Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optical properties of photodynamic therapy drugs in different environments: the paradigmatic case of temoporfin†
Busenur
Aslanoglu
a,
Ilya
Yakavets
 bcd,
Vladimir
Zorin
bcd,
Vladimir
Zorin
 be,
Henri-Pierre
Lassalle
be,
Henri-Pierre
Lassalle
 cd,
Francesca
Ingrosso
cd,
Francesca
Ingrosso
 f,
Antonio
Monari
f,
Antonio
Monari
 *f and
Saron
Catak
*f and
Saron
Catak
 *a
*a
aBogazici University, Department of Chemistry, Bebek 34342, Istanbul, Turkey. E-mail: saron.catak@boun.edu.tr
bLaboratory of Biophysics and Biotechnology, Belarussian State University, 220030, Minsk, Belarus
cUniversité de Lorraine and CNRS, CRAN, UMR 7039, F-54000, Nancy, France
dUniversité de Lorraine, Institut de Cancérologie de Lorraine, F-54000, Nancy, France
eInternational Sakharov Environmental Institute, 220030, Minsk, Belarus
fUniversité de Lorraine and CNRS, LPCT, UMR 7019, F-54000, Nancy, France. E-mail: antonio.monari@univ-lorraine.fr
First published on 16th July 2020
Abstract
Computational tools have been used to study the photophysical and photochemical features of photosensitizers in photodynamic therapy (PDT) – a minimally invasive, less aggressive alternative for cancer treatment. PDT is mainly based on the activation of molecular oxygen through the action of a photoexcited sensitizer (photosensitizer). Temoporfin, widely known as mTHPC, is a second-generation photosensitizer, which produces the cytotoxic singlet oxygen when irradiated with visible light and hence destroys tumor cells. However, the bioavailability of the mostly hydrophobic photosensitizer, and hence its incorporation into cells, is fundamental to achieve the desired effect on malignant tissues via PDT. In this study, we focus on the optical properties of the temoporfin chromophore in different environments – in vacuo, in solution, encapsulated in drug delivery agents, namely cyclodextrin, and interacting with a lipid bilayer.
Introduction
The exploitation of light as a therapeutic agent for the treatment of diverse and problematic diseases has gained large popularity in the last few decades,1 paving the way to the definition of novel and less invasive, yet efficient, therapeutic strategies.2 This has most notably involved the treatment of bacterial and viral diseases,3,4 as well as more debilitating conditions such as some type of cancers.5–7 However, the use of light for therapeutic purposes may be traced back to Ancient Greece, where exposure to sunlight, termed heliotherapy, was commonly used to treat a variety of conditions, including muscular weakness and skin diseases.7,8 The importance of such practices is also reflected in the fact that the god Apollon was associated with both medicine and the sun.Today the therapeutic use of light has evolved considerably, particularly owing to the milestone discovery and application of photodynamic therapy (PDT).9,10 In this context, a drug that is inert in the dark is administered to a patient, either topically or systemically, and the region in which the lesion is localized is then irradiated, usually with visible or infrared light.11 The absorption of light by the drug – the photosensitizer – triggers photophysical pathways leading to the disruption of biological macromolecules (nucleic acids, proteins, or lipid membranes) and, consequently, to cell death, and ultimately to the eradication of the lesion.12
Although photophysical or photochemical pathways subsequent to light absorption may be diverse, two main families of events can be underlined: type I reactions involve either hydrogen abstraction or electron transfer from the excited photosensitizer (PS) to a substrate molecule to produce free radicals or radical ions.13 Type II reactions transfer energy to molecular oxygen to produce singlet oxygen (1O2).14 In particular, type II pathways usually proceed through intersystem crossing (ISC) populating the photosensitizer's triplet state manifold allowing the activation of the molecular oxygen from its triplet (3O2) to singlet (1O2) state.15 Subsequently, the action of 1O2 will induce the oxidative stress responsible for cell death.16 PDT has proven efficient in the treatment of a variety of diseases, including psoriasis17 as well as certain types of cancer,18 such as esophageal19 and cervical,20 greatly limiting the side-effect of conventional therapy due to the spatial-selectivity of irradiation.21,22
There are several requirements that should be met by PDT agents to increase their efficiency and guarantee their therapeutic effects.23 These include: facile ISC, high quantum yield for the production of 1O2,24 absorption in the red or infrared portion of the electromagnetic spectrum to cover the therapeutic window for which penetration into tissues is maximal,25 as well as enhanced selectivity towards bacterial26 or cancer cells.27 However, the need for oxygen activation in PDT may present an obstacle for the treatment of hypoxic solid tumors.28 Hence, the exploitation of different photochemical pathways, such as photodissociation, have been proposed under the general name of light-assisted chemotherapy (LAC).29–32 The displacