Open Access Article
Open Access ArticleFatty acid epoxidation by Collariella virescens peroxygenase and heme-channel variants†
Alejandro
González-Benjumea‡
 a,
Juan
Carro‡
b,
Chantal
Renau-Mínguez
a,
Juan
Carro‡
b,
Chantal
Renau-Mínguez
 b,
Dolores
Linde
b,
Elena
Fernández-Fueyo§
b,
Dolores
Linde
b,
Elena
Fernández-Fueyo§
 b,
Ana
Gutiérrez
b,
Ana
Gutiérrez
 *a and
Angel T.
Martínez
*a and
Angel T.
Martínez
 *b
*b
aInstituto de Recursos Naturales y Agrobiología de Sevilla, CSIC, Reina Mercedes 10, E-41012 Sevilla, Spain. E-mail: anagu@irnase.csic.es
bCentro de Investigaciones Biológicas, CSIC, Ramiro de Maeztu 9, E-28040 Madrid, Spain. E-mail: atmartinez@cib.csic.es
First published on 10th January 2020
Abstract
Enzyme-driven oxygenation reactions are in the spotlight for organic synthesis. In this regard, a heme-thiolate unspecific peroxygenase (UPO) from the fungus Chaetomium globosum has recently proven to be a suitable catalyst for selective epoxidation of unsaturated fatty acids in the context of the bio-based industry, but this enzyme could not be expressed in Escherichia coli for directed mutagenesis studies. Here, a previously unknown UPO from the related Collariella virescens (synonym: Chaetomium virescens) was obtained by E. coli expression of a putative upo gene. The activity of the purified enzyme on unsaturated fatty acids with different lengths and unsaturation degrees was tested. The ability of C. virescens UPO to epoxidize these compounds increases in the order myristoleic acid (C14:1) < palmitoleic acid (C16:1) < oleic acid (C18:1) differing from that observed for the C. globosum UPO, which also forms less hydroxylated derivatives. Given the possibility to produce the C. virescens UPO in E. coli as a recombinant enzyme and its oxyfunctionalization ability, some mutated variants were obtained mimicking the active-site of C. globosum UPO and evaluated on 18-carbon unsaturated fatty acids. Results revealed that widening the heme-access channel of C. virescens UPO by substituting a phenylalanine residue (in a F88L variant) maintains the enzyme epoxidation activity, and reduces undesired hydroxylation side-reactions (from 34% to only 7% of linoleic acid products) approaching the oxyfunctionalization pattern obtained with C. globosum UPO, although maintaining the absence of diepoxides. Conversely, its partial occlusion by introducing a second phenylalanine residue (in a T158F variant) resulted in partial selective epoxidation of linoleic acid (C18:2), while the oleic acid epoxidation was prevented. The above results show how E. coli expression can speed up the availability of new UPOs, and the design of ad hoc variants of these self-sufficient monooxygenases.
Introduction
Unspecific peroxygenases (UPOs) have been described as “dream” biocatalysts for a variety of oxyfunctionalization reactions of industrial interest that are difficult (sometimes impossible) to be carried out only by chemical means.1 Although the natural substrates and functions of UPOs are still to be identified, the known reactions of these promising biocatalysts comprise alkane hydroxylation, epoxidation of alkenes and aromatics, heteroatom oxygenation, O- and N-dealkylation and one-electron oxidation, among others.2 Previously described heme-thiolate peroxidases/peroxygenases include, after the classical chloroperoxidase of Leptoxyphium fumago,3 the wild (non-recombinant) UPOs successively reported from Agrocybe aegerita,4Coprinellus radians,5Marasmius rotula,6Chaetomium globosum,7 and Marasmius wettsteinii,8 and the recombinant UPOs of Coprinopsis cinerea9 and Humicola insolens produced by Novozymes A/S (Bagsvaerd, DK).7 Although with a fully different phylogenetic origin, UPOs share active-site characteristics (such as the cysteine sulphur atom as a heme iron ligand) and common reaction chemistry with P450 monooxygenases (P450s).10 However, UPOs exhibit important biotechnological advantages due to their higher stability – they are secreted enzymes while P450s are intracellular proteins – and self-sufficient nature, in the sense that they are directly oxidized by H2O2 while P450s often require an auxiliary flavin-containing enzyme of the protein domain and a source of reducing power.2,11,12 UPOs also present advantages for oxygenation reactions over: i) non-heme monooxygenases that share disadvantages with P450s due to their intracellular nature;13 and ii) lipases that generate aggressive peracids in chemo-enzymatic epoxidation.14A new member of the heme-thiolate peroxidase superfamily has been obtained by the authors, by Escherichia coli expression of a putative upo gene of Collariella virescens (syn. Chaetomium virescens). C. virescens is a chaetomium-like fungus11 that, together with C. globosum and Humicola insolens, belongs to the Chaetomiaceae family of ascomycetes. In contrast, the other fungal species cited above (except L. fumago) are basidiomycetes. Interestingly, putative upo genes (up to several thousand in sequenced genomes) are unique to ascomycete and basidiomycete species, with only a few exceptions due to horizontal gene transfer.15 The C. virescens UPO has not been isolated from the corresponding fungus, being therefore only known as a recombinant protein. Heterologous expression in E. coli provides a way for expanding the limited repertoire of currently available UPO enzymes by exploring the abundant putative upo genes identified in sequenced fungal genomes and other databases.15,16 In a similar way, E. coli expression paves the way for investigating structural–functional relationships and engineering UPOs, as recently shown17 for the previously described6 UPO of M. rotula that catalyzes several unique reactions.8,18–23
Here, we report C. virescens UPO (produced in E. coli) as a new model enzyme for epoxidation reactions. Epoxides are among the most widely used intermediates in organic synthesis as precursors to complex molecules.24 Here, we focus on fatty-acid epoxidation, recently reported for C. globosum and M. rotula UPOs but absent from the reaction repertoire of the best known UPO of A. aegerita.19 Reactive fatty-acid epoxides from lipid feedstocks are of interest for the production of bio-based adhesives and other chemicals.25 This is a new area of potential UPO application, in the frame of circular economy, together with the stereo/regio-selective syntheses of specialty chemicals, pharmaceuticals and other molecules of different origins.1,12,26 With the above purpose, unsaturated fatty acids representative of different plant oils were treated with the C. virescens UPO. The results were compared to those reported for the C. globosum UPO.19 Moreover, the key importance of the heme-access channel architecture for the epoxidation of unsaturated fatty acids is analyzed here by site-directed mutagenesis.
Material and methods
UPO production
The protein sequence of C. virescens UPO was obtained from the patent of Lund et al.27 and the wild-type recombinant (hereinafter native) enzyme was produced in E. coli as summarized below. DNA sequences were optimized for E. coli expression,28 synthesized by ATG:biosynthetics GmbH (Germany) and cloned into the pET23a plasmid under the control of the T7lac promoter. The plasmid was transformed into E. coli C41 (DE3) competent cells, which were grown in lactose-containing auto-induction medium ZYM-505229 for 4–5 days, supplemented with hemin at 16 °C and 150 rpm to obtain the UPO as a soluble and active enzyme.17,30 Cells were harvested and resuspended in 10 mM tris(hydroxymethyl)-aminomethane (Tris, pH 7.4), 5 mM dithiothreitol and 20 mM ethylenediaminetetraacetic acid. Lysozyme (2 mg ml−1), DNase and sonication were used to lyse the cells, and the soluble fraction containing the recombinant UPO was recovered after debris removal.UPO purification
The recombinant enzyme was purified through two chromatographic steps in an ÄKTA (GE Healthcare) fast protein liquid chromatography system. Cation-exchange chromatography was firstly applied using a HiTrap SPFF column (GE Healthcare, USA) in 10 mM Tris (pH 7.4). The retained proteins were eluted with a gradient of 1 M NaCl (in the same buffer) and concentrated in Amicon 3 K (Sigma-Aldrich, USA). The final purification step was isocratic size-exclusion chromatography (SEC) with a Superdex 75 column 10/300 GL (GE Healthcare, USA), in 10 mM Tris (pH 7.4) with 0.15 M NaCl. Protein elution was monitored at 410 nm (heme proteins) and 280 nm (all proteins).At the different purification steps, the yield was monitored using a Thermo Scientific NanoDrop 2000 at 280 nm, and activity by measuring 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) and veratryl alcohol oxidation (in 0.1 M Tris, pH 7.4, containing 5 mM ABTS or veratryl alcohol and 1 mM H2O2) to the ABTS cation radical (ε436 nm 29.30 mM−1 cm−1) and veratraldehyde (ε310 nm 9.30 mM−1 cm−1), respectively. Enzyme purification was confirmed by 12% polyacrylamide gel electrophoresis (PAGE) in the presence of 0.1% sodium dodecylsulfate (SDS) and ∼1% mercaptoethanol (with 5% concentration in the loading buffer). SDS-PAGE yielded the molecular mass of the protein monomer, while the native protein mass was provided by Superdex 75 SEC under the conditions described above.
Proper folding and binding of the cofactor was evaluated by inspecting the UV-vis spectrum of the resting state of the enzyme in 10 mM phosphate (pH 7.4) using a Cary 60 spectrophotometer. Formation of a complex between the chemically-reduced enzyme (ferrous form) and CO, characteristic of active heme-thiolate enzymes,31 was confirmed in 0.2 M phosphate (pH 8) after addition of Na2S2O4 and CO flushing.
Molecular modeling and directed mutagenesis
The molecular structure of C. virescens UPO was examined after its homology modeling at the Swiss-Model server (https://swissmodel.expasy.org),32 using the crystal structures of A. aegerita and M. rotula UPOs available in the Protein Data Bank (PDB 2YP1 and 5FUJ, respectively) as templates. PyMOL ver 2.033 and Swiss-PdbViewer ver 4.134 were used for examining the molecular structures and in silico designing UPO mutated variants.Simple (F88L and T158F) and double (F88L/T158F) mutations, mimicking the C. globosum UPO active site, were introduced in the upo gene of C. virescens using the Expand Long Template PCR kit from Roche (Basel, Switzerland). PCR reactions were run using the following DNA oligos harboring the desired mismatches (substituted nucleotides underlined): i) F88L mutation, 5′-AAC CGC CAT AAC CTG TT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) GAA CAT GAT GCA TCT C-3′; and ii) T158F mutation, 5′-ACT TAC ACC GTT CAG CAG CGT ATC
GAA CAT GAT GCA TCT C-3′; and ii) T158F mutation, 5′-ACT TAC ACC GTT CAG CAG CGT ATC ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) AGT TAC GGT GAA ACG-3′ (along with their reverse complementary counterparts). To obtain the simple mutated gene, the CvirUPO-pET23a plasmid was used as a template, while the plasmid harboring the T158F mutation, combined with the F88L oligos, was used to obtain the doubly mutated gene.
AGT TAC GGT GAA ACG-3′ (along with their reverse complementary counterparts). To obtain the simple mutated gene, the CvirUPO-pET23a plasmid was used as a template, while the plasmid harboring the T158F mutation, combined with the F88L oligos, was used to obtain the doubly mutated gene.
The PCR reactions (50 μL volume) were carried out in an Eppendorf (Hamburg, Germany) Mastercycler pro-S using 30 ng of template DNA, 0.5 mM each dNTP, 125 ng of direct and reverse primers, 5 units of Expand Long Template PCR System polymerase mix (Roche), and the manufacturer's buffer. Reaction conditions included: i) the initial denaturation step of 1 min at 95 °C; ii) 22 cycles of 30 s at 95 °C, 30 s at 60 °C, and 7 min at 68 °C, each; and iii) the final elongation step of 7 min at 68 °C. The mutated variants were produced in E. coli and purified as described for the native enzyme.
Substrate and product standards
The following unsaturated fatty acids (cis isomers) were obtained from Sigma-Aldrich to be used as UPO substrates: myristoleic (cis-9-tetradecenoic), palmitoleic (cis-9-hexadecenoic), oleic (cis-9-octadecenoic), linoleic (cis,cis-9,12-octadecadienoic) and α-linolenic (cis,cis,cis-9,12,15-octadecatrienoic) acids. Saturated myristic (tetradecanoic) acid, analyzed for comparative purposes, was also from Sigma-Aldrich.The following epoxide standards were used to identify the reaction products: (±)-cis-9,10-epoxyoctadecanoic acid and (±)-9(10)-EpOME (coronaric acid, corresponding to 9,10-cis-epoxidized linoleic acid) from Santa Cruz Biotechnology, (±)-12(13)-EpOME (vernolic acid, corresponding to 12,13-cis-epoxidized linoleic acid) from Cayman, and (±)-9,10−12,13-diepoxyoctadecanoic acid from Larodan. Since standards of epoxidized α-linolenic acid are not available, they were chemically synthesized by adding a solution of peracetic acid (1.8 mmol) and sodium acetate (0.7 mmol) to α-linolenic acid (0.5 mmol) using a syringe pump for 1 h at 0 °C. The reaction was stirred for an additional hour, and a mixture of monoepoxides and diepoxides was recovered by liquid–liquid extraction with methyl tert-butyl ether.
Oxyfunctionalization by native UPO and its mutated variants
The above monounsaturated and polyunsaturated fatty acids were used as substrates to evaluate the epoxidation ability of the native C. virescens UPO, as previously described for the C. globosum UPO.19 Moreover, its action on saturated vs. unsaturated fatty acids was investigated using 14-carbon myristic and myristoleic acids. Up to 30 min reactions were performed with 0.1 mM substrate, 0.25 μM enzyme and 1.25 mM H2O2 in 50 mM phosphate (pH 7) containing 20% acetone. In control experiments, substrates were treated under the same conditions but without the enzyme.The effect of the F88L and T158F mutations on the epoxidation ability of the C. virescens UPO was evaluated on the 18-carbon mono-, di- and tri-unsaturated fatty acids (oleic, linoleic and α-linolenic acids, respectively). For this comparison, 30 min reactions were performed under the above conditions using 0.25–0.40 μM enzyme.
Product recovery and GC-MS analyses
Products from the above reactions were liquid–liquid extracted with methyl tert-butyl ether, and dried under N2. N,O-Bis(trimethylsilyl)trifluoroacetamide (Supelco) was used to prepare trimethylsilyl derivatives. Their GC-MS analyses were performed with GC-MS QP2010 Ultra equipment using a fused-silica DB-5HT 30-m capillary column from J&W Scientific (USA). The oven was heated from 120 °C (1 min) to 300 °C (15 min) at 5 °C min−1. The injector and transfer line were kept at 300 °C. Compounds were identified by mass fragmentography and by comparison of their mass spectra with those of authentic standards. Quantifications were obtained from total-ion peak areas using external standard curves and molar response factors of the same or similar compounds.Kinetic analyses
Kinetic curves for oleic acid oxidation by the native enzyme and the two variants (75 nM) were obtained by varying the concentration of the substrate from 6.25 μM to 1.6 mM, in 50 mM phosphate (pH 7) containing 20% acetone (v/v) at 30 °C. Reactions (1 mL volume) were triggered by the addition of 0.5 mM H2O2, and stopped after 1 min by vigorous shaking with 100 μL of 0.1 M sodium azide. The products were extracted and analyzed by GC-MS, as described above. Reactions were carried out in triplicate. After analyzing the kinetic curves obtained, means and standard errors for the constant values were obtained by nonlinear-least-squares fitting to the Michaelis–Menten model.Results
New C. virescens UPO
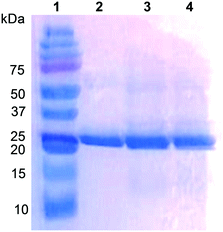
The UPO from the ascomycete C. virescens was produced in E. coli as an active and soluble enzyme, and purified by two steps of cation exchange (HiTrap SPFF column) and SEC (Superdex 75 column) to electrophoretic homogeneity (Fig. 1, lane 2). Proper folding and heme incorporation were shown by the Reinheitszahl (A420/A280 ratio) of 1.6 and the characteristic displacement of the Soret band in the CO complex. The dimeric nature of this UPO was revealed by the two-fold higher molecular mass from Superdex 75 SEC (∼52 kDa, Fig. S1†) compared with the mass estimated from SDS-PAGE under reducing conditions (∼25 kDa, Fig. 1).Typically, 3 L of E. coli culture yielded 292 U total UPO activity estimated with ABTS (or 36 U estimated with veratryl alcohol) after cation-exchange chromatography of the cell lysate (yield 60%), corresponding to 59 mg of protein with a specific activity of 4.9 U mg−1 estimated with ABTS (or 0.6 U mg−1 estimated with veratryl alcohol) that should be sufficient for oxyfunctionalization reactions. The specific activity increases to 38.2 U mg−1 (estimated with ABTS) after the subsequent SEC step, but the yield decreases to ∼25%.
Fatty-acid epoxidation by native C. virescens UPO
The ability of C. virescens UPO to epoxidize unsaturated fatty acids of different chain lengths (from 14 to 18 carbon atoms) and unsaturation degrees (18-carbon acids with 1–3 double bonds) was investigated, and a GC-MS comparison is provided in Fig. 2A–E. Firstly, a positive effect of increasing chain length is observed, since oleic and myristoleic acids are the best and the worst UPO mono-unsaturated substrates, respectively (as shown by the decrease of the peak of the unreacted substrate in Fig. 2A–C). The epoxy derivative was the most abundant product of the reactions with the three mono-unsaturated substrates, followed by the hydroxy derivative at the more terminal allylic positions: ω-3, ω-5, and ω-7 in Fig. 2A to C, respectively (with small amounts of more terminal hydroxy-fatty acids). Interestingly, although (sub)terminal hydroxylation is the preferred reaction on saturated fatty acids, such a reaction was insignificant with mono-unsaturated fatty acids (as shown here for usual Δ9 fatty acids). The above is illustrated in the reactions of C. virescens UPO with myristoleic and myristic acids yielding as the main products the epoxide and (ω-1)-hydroxy derivatives, respectively (Fig. S2A and B,† respectively).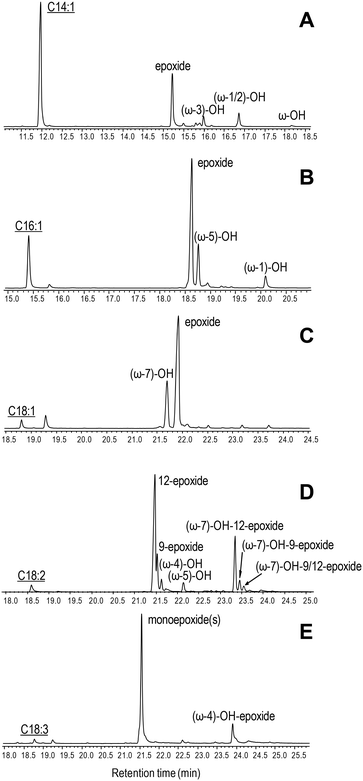 | ||
| Fig. 2 GC-MS comparison of C. virescens UPO reactions with myristoleic (A), palmitoleic (B), oleic (C), linoleic (D) and α-linolenic (E) acids. The reactions shown were performed with 0.1 mM fatty acid, 0.25–0.40 μM enzyme and 1.25 mM H2O2, in 50 mM phosphate (pH 7) at 30 °C, for 30 min. Peaks of hydroxy, epoxy and hydroxy–epoxy derivatives are shown, together with the remaining substrate (underlined). The small peaks after the substrate are fatty-acid impurities. See Table 2 for peak quantification. | ||
Regarding di- and tri-unsaturated fatty acids (linoleic and α- linolenic acids, respectively) (Fig. 2D and E), the epoxidation degree was higher than that found for mono-unsaturated oleic acid, with (non-epoxidized) hydroxy-fatty acids appearing as very small peaks. In fact, nearly complete epoxidation of the two polyunsaturated fatty acids was achieved if we consider both epoxides and hydroxy-epoxides, such as (ω-7)-hydroxy-12-epoxide and (ω-7)-hydroxy-9-epoxide in Fig. 2D and (ω-4)-hydroxy-epoxide in Fig. 2E. Epoxides at the more terminal double bond are the main products from the above polyunsaturated fatty acids, followed by other monoepoxides. No diepoxides or triepoxides could be identified in spite of the use of different reagent concentrations and reaction conditions (up to 1 μM enzyme, 5 mM H2O2 and 1 h incubation time).
Molecular structure of the C. virescens UPO
Crystal structures are available for the wild UPOs of A. aegerita (PDB 2YP1) and M. rotula (PDB 5FUJ and 5FUK), and a homology model has been reported for the C. globosum UPO.20 In a similar way, a molecular model was generated here for the C. virescens UPO, as the basis for investigating its structural–functional relationships.The C. virescens UPO would be a predominantly helical protein (Fig. S3A†), with the heme cofactor buried in a central cavity connected to the solvent by a main access channel (Fig. S3B†). Such a channel enables substrate access directly over the iron ion of the cofactor, at the so-called distal side of the heme (opposed to the proximal side, where the sulfur atom of a cysteine residue occupies the fifth coordination position of the heme iron).
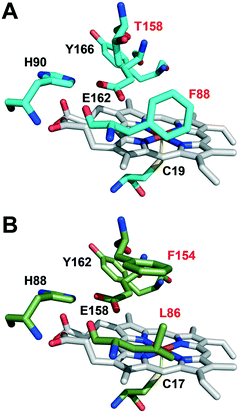
The active-site residues in C. virescens UPO were compared to those of UPO from the related C. globosum (Fig. 3). In the enzymes of C. virescens/C. globosum, His90/88, Glu162/158 and Tyr 166/162 were conserved at the upper (distal) side of the heme, where catalysis takes place, together with the proximal Cys19/17 at the lower side. As suggested for other UPOs,2 Glu162 would act as a catalytic acid/base in the heterolytic cleavage of H2O2 for enzyme activation, and His90 would contribute to the reaction.
However, the positions corresponding to distal Leu86 and Phe154 in C. globosum are occupied by Phe88 and Thr158, respectively, in the C. virescens enzyme. The two latter residues, which are located near the heme access channel with Phe88 fully exposed to the solvent (Fig. S3B†), were mutated here to mimic the C. globosum UPO active site. Unfortunately, an active double variant (F88L/T158F) could not be obtained, but the two simple variants (F88L and T158F) were purified to homogeneity (Fig. 1, lanes 3 and 4, respectively) and the reactions were investigated to understand the differences observed in the catalytic properties of these two UPOs, despite their common origin from chaetomium-like fungi.
Comparison of native UPO and its mutated variants
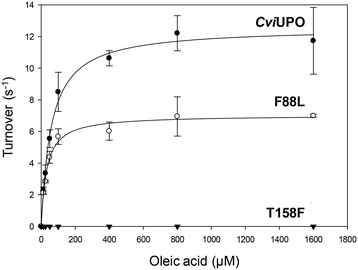
The oxygenation ability of the native C. virescens UPO and its F88L and T158F variants was evaluated using 18-carbon oleic, linoleic and α-linolenic acids as substrates. Given the solubility limitations of polyunsaturated fatty acids, kinetic curves could be only obtained for oleic acid (Fig. 4), whose catalytic constants are provided in Table 1. The activity on oleic acid, which was fully abolished in the T158F variant, slightly increased in terms of the catalytic efficiency in F88L (despite a lower turnover number) due to a lower Km value revealing a higher affinity for the substrate. This affinity was even higher for the wild C. globosum UPO, resulting in the highest catalytic efficiency despite the higher turnover number of the C. virescens enzyme.Due to the above-mentioned solubility limitations, a comparison of UPO activities on polyunsaturated fatty acids was based on chromatographic analyses at fixed incubation times, in agreement with previous studies.19 The effect of mutations, as shown by the GC-MS profiles of the two variants compared with the native enzyme (Fig. S4–S6†) and the product quantification provided in Table 2, included: i) the decrease of epoxidation by the T158F variant in the order α-linolenic acid (C18:3) > linoleic acid (C18:2) > oleic acid (C18:1), with full loss of activity on the latter acid (Fig. 5) and 11% and 85% total conversion of linoleic and α-linolenic acids, respectively (Table 2); and ii) the increase in selectivity, with a lower amount of undesired hydroxylated side-products, by the F88L variant, which was more evident in the reaction of linoleic acid with 9-epoxide and 12-epoxide as the main products (Fig. 6). Indeed, the total amount of linoleic acid hydroxylated derivatives (hydroxy and hydroxy-epoxides included) decreased from 34% of the identified products in the native UPO reaction to only 7% in the F88L reaction (Table 2). Such a decrease was not produced by the T158F variant that even increased the amount of hydroxy α-linolenic acid (Table 2) due to the formation of the new (ω-6)-hydroxy-derivative by double bond displacement (Fig. S6C†).
