the sp3 hybrid. Given the fact that two hydrogen atoms bind to N3, a positive charge is assumed for the ([–NH2–])+ group. Therefore, the charge distribution of the zwitterionic anhydrous HCIP is shown in Scheme 2.
Hydrogen bonds N3[H3]⋯O2 = 2.623(3) Å and N3[H3]⋯O1 = 2.882(3) Å lead to the 1D ribbon assembly (Fig. 2B).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, νasym(COO) and νsym(COO) vibrations, respectively (Fig. S1†). The absence of the vibration band at 1710 cm−1 in the spectrum of CIPTIN suggests the coordination of the ligand through the oxygen atom of the keto C
O, νasym(COO) and νsym(COO) vibrations, respectively (Fig. S1†). The absence of the vibration band at 1710 cm−1 in the spectrum of CIPTIN suggests the coordination of the ligand through the oxygen atom of the keto C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group (Fig. S1†). The νasym(COO) and νsym(COO) vibration bands are shifted at 1622 cm−1 and 1381 cm−1, respectively in the spectrum of CIPTIN. The presence of the C
O group (Fig. S1†). The νasym(COO) and νsym(COO) vibration bands are shifted at 1622 cm−1 and 1381 cm−1, respectively in the spectrum of CIPTIN. The presence of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond of CIPTIN at 1272 cm−1 suggests that this N atom is not involved in the coordination sphere of the metal. The absence of the (O–H) vibration at 3520 cm−1 in the CIPTIN spectrum indicates the deprotonation of the carboxyl group. Therefore, ciprofloxacin is coordinated to the tin metal ion via the keto and carboxyl oxygen atom (Fig. S1†).
N bond of CIPTIN at 1272 cm−1 suggests that this N atom is not involved in the coordination sphere of the metal. The absence of the (O–H) vibration at 3520 cm−1 in the CIPTIN spectrum indicates the deprotonation of the carboxyl group. Therefore, ciprofloxacin is coordinated to the tin metal ion via the keto and carboxyl oxygen atom (Fig. S1†).
Solution studies
Antimicrobial studies
The broth dilution method was used for the determination of the MIC values of CIPTIN, HCIP·HCl, HCIP and DPTD, upon incubation of Gram-negative (P. aeruginosa and E. coli) or Gram-positive (S. aureus and S. epidermidis) bacteria for 20 h (Table 1).
| Compound | P. aeruginosa | E. coli | S. aureus | S. epidermidis | Ref. |
|---|---|---|---|---|---|
| a This study, PenNa = sodium salt of penicillin. | |||||
| MIC (μM) | |||||
| Ph 2 Sn(CIP) 2 CIPTIN | 0.741 ± 0.035 | 0.301 ± 0.062 | 1.179 ± 0.083 | 0.255 ± 0.041 | |
| Me 3 SnCIP | 0.253 | 1.012 | 0.125 | — | 13 |
| Cy 3 SnCIP | 0.089 | 1.432 | 0.179 | — | 13 |
| Ag(CIP) 2 NO 3 CIPAG | 0.612 ± 0.142 | — | 0.537 ± 0.067 | 0.458 ± 0.083 | 8 |
| [Ag(Pen)(DMSO)] 2 PenAg | 23.0 ± 2.290 | — | 0.080 ± 0.020 | 2.410 ± 0.880 | 9 |
| HCIP·HCl | 1.174 ± 0.221 | 0.938 ± 0.347 | 1.454 ± 0.123 | 1.079 ± 0.121 | 8 |
| HCIP | 1.048 ± 0.037 | 0.443 ± 0.041 | 1.459 ± 0.013 | 0.699 ± 0.025 | |
| PenNa | >2000 | — | 0.140 ± 0.020 | 3.760 ± 1.140 | 9 |
| Ph 2 SnCl 2 DPTD | > 100 | 25.713 ± 1.535 | 47.529 ± 8.761 | 13.743 ± 0.263 | |
| AgNO 3 | 60.0 | — | 79.500 | 39.400 | 8 |
| MBC (μM) | |||||
| Ph 2 Sn(CIP) 2 CIPTIN | 0.800 ± 0.195 | 0.445 ± 0.040 | 1.600 ± 0.110 | 0.667 ± 0.045 | |
| Me 3 SnCIP | 0.506 | 1.012 | 0.506 | — | 13 |
| Cy 3 SnCIP | 0.716 | 2.863 | 0.358 | — | 13 |
| Ag(CIP) 2 NO 3 CIPAG | 0.700 | — | 1.000 | 0.800 | 8 |
| [Ag(Pen)(DMSO)] 2 PenAg | 46.6 | — | — | — | 9 |
| HCIP·HCl | 1.6 | 1.624 ± 0.127 | 2.000 | 1.600 | 8 |
| HCIP | 1.280 ± 0.096 | 1.250 ± 0.098 | 2.225 ± 0.050 | 1.200 ± 0.160 | |
| PenNa | 2000 | — | — | — | 9 |
| Ph 2 SnCl 2 DPTD | > 100 | > 100 | > 100 | > 100 | |
| AgNO 3 | 91.5 | 95.0 | 140.0 | 8 | |
| MBC/MIC | |||||
| Ph 2 Sn(CIP) 2 CIPTIN | 1.08 | 1.48 | 1.36 | 2.62 | |
| Me 3 SnCIP | 2.0 | 1.0 | 4.05 | — | 13 |
| Cy 3 SnCIP | 8.04 | 2.0 | 2.0 | — | 13 |
| Ag(CIP) 2 NO 3 CIPAG | 1.14 | — | 1.86 | 1.75 | 8 |
| HCIP·HCl | 1.36 | 1.73 | 1.38 | 1.48 | 8 |
| HCIP | 1.22 | 2.82 | 1.53 | 1.72 | |
| AgNO 3 | 1.53 | 1.19 | 3.55 | 8 | |
| IZ (mm) | |||||
| Ph 2 Sn(CIP) 2 CIPTIN | 40.8 ± 1.5 | 34.0 ± 0.8 | 36.0 ± 1.1 | 42.7 ± 0.8 | |
| Ag(CIP) 2 NO 3 CIPAG | 32 | — | 28 | 34 | 8 |
| [Ag(Pen)(DMSO)] 2 PenAg | 17 | — | 57 | 34 | 9 |
| HCIP·HCl | 32.0 | 32.0 ± 0.8 | 24.0 | 36.0 | 8 |
| HCIP | 35.5 ± 0.6 | 33.0 ± 0.8 | 30.5 ± 0.6 | 39.2 ± 0.9 | |
| PenNa | 10 | — | 60 | 34 | 9 |
| Ph 2 SnCl 2 DPTD | 10.0± 0.0 | 18.7 ± 0.9 | 13.1 ± 0.6 | 15.8± 0.4 | |
| AgNO 3 | 13 | 12 | 14 | 8 | |
The MIC values of CIPTIN lie in the range of nanomolar concentrations against P. aeruginosa, E. coli, S. aureus and S. epidermidis, (0.741 ± 0.035, 0.301 ± 0.062, 1.179 ± 0.083 and 0.255 ± 0.041 μM, respectively) (Table 1, Fig. S9†). The corresponding MIC values of the drug in HCIP·HCl and HCIP are: 1.174 ± 0.221 μM and 1.048 ± 0.037 μM (P. aeruginosa), 0.938 ± 0.347 μM and 0.443 ± 0.041 μM (E. coli), 1.454 ± 0.123 μM and 1.459 ± 0.013 μM (S. aureus) and 1.079 ± 0.121 μM and 0.699 ± 0.025 μM (S. epidermidis), respectively (Fig. S10 and S11†).8 The MIC values of DPTD, on the other hand, lie in the micromolar range of concentrations for the tested bacteria: >100 μM, 25.713 ± 1.535 μM, 47.529 ± 8.761 μM and 13.743 ± 0.263 μM towards P. aeruginosa, E. coli, S. aureus and S. epidermidis, respectively (Fig. S12†). Thus, CIPTIN shows 1.2 to 4.2-fold stronger antimicrobial activity than those of HCIP·HCl or HCIP towards both Gram negative and positive bacteria. The strongest antimicrobial effect of CIPTIN is observed towards S. epidermidis, which is 4.2-fold higher than that of HCIP·HCl and 2.7-fold higher than that of HCIP. Moreover, CIPTIN activity rises up to 135-fold stronger than DPTD activity against these bacteria. Therefore, CIPTIN exhibits superior activity to that of both its ingredients (antibiotic and DPTD).
Microbes are classified into susceptible (MIC <50 μM) or resistant (MIC >100 μM) towards an antimicrobial agent by their MIC values.8,10 Thus, all tested strains here are considered as susceptible to CIPTIN.
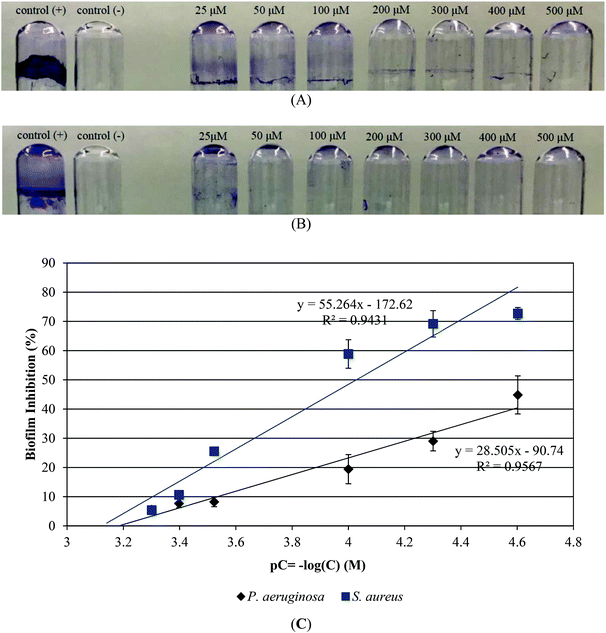
The MBC values of CIPTIN for Gram negative bacteria P. aeruginosa and E. coli are 0.800 ± 0.195 μM and 0.445 ± 0.040 μM respectively, and against Gram positive bacteria S. aureus and S. epidermidis are 1.600 ± 0.110 μM and 0.667 ± 0.045 μM, respectively (Table 1, Fig. S13†). The corresponding MBC values of HCIP·HCl and HCIP are 1.200–2.225 μM against both Gram negative and Gram positive bacteria (Table 1, Fig. S14 and S15†). In the case of DPTD, the MBC values are higher than 100 μM (Fig. S16†). Thus, CIPTIN exhibits higher bactericidal activity than those of free drugs which rises up to 3.6-fold in respect of HCIP·HCl or up to 2.8-fold for HCIP. DPTD has no bactericidal activity.
Since the MBC/MIC values of CIPTIN lie between 1.08 and 2.62, against the tested microbial strains, (Table 1) CIPTIN is classified into bactericidal agents. This is because when the MBC/MIC value is ≤2, an agent is considered as a bactericidal agent, while when the MBC/MIC value is ≥4, as a bacteriostatic agent.8,10,14–17 The MBC/MIC values of HCIP·HCl and HCIP varied between 1.22 and 2.82 and they are bactericidal drugs.
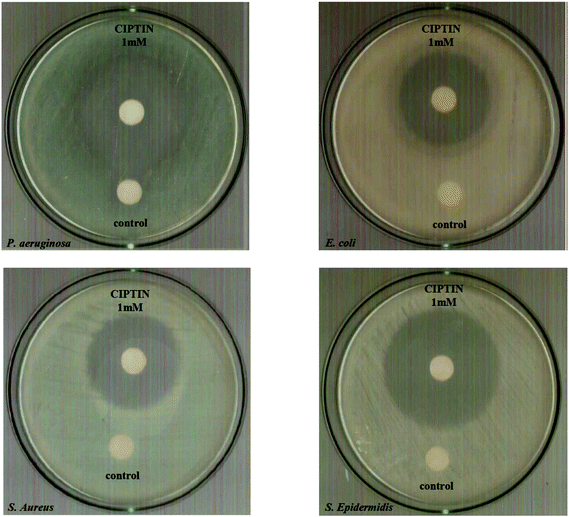
The IZs caused by CIPTIN determined against P. aeruginosa, E. coli, S. aureus and S. epidermidis are 40.8 ± 1.5, 34.0 ± 0.8, 36.0 ± 1.1 and 42.7 ± 0.8 mm, respectively classifying these strains into susceptible ones towards CIPTIN (Fig. 5). The ranges of IZs, which were caused by HCIP·HCl and HCIP, lie between 32.0 and 35.5 mm (against Gram negative bacteria) and 24.0–39.2 mm (against Gram positive bacteria). These values classify the tested bacteria into susceptible ones against HCIP·HCl and HCIP (Table 1, Fig. S17†). The corresponding IZs developed by DPTD lie between 10.0 and 18.7 mm against the tested bacteria classifying them into resistant or intermediate ones towards DPTD. Generally, CIPTIN causes greater inhibition zones than those of the commercial antibiotic or DPTD, against both negative and positive bacterial strains.
The therapeutic window of an agent is defined by the selectivity index (SI):8,26
| SI = IC50(against HaCaT cells)/MIC(against tested bacteria) |
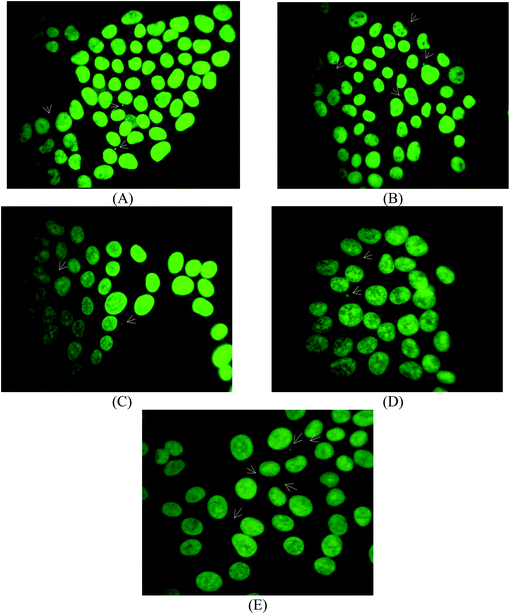
In order to evaluate the genotoxicity of CIPTIN,·HCIP·HCl, HCIP and DPTD, HaCaT cells were treated using the agents at their IC50 values (Fig. 7). The corresponding frequency of micronucleus after treatment of HaCaT cells using CIPTIN is 2.60 ± 0.15%, while the frequency without treatment of the cells is 2.17 ± 0.05%. Therefore, CIPTIN exhibits negligible genotoxicity. The micronucleus frequencies of HaCaT cells after treatment using HCIP·HCl (2.44 ± 0.2)% or HCIP (2.48 ± 0.1)% are close to that of CIPTIN. The MN formed upon treatment of HaCaT cells using DPTD is 3.05 ± 0.2%, which is significantly higher than that of CIPTIN.
Conclusions
The design and the development of antimicrobial agents are of great importance. The new metalloantibiotic CIPTIN has been synthesized and characterized within this work. The crystal structure reveals that the geometry around the metal center is octahedral (Fig. 1B). The stereoselective synthesis results in the Δ-cis-[Ph2Sn(CIP)2] isomer only (Fig. 1B). 2D layer supramolecular assemblies are established based on strong hydrogen bonding interactions (Fig. 3). Four molecules form square planes with their tin atoms placed on the edges.These constitute the building blocks of the layers (Fig. 3). The area covered by each square is 195.133 Å2.CIPTIN has been evaluated towards the Gram negative microbes P. aeruginosa and E. coli and Gram positive microbes S. aureus and S. epidermidis. CIPTIN is classified as a bactericidal agent against P. aeruginosa, E. coli, S. aureus and S. epidermidis (Table 1), while these microbes are ranked among those susceptible to it (IZ values) (Table 1).
The antibacterial activity of CIPTIN (Δ-cis-[Ph2Sn(CIP)2]) is compared to that of CIPAG with the formula ({[Ag(HCIP)2]NO3˙}),8 [Me3Sn(CIP)], and [Cy3Sn(CIP)]13 and PenAg with the formula [Ag(Pen)(CH3OH)]2, (HPen = penicillin)9 and their ingredients HCIP·HCl, HCIP, PenNa, DPTD and AgNO3 (Table 1, Fig. 8).
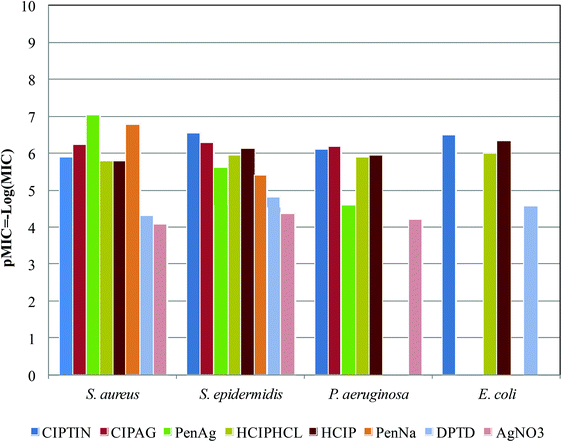 | ||
| Fig. 8 Histogram of pMIC (−log(MIC)) values of CIPTIN (Δ-cis-[Ph2Sn(CIP)2]), ({[Ag(HCIP)2]NO3˙}),8PenAg [Ag(Pen)(CH3OH)]2, (HPen = penicillin),9HCIP·HCl, HCIP, PenNa, DPTD and AgNO3, against S. aureus, S. epidermidis, P. aeruginosa and E. coli. | ||
CIPTIN exhibits stronger activity compared to ciprofloxacin against both the tested Gram positive and Gram negative bacteria as well as [Me3Sn(CIP)] and [Cy3Sn(CIP)],13 suggesting that the presence of the tin(IV) ion enhances the effectiveness of ciprofloxacin even more (Table 1, Fig. 8). Although the activity of DPTC is significantly lower than that of CIPTIN, this should be attributed to the synergistic effect of both the organotin moiety and ciprofloxacin. CIPTIN exhibits superior activity to penicillin against Gram negative microbes, as expected since penicillin is inactive against Gram negative bacteria microbes (Fig. 8). Against Gram positive microbes, CIPTIN exhibits stronger activity against S. epidermidis, while penicillin shows higher activity against S. aureus (Fig. 8). Given the fact that ciprofloxacin exhibits stronger activity than penicillin against S. epidermidis, while penicillin against S. aureus, CIPTIN follows the relative efficiency of ciprofloxacin towards penicillin. Among the compounds of ciprofloxacin, CIPTIN exhibits higher activity compared to CIPAG against S. epidermidis, which however is more effective against S. aureus and P. aeruginosa (Fig. 8). Therefore, the antibiotic activity is differentiated by the type of metal ion which coordinates with the drug. Moreover, among organotins, [Me3Sn(CIP)] and [Cy3Sn(CIP)]13 show stronger activity compared to CIPTIN against P. aeruginosa and S. aureus (Table 1). However, this should be assigned to the higher toxicity of tri-organotin moieties than to that of di-organotin moieties. Although PenAg shows better activity against S. aureus, CIPTIN, however is more drastic against S. epidermidis and P. aeruginosa, where ciprofloxacin itself shows better activity compared to free penicillin (Fig. 8). Therefore, the activity of the metallodrug follows the corresponding efficiency of the antibiotic, regardless of the type of metal ion Sn(IV) or Ag(I). Moreover, CIPTIN shows superior activity to that of silver(I) nitrate against all the tested microbes (Fig. 8), confirming that the conclusion is already drawn for the synergy of the metal ion with the drug on the activity of the metallodrug. CIPTIN eliminates the biofilm in a similar manner to that of the drug (Table 1). The in vitro toxicity of CIPTIN against HaCaT cells shows a selectivity index (SI) higher than 1 suggesting a wide therapeutic window, while its in vitro genotoxicity is even lower than that of HCIP·HCl or HCIP.
In conclusion, the coordination of a drug on a metal ion strongly enhances its activity. The antimicrobial information which has been stored in a drug affects the activity of the metallodrug and it is affected by the coordination demands stored in the metal ion, creating a combination of a new entity with enhanced and novel biological properties.
Experimental
Materials and instruments
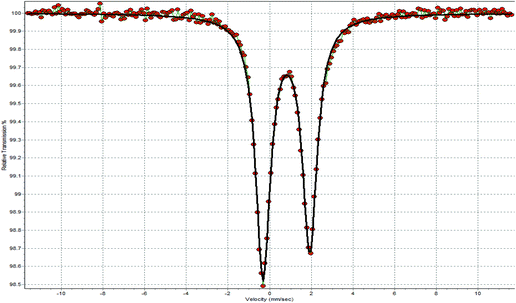
All solvents used were of reagent grade. Tryptone tryptophan medium, beef extract powder, peptone bacteriological, and soy peptone were purchased from Biolife. Agar and yeast extract were purchased from Fluka Analytical. Sodium chloride, D(+)-glucose, and di potassium hydrogen phosphate trihydrate were purchased from Merck. Dulbecco's modified Eagle's medium, (DMEM), fetal bovine serum, glutamine and trypsin were purchased from Gibco, Glasgow, UK. Phosphate buffer saline (PBS) was purchased from Sigma-Aldrich. Melting points were measured in open tubes with Stuart Scientific apparatus and are uncorrected. IR spectra in the region of 4000–370 cm−1 were obtained using a Cary 670 FTIR spectrometer, Agilent Technologies. A UV-1600 PC series spectrophotometer of VWR was used to obtain electronic absorption spectra. The 1H NMR spectra were recorded using a Bruker AC 250, 400 MHFT-NMR instrument in DMSO-d6. ESI-MS spectra were recorded with an Agilent 1100/LC–MS system. 119Sn Mössbauer spectra were recorded at various sample temperatures (85 K), with a constant-acceleration spectrometer equipped with a CaSnO3 source kept at room temperature. The calibration of the spectrometer was carried out with a 57Co source and an Fe absorber at room temperature. The line widths were very close to the natural width (0.28 mm s−1). The content of the tin samples was calculated to be approximately 4 mg Sn in the 2-cm2 area of the sample holder. XRF measurements were carried out using an Am-241 radio isotopic source (exciting radiation 59.5 keV). Thermogravimetry–Differential Thermal Analysis (TG–DTA) of CIPTIN was carried out using Seiko SII TG/DTA 7200 apparatus under a N2 flow (40 cm3 min−1) with a heating rate of 10 K min−1.Synthesis and crystallization of CIPTIN
A clear solution of 0.5 mmol ciprofloxacin hydrochloride (HCIP·HCl) (0.184 g) in dd water (8 mL) was treated with 0.75 mmol KOH (750 μL 1 N). A solution of 0.25 mmol diphenyltin dichloride (DPTD) (0.086 g) in methanol (3 mL) was added to the previous one under continuous stirring for 30 min and the resulting white precipitation was immediately filtered off. Crystals of CIPTIN suitable for X-ray analysis were grown from slow evaporation of the solution after one day.
CIPTIN: Yellowish crystal, melting point: > 300 °C; elemental analysis found: C: 59.55; H: 4.92; N: 9.23%; calculated for C46H44N6O6F2Sn: C: 59.19; H: 4.75; N: 9.00%. IR (cm−1), (KBr): 3906w, 2965w, 2698w, 2487w, 2111w, 1918w, 1604w, 1474w, 1265s, 1142s, 1025s, 944vs, 809s, 739vs, 628s, 446s; 1H NMR (ppm) in DMSO-d6: 8.87 (s, H[Npiperazine ring]), 8.67 (s, H[2C]), 7.95–7.92 (d, H[5C]), 7.79–7.81 (d, H[8C]), 7.72–7.58 (m, Ph-), 7.31–7.16 (m, Ph-), 3.85 (s, H[aC]), 3.47 (d, H[cC]), 1.33 (m, cyclopropyl CH2), 1.19 (m, cyclopropyl CH2); UV-vis (DMSO): λ = 284 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε = 4.68), λ = 320 nm (log
ε = 4.68), λ = 320 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε = 4.21), 331 nm, (log
ε = 4.21), 331 nm, (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε = 4.20).
ε = 4.20).