| Products (%) | Conversion (%) | |||||
|---|---|---|---|---|---|---|
| 15-Epoxy | 12-Epoxyb | 9-Epoxyb | Hydroxy | Hydroxy–epoxy | ||
| a In 50 mM phosphate (pH 7) containing 0.1 mM substrate, 1.25 mM H2O2, 20% acetone, and 0.25–0.40 μM enzyme, at 30 °C. b Corresponding to different products from each fatty acid, as shown in Fig. S7. c —, not detected. | ||||||
| Oleic: | ||||||
| CviUPO | —c | — | 70.0 | 28.6 | 1.4 | 96 |
| F88L | — | — | 85.6 | 14.4 | — | 77 |
| T158F | — | — | — | — | 0 | |
| Linoleic: | ||||||
| CviUPO | — | 55.8 | 10.2 | 8.2 | 25.8 | 96 |
| F88L | — | 56.6 | 36.3 | 7.1 | — | 93 |
| T158F | — | 89.0 | — | 11.0 | — | 11 |
| α-Linolenic: | ||||||
| CviUPO | 77.4 | 5.9 | 1.7 | — | 15.0 | 97 |
| F88L | 78.0 | 4.8 | 15.9 | 1.3 | — | 99 |
| T158F | 58.4 | 25.9 | — | 15.7 | — | 85 |
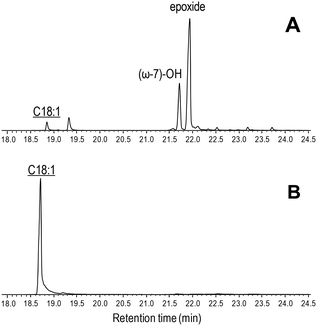 | ||
| Fig. 5 GC-MS comparison of the reactions of C. virescens UPO (A) and its T158F variant (B) with oleic acid. Peaks of hydroxy and epoxy derivatives are shown, together with the remaining substrate (underlined). The small peak after the substrate is a fatty-acid impurity. See Fig. 2 for the reaction conditions and Table 2 for peak quantification. | ||
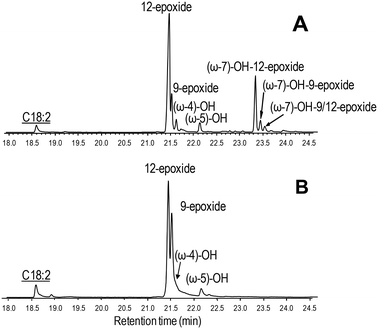 | ||
| Fig. 6 GC-MS comparison of the reactions of C. virescens UPO (A) and its F88L variant (B) with linoleic acid. Peaks of epoxy, hydroxy and hydroxy-epoxide derivatives are shown, together with the remaining substrate (underlined). See Fig. 2 for the reaction conditions and Table 2 for peak quantification. | ||
Discussion
C. globosum UPO catalyzes oxygenation reactions of interest,7,20,35 fatty-acid selective epoxidation included,19 but has not been expressed in E. coli to date. On the other hand, the sequence of a putative UPO of the chaetomium-like fungus C. virescens was reported by Novozymes A/S,27,36 but no information on its production or ability to epoxidize fatty acids had been reported till its current expression in E. coli. Simple (i.e. non-hydroxylated) epoxides are the main products of the reaction of unsaturated fatty acids with the UPOs from these two related ascomycete species, in contrast to the basidiomycete UPOs that lack epoxidation activity on oleic and linoleic acids (such as A. aegerita and C. cinerea UPOs),9,37 or those yielding abundant hydroxylated epoxides from oleic and other unsaturated fatty acids (such as M. rotula UPO).19 Nevertheless, interesting differences between the C. globosum and C. virescens UPOs are also observed. Such dissimilarities mainly concern the reaction with polyunsaturated fatty acids, where diepoxides and diepoxide derivatives are formed by the C. globosum enzyme, while we did not detect them in the C. virescens UPO reactions. Therefore, using these two chaetomium-like UPOs, the balance between monoepoxides and diepoxides could be controlled in the oxygenation of polyunsaturated fatty acids, such as α-linolenic acid.The availability of recombinant C. virescens UPO, with higher production levels than those reported for E. coli-expressed M. rotula UPO,17 opens the possibility of (i) engineering ascomycete UPOs for ad hoc reactions, and (ii) further investigating the differences between the chaetomium-like and other UPOs. In this context, the mutations introduced in the C. virescens UPO were based on the heme environment differences with the C. globosum enzyme. Although the oxygenation properties of these first two variants did not reproduce those of the latter UPO (an active double variant with the same residues of the C. globosum enzyme could not be obtained), significant changes were observed that, together with previous results from site-directed mutagenesis of M. rotula UPO,17 contribute to our understanding of the structural–functional relationships in UPO enzymes and will help to obtain ad hoc oxygenation biocatalysts.
Thus, it was observed that the T158F mutation reduced the fatty-acid epoxidation ability of the enzyme, which was completely absent when oleic acid was used as a substrate. However, some epoxidation was obtained on linoleic acid, and α-linolenic acid was largely epoxidized by this variant, yielding also a new hydroxy-fatty acid.
The effect of the T158F mutation on the solvent access surface of the enzyme was examined to explain such very different activities. After in silico mutation, the main heme access channel of the native UPO (Fig. 7A, arrow) was completely occluded in the T158F variant (by the phenylalanine side chain) and only a small accessory channel remained (Fig. 7C, arrowhead). This would cause significant limitations in the substrate access that, in agreement with the epoxidation experimental results, were: i) the most important for oleic acid that needs a wider channel to access in the bent position required for approaching its ω-8 (Δ9) carbon to the heme Fe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of the H2O2-activated enzyme,17 and ii) the least important for α-linolenic acid that only needs to approach its subterminal ω-2 (Δ15) carbon to be epoxidized. Additional study is required to confirm if the above residual epoxidation is produced at the small secondary channel previously mentioned.
O of the H2O2-activated enzyme,17 and ii) the least important for α-linolenic acid that only needs to approach its subterminal ω-2 (Δ15) carbon to be epoxidized. Additional study is required to confirm if the above residual epoxidation is produced at the small secondary channel previously mentioned.
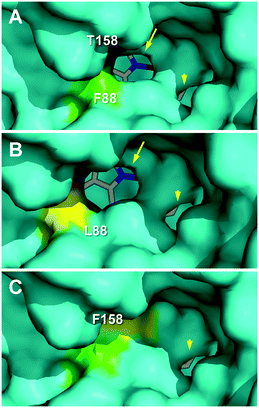 | ||
| Fig. 7 Main heme access channel (arrows) in C. virescens UPO (A) and its F88L variant (B), being largely occluded in the T158F variant (C), together with a small secondary channel (arrowheads). Solvent access surfaces of homology models obtained using available UPO crystal structures (PDB entries 2YP1 and 5FUJ) as templates, with colored regions corresponding to the selected residues and heme cofactor as sticks. | ||
The above considerations would also explain the higher epoxidation (vs. hydroxylation) by the F88L variant, similar to that obtained with the C. globosum UPO.19 Its slightly enlarged heme access channel (Fig. 7B, arrow) compared with the native enzyme (Fig. 7A) would facilitate the approach of the ω-8 (Δ9) carbon of oleic and linoleic acids promoting their epoxidation and decreasing the ω-7 hydroxylation, as observed experimentally. In the case of α-linolenic acid, the scarce experimental differences between epoxidation by the native enzyme and the F88L variant suggest that no channel enlargement is required for oxygenation. Interestingly, in contrast with the C. globosum UPO that largely diepoxidized linoleic acid (and α-linolenic acid), the C. virescens ability to selectively produce monoepoxides was maintained in the two mutated variants.
Finally, despite the differences in active-site architecture and effect of mutations, the results presented here confirm the feasibility of modulating UPO activities by engineering the heme access channel, as proposed for M. rotula UPO.17
Conclusions
In the present study, the repertoire of UPO enzymes is expanded by E. coli expression of a putative upo gene from C. virescens, while the selective fatty-acid epoxidizing UPO of related C. globosum cannot be expressed in E. coli for directed mutagenesis and other studies. Besides, the key importance of heme-channel architecture for selective epoxidation of unsaturated fatty acids is confirmed. A comparison of the catalysis by the native UPO and two mutated variants, in which the substrate access channel is either widened or constrained, is reported. Such subtle modifications exert dramatic effects on the reactivity of the enzyme with (poly)unsaturated fatty acids, as revealed by the abolishment of the activity on oleic acid in the T158F variant, which in turn fully maintains its epoxidative activity on α-linolenic acid. Moreover, a slight enlargement of the channel brings about a more selective epoxidation of unsaturated fatty acids in the F88L variant that gives rise to fewer undesired hydroxylated by-products, as found for the C. globosum UPO.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been funded by the SusBind (“Sustainable biobinders”; H2020-BBI-JTI-2017-792063; https://susbind.eu) project of the European BBI-JU (http://www.bbi-europe.eu) in the frame of EU Horizon 2020.References
- Y. Wang, D. Lan, R. Durrani and F. Hollmann, Curr. Opin. Chem. Biol., 2017, 37, 1–9 CrossRef PubMed.
- M. Hofrichter, H. Kellner, M. J. Pecyna and R. Ullrich, Adv. Exp. Med. Biol., 2015, 851, 341–368 CrossRef CAS PubMed.
- D. R. Morris and L. P. Hager, J. Biol. Chem., 1966, 241, 1763–1768 CAS.
- R. Ullrich, J. Nuske, K. Scheibner, J. Spantzel and M. Hofrichter, Appl. Environ. Microbiol., 2004, 70, 4575–4581 CrossRef CAS PubMed.
- D. H. Anh, R. Ullrich, D. Benndorf, A. Svatos, A. Muck and M. Hofrichter, Appl. Environ. Microbiol., 2007, 73, 5477–5485 CrossRef CAS PubMed.
- G. Gröbe, M. Ullrich, M. Pecyna, D. Kapturska, S. Friedrich, M. Hofrichter and K. Scheibner, AMB Express, 2011, 1, 31–42 CrossRef PubMed.
- J. Kiebist, K. U. Schmidtke, J. Zimmermann, H. Kellner, N. Jehmlich, R. Ullrich, D. Zänder, M. Hofrichter and K. Scheibner, ChemBioChem, 2017, 18, 563–569 CrossRef CAS PubMed.
- R. Ullrich, M. Poraj-Kobielska, S. Scholze, C. Halbout, M. Sandvoss, M. J. Pecyna, K. Scheibner and M. Hofrichter, J. Inorg. Biochem., 2018, 183, 84–93 CrossRef CAS PubMed.
- E. D. Babot, J. C. del Río, L. Kalum, A. T. Martínez and A. Gutiérrez, Biotechnol. Bioeng., 2013, 110, 2332 CrossRef PubMed.
- E. G. Hrycay and S. M. Bandiera, Monooxygenase, Peroxidase and Peroxygenase Properties and Reaction Mechanisms of Cytochrome P450 Enzymes, 2015 Search PubMed.
- X. W. Wang, J. Houbraken, J. Z. Groenewald, M. Meijer, B. Andersen, K. F. Nielsen, P. W. Crous and R. A. Samson, Stud. Mycol., 2016, 84, 145–224 CrossRef CAS PubMed.
- M. Hofrichter and R. Ullrich, Curr. Opin. Chem. Biol., 2014, 19, 116–125 CrossRef CAS PubMed.
- D. E. T. Pazmino, M. Winkler, A. Glieder and M. W. Fraaije, J. Biotechnol., 2010, 146, 9–24 CrossRef PubMed.
- C. Tiran, J. Lecomte, E. Dubreucq and P. Villeneuve, OCL, 2008, 15, 179–183 CrossRef CAS.