X-ray structure determination
Intensity data of the crystals of CIPTIN and HCIP were collected using an Oxford Diffraction CCD instrument, using graphite monochromated Mo radiation (λ = 0.71073 Å). Cell parameters were determined by least-squares refinement of the diffraction data from 25 reflections. All data were corrected for Lorentz-polarization effects and absorption.29 The structures were solved with direct methods with SHELXS97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 and refined by full-matrix least-squares procedures on F2 with SHELXL97.31 All non-hydrogen atoms were refined anisotropically, and hydrogen atoms were located at calculated positions and refined via the “riding model” with isotropic thermal parameters fixed at 1.2 (1.3 for CH3 groups) times the Ueq value of the appropriate carrier atom.
30 and refined by full-matrix least-squares procedures on F2 with SHELXL97.31 All non-hydrogen atoms were refined anisotropically, and hydrogen atoms were located at calculated positions and refined via the “riding model” with isotropic thermal parameters fixed at 1.2 (1.3 for CH3 groups) times the Ueq value of the appropriate carrier atom.
CIPTIN: C46H44N6O6F2Sn, MW = 933.612, monoclinic, space group I2/a, a = 14.7410(6), b = 20.2602(5), c = 18.8558(7) Å, α = 90, β = 111.400(4), γ = 90°, V = 5243.1(3) Å3, Z = 4, T = 100 K, ρ (calc) = 1.183 g cm−3, μ = 4.320 mm−1, F(000) = 1912. 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 180 reflections measured, 4682 unique (Rint = 0.052). The final R1 = 0.0523 (for 3818 reflections with I > 2σ(I)) and wR(F2) = 0.1428 (all data) S = 0.99.
180 reflections measured, 4682 unique (Rint = 0.052). The final R1 = 0.0523 (for 3818 reflections with I > 2σ(I)) and wR(F2) = 0.1428 (all data) S = 0.99.
HCIP: C17H18FN3O3, MW = 331.34, triclinic, space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , a = 7.8945(7), b = 8.5010(7), c = 10.7522(9) Å, α = 87.440(7), β = 84.496(7), γ = 88.912(7)°, V = 717.47(11) Å3, Z = 2, T = 100 K, ρ (calc) = 1.534 g cm−3, μ = 0.967 mm−1, F(000) = 348. 4189 reflections measured, 2531 unique (Rint = 0.041). The final R1 = 0.0511 (for 1996 reflections with I > 2σ(I)) and wR(F2) = 0.1440 (all data) S = 1.07.
, a = 7.8945(7), b = 8.5010(7), c = 10.7522(9) Å, α = 87.440(7), β = 84.496(7), γ = 88.912(7)°, V = 717.47(11) Å3, Z = 2, T = 100 K, ρ (calc) = 1.534 g cm−3, μ = 0.967 mm−1, F(000) = 348. 4189 reflections measured, 2531 unique (Rint = 0.041). The final R1 = 0.0511 (for 1996 reflections with I > 2σ(I)) and wR(F2) = 0.1440 (all data) S = 1.07.
Crystallographic data (excluding structure factors) for the structure reported in this paper has been deposited with the Cambridge Crystallographic Data Centre as supplementary publication no. CCDC 1993746 (CIPTIN), CCDC 2002394 (HCIP).†
Biological tests
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM. The growth was assessed after incubation for 20 h. The MIC value was determined as the concentration of the compound which inhibits the visible growth of the bacterium being investigated, while it is confirmed by measuring the solution optical density at 620 nm vs. concentrations.8–10,14–17
000 nM. The growth was assessed after incubation for 20 h. The MIC value was determined as the concentration of the compound which inhibits the visible growth of the bacterium being investigated, while it is confirmed by measuring the solution optical density at 620 nm vs. concentrations.8–10,14–17
In vitro toxicity evaluation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) in glass cover slips which were afterwards placed in six-well plates, with 3 mL of cell culture medium and incubated for 24 h. HaCaT cells were exposed with compounds in IC50 values for a period of 48 h. The number of micronucleated cells per 1000 cells was determined.
000 cells per well) in glass cover slips which were afterwards placed in six-well plates, with 3 mL of cell culture medium and incubated for 24 h. HaCaT cells were exposed with compounds in IC50 values for a period of 48 h. The number of micronucleated cells per 1000 cells was determined.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
[i] This research has been co-financed by the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH – CREATE – INNOVATE (project code: T1EDK- 02990). [ii] This research is also co-financed by Greece and the European Union (European Social Fund- ESF) through the Operational Programme “Human Resources Development, Education and Lifelong Learning” in the context of the project “Strengthening Human Resources Research Potential via Doctorate Research” (MIS-5000432), implemented by the State Scholarships Foundation (IKϒ). (Project No. 2018-050-0502-14406) [iii] Professor T. Vaimakis is thanked for the useful discussion and the recording of TG-DTA/DSC diagrams. [iv] The COST Action CA15114 “Anti-Microbial Coating Innovations to prevent infectious diseases (AMICI)” is thanked for stimulating discussion. (v) S.K.H. acknowledges the Oncology Department of Novartis Hellas S.A.C.I. for the financial support (Project No. 82819).References
- N. R. Cozzarelli, DNA gyrase and the supercoiling of DNA, Science, 1980, 207, 953–960 CrossRef CAS PubMed.
- M. Brunner, O. Langer, G. Dobrozemsky, U. Muller, M. Zeitlinger, M. Mitterhauser, W. Wadsak, R. Dudczak, K. Kletter and M. Müller, [18F] Ciprofloxacin, a new positron emission tomography tracer for noninvasive assessment of the tissue distribution and pharmacokinetics of ciprofloxacin in humans, Antimicrob. Agents Chemother., 2004, 48, 3850–3857 CrossRef CAS PubMed.
- F. Silva, O. Lourenço, J. A. Queiroz and F. C. Domingues, Bacteriostatic versus bactericidal activity of ciprofloxacin in Escherichia coli assessed by flow cytometry using a novel far-red dye, J. Antibiot., 2011, 64, 321–325 CrossRef CAS PubMed.
- M. Mulder, J. C. Kiefte-de Jong, W. H. Goessens, H. de Visser, A. Hofman, B. H. Stricker and A. Verbon, Risk factors for resistance to ciprofloxacin in community-acquired urinary tract infections due to Escherichia coli in an elderly population, J. Antimicrob. Chemother., 2017, 72, 281–289 CrossRef CAS PubMed.
- J. Overgaard, I. Turel and D. E. Hibbs, Experimental electron density study of a complex between copper(II) and the antibacterial quinolone family member ciprofloxacin, Dalton Trans., 2007, 2171–2178 RSC.
- P. Drevensek, T. Zupancic, B. Pihlar, R. Jerala, U. Kolitsch, A. Plaper and I. Turel, Mixed-valence Cu(II)/Cu(I) complex of quinolone ciprofloxacin isolated by a hydrothermal reaction in the presence of L-histidine: comparison of biological activities of various copper-ciprofloxacin compounds, J. Inorg. Biochem., 2005, 99, 432–442 CrossRef CAS PubMed.
- P. Drevensek, N. Poklar Ulrih, A. Majerle and I. Turel, Synthesis, characterization and DNA binding of magnesium-ciprofloxacin (cfH) complex [Mg(cf)2] * 2.5H2O, J. Inorg. Biochem., 2006, 100, 1705–1713 CrossRef CAS PubMed.
- I. Milionis, C. N. Banti, I. Sainis, C. P. Raptopoulou, V. Psycharis, N. Kourkoumelis and S. K. Hadjikakou, Silver ciprofloxacin (CIPAG): a successful combination of chemically modified antibiotic in inorganic–organic hybrid, JBIC, J. Biol. Inorg. Chem., 2018, 23, 705–723 CrossRef CAS PubMed.
- I. Ketikidis, C. N. Banti, N. Kourkoumelis, C. G. Tsiafoulis, C. Papachristodoulou, A. G. Kalampounias and S. K. Hadjikakou, Conjugation of Penicillin-G with Silver(I) Ions Expands Its Antimicrobial Activity against Gram Negative Bacteria, Antibiotics, 2020, 9, 25 CrossRef CAS PubMed.
- M. P. Chrysouli, C. N. Banti, I. Milionis, D. Koumasi, C. P. Raptopoulou, V. Psycharis, I. Sainis and S. K. Hadjikakou, A water-soluble silver(I) formulation as an effective disinfectant of contact lenses cases, Mater. Sci. Eng., C, 2018, 93, 902–910 CrossRef CAS PubMed.
- P. J. Smith, Chemistry of Tin, Blackie Academic & Professional, London, 2nd edn, 1997, ISBN 0-7514-0385-7 Search PubMed.
- A. G. Davies, Organotin Chemistry, Wiley-VCH Verlag GmbH & Co. KGaA, 2nd edn, 2004 Search PubMed.
- R. Joshi, S. K. Yadav, H. Mishra, N. Pandey, R. Tilak and S. Pokharia, Interaction of triorganotin(IV) moiety with quinolone antibacterial drug ciprofloxacin: Synthesis, spectroscopic investigation, electronic structure calculation, and biological evaluation, Heteroat. Chem., 2018, 29, 21433 CrossRef.
- I. Sainis, C. N. Banti, A. M. Owczarzak, L. Kyros, N. Kourkoumelis, M. Kubicki and S. K. Hadjikakou, New antibacterial, non-genotoxic materials, derived from the functionalization of the anti-thyroid drug methimazole with silver ions, J. Inorg. Biochem., 2016, 160, 114–124 CrossRef CAS PubMed.
- M. K. Stathopoulou, C. N. Banti, N. Kourkoumelis, A. G. Hatzidimitriou, A. G. Kalampounias and S. K. Hadjikakou, Silver complex of salicylic acid and its hydrogel- cream in wound healing chemotherapy, J. Inorg. Biochem., 2018, 181, 41–55 CrossRef CAS PubMed.
- A. Papadimitriou, I. Ketikidis, M.-E. K. Stathopoulou, C. N. Banti, C. Papachristodoulou, L. Zoumpoulakis, S. Agathopoulos, G. V. Vagenas and S. K. Hadjikakou, Innovative material containing the natural product curcumin, with enhanced antimicrobial properties for active packaging, Mater. Sci. Eng., C, 2018, 84, 118–122 CrossRef CAS PubMed.