- M. Hofrichter, H. Kellner, R. Herzog, A. Karich, C. Liers, K. Scheibner, V. Wambui and R. Ullrich, in Grand challenges in fungal biotechnology, ed. H. Nevalainen, Springer, Berlin, ch. 14, 2019, p. 52 Search PubMed.
- M. Faiza, S. Huang, D. Lan and Y. Wang, BMC Evol. Biol., 2019, 19, 76 CrossRef PubMed.
- J. Carro, A. González-Benjumea, E. Fernández-Fueyo, C. Aranda, V. Guallar, A. Gutiérrez and A. T. Martínez, ACS Catal., 2019, 9, 6234–6242 CrossRef CAS.
- A. Olmedo, J. C. del Río, J. Kiebist, K. Scheibner, A. T. Martínez and A. Gutiérrez, Chem. – Eur. J., 2017, 23, 16985–16989 CrossRef CAS PubMed.
- C. Aranda, A. Olmedo, J. Kiebist, K. Scheibner, J. C. del Río, A. T. Martínez and A. Gutiérrez, ChemCatChem, 2018, 10, 3964–3968 CrossRef CAS.
- C. Aranda, R. Ullrich, J. Kiebist, K. Scheibner, J. C. del Río, M. Hofrichter, A. T. Martínez and A. Gutiérrez, Catal. Sci. Technol., 2018, 8, 2394–2401 RSC.
- J. Kiebist, W. Holla, J. Heidrich, M. Poraj-Kobielska, M. Sandvoss, R. Simonis, G. Groebe, J. Atzrodt, M. Hofrichter and K. Scheibner, Bioorg. Med. Chem., 2015, 23, 4324–4332 CrossRef CAS PubMed.
- A. Karich, R. Ullrich, K. Scheibner and M. Hofrichter, Front. Microbiol., 2017, 8, 1463 CrossRef PubMed.
- A. Olmedo, C. Aranda, J. C. del Río, J. Kiebist, K. Scheibner, A. T. Martínez and A. Gutiérrez, Angew. Chem., Int. Ed., 2016, 55, 12248–12251 CrossRef CAS PubMed.
- A. K. Yudin, Aziridines and epoxides in organic synthesis, Wiley, New Jersey, 2006 Search PubMed.
- A. Corma, S. Iborra and A. Velty, Chem. Rev., 2007, 107, 2411–2502 CrossRef CAS PubMed.
- S. Bormann, A. G. Baraibar, Y. Ni, D. Holtmann and F. Hollmann, Catal. Sci. Technol., 2015, 5, 2038–2052 RSC.
- H. Lund, L. Kalum, M. Hofrichter and S. Peter, US Pat., US 9908860 B2, 2018 Search PubMed.
- P. Puigbò, E. Guzmán, A. Romeu and S. Garcia-Vallvé, Nucleic Acids Res., 2007, 35, W126–W131 CrossRef PubMed.
- F. W. Studier, Protein Express. Purif., 2005, 41, 207–234 CrossRef CAS PubMed.
- E. Fernández-Fueyo, C. Aranda, A. Gutiérrez and A. T. Martínez, Patent (European), 2018, EP18382514.0.
- C. R. Otey, Methods Mol. Biol., 2003, 230, 137–139 CAS.
- A. Waterhouse, M. Bertoni, S. Bienert, G. Studer, G. Tauriello, R. Gumienny, F. T. Heer, T. A. P. de Beer, C. Rempfer, L. Bordoli, R. Lepore and T. Schwede, Nucleic Acids Res., 2018, 46, W296–W303 CrossRef CAS PubMed.
- Schrödinger, The PyMOL molecular graphics system, version 2.0, Schrödinger, LLC, New York, 2017, https://pymol.org Search PubMed.
- M. Biasini, S. Bienert, A. Waterhouse, K. Arnold, G. Studer, T. Schmidt, F. Kiefer, T. G. Cassarino, M. Bertoni, L. Bordoli and T. Schwede, Nucleic Acids Res., 2014, 42, W252–W258 CrossRef CAS PubMed.
- C. Aranda, M. Municoy, V. Guallar, J. Kiebist, K. Scheibner, R. Ullrich, J. C. del Río, M. Hofrichter, A. T. Martínez and A. Gutiérrez, Catal. Sci. Technol., 2018, 9, 1398–1405 RSC.
- L. Kalum, M. D. Morant, H. Lund, J. Jensen, I. Lapainaite, N. H. Soerensen, S. Pedersen, L. Østergaard and F. Xu, Patent (International),WO2014-015256A2, 2014 Search PubMed.
- A. Gutiérrez, E. D. Babot, R. Ullrich, M. Hofrichter, A. T. Martínez and J. C. del Río, Arch. Biochem. Biophys., 2011, 514, 33–43 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supporting information is available including Sephadex 75 chromatography (Fig. S1), GC-MS of the reactions with unsaturated and saturated fatty acids (Fig. S2), a molecular model of C. virescens UPO (Fig. S3), GC-MS of the reactions of C. virescens UPO and variants with oleic (Fig. S4), linoleic (Fig. S5) and α-linolenic (Fig. S6) acids, and formulae of fatty acid derivatives (Fig. S7). See DOI: 10.1039/c9cy02332a |
| ‡ These two authors contributed equally to this work. |
| § Present Address: Technical University of Delft, van der Maasweg 9, 2629HZ Delft, The Netherlands. |
| This journal is © The Royal Society of Chemistry 2020 |