- V. A. Karetsi, C. N. Banti, N. Kourkoumelis, C. Papachristodoulou, C. D. Stalikas, C. P. Raptopoulou, V. Psycharis, P. Zoumpoulakis, T. Mavromoustakos, I. Sainis and S. K. Hadjikakou, An Efficient Disinfectant, Composite Material {SLS@[Zn3(CitH)2]} as Ingredient for Development of Sterilized and Non Infectious Contact Lens, Antibiotics, 2019, 8, 213 CrossRef CAS PubMed.
- F. P. A. Fabbiani, B. Dittrich, A. J. Florence, T. Gelbrich, M. B. Hursthouse, W. F. Kuhs, N. Shankland and H. Sowa, Crystal structures with a challenge: high-pressure crystallisation of ciprofloxacin sodium salts and their recovery to ambient pressure, CrystEngComm, 2009, 11, 1396–1406 RSC.
- C. N. Banti, E. I. Gkaniatsou, N. Kourkoumelis, M. J. Manos, A. J. Tasiopoulos, T. Bakas and S. K. Hadjikakou, Assessment of organotins against the linoleic acid, glutathione and CT-DNA, Inorg. Chim. Acta, 2014, 423, 98–106 CrossRef CAS.
- S. K. Hadjikakou, M. A. Abdellah, N. Hadjiliadis, M. Kubicki, T. Bakas, N. Kourkoumelis, Y. V. Simos, S. Karkabounas, M. M. Barsan and I. S. Butler, Synthesis, characterization, and biological studies of organotin(IV) derivatives with o- or p-hydroxybenzoic acids, Bioinorg. Chem. Appl., 2009, 542979 Search PubMed.
- M. N. Xanthopoulou, S. K. Hadjikakou, N. Hadjiliadis, M. Kubicki, S. Skoulika, T. Bakas, M. Baril and I. S. Butler, Synthesis, structural characterization, and biological studies of six- and five-coordinate organotin(IV) complexes with the thioamides 2- mercaptobenzothiazole, 5-chloro-2-mercaptobenzothiazole, and 2-mercaptobenzoxazole, Inorg. Chem., 2007, 46, 1187 CrossRef CAS PubMed.
- M. N. Xanthopoulou, S. K. Hadjikakou, N. Hadjiliadis, M. Schürmann, K. Jurkschat, A. Michaelides, S. Skoulika, T. Bakas, J. Binolis, S. Karkabounas and K. Charalabopoulos, Synthesis, structural characterization and in vitro cytotoxicity of organotin(IV) derivatives of heterocyclic thioamides, 2-mercaptobenzothiazole, 5-chloro-2-mercaptobenzothiazole, 3-methyl-2-mercaptobenzothiazole and 2-mercaptonicotinic acid, J. Inorg. Biochem., 2003, 96, 425–434 CrossRef CAS PubMed.
- P. Pachori, R. Gothalwal and P. Gandhi, Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review, Genes Dis., 2019, 6, 109–119 CrossRef PubMed.
- M. Otto, Staphylococcus epidermidis – the “accidental” pathogen, Nat. Rev. Microbiol., 2009, 7, 555–567 CrossRef CAS PubMed.
- D. L. Shungu, E. Weinberg and H. H. Gadebusch, Tentative interpretive standards for disk diffusion susceptibility testing with norfloxacin (MK-0366, AM-715), Antimicrob. Agents Chemother., 1983, 23, 256–260 CrossRef CAS PubMed.
- M. P. Pereira and S. O. Kelley, Maximizing the therapeutic window of an antimicrobial drug by imparting mitochondrial sequestration in human cells, J. Am. Chem. Soc., 2011, 133, 3260–3263 CrossRef CAS PubMed.
- C. N. Banti and S. K. Hadjikakou, Evaluation of Genotoxicity by Micronucleus Assay in vitro and by Allium cepa Test in vivo, Bio-Protoc., 2019, 9, e3311 Search PubMed.
- M. Fenech, The micronucleus assay determination of chromosomal level DNA damage, Methods Mol. Biol., 2008, 410, 185–216 CrossRef CAS PubMed.
- CrysAlis RED, version 1.171.31.5, Oxford Diffraction Ltd., 2006 (release 28-08-2006 CrysAlis171.NET), Oxford Diffraction, Crysalis CCD and Crysalis RED, Version p171.29.2, Oxford Diffraction Ltd., Abingdon, Oxford, England, 2006 Search PubMed.
- G. M. Sheldrick, Phase annealing in SHELX-90: direct methods for larger structures, ActaCrystallogr., Sect.A: Found. Crystallogr., 1990, 46, 467 CrossRef.
- G. M. Sheldrick, SHELXL-97, Program for the Refinement of Crystal Structures, University of Göttingen, Göttingen, Germany, 1997 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1993746 and 2002394. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/D0DT01665A |
| This journal is © The Royal Society of Chemistry 2020 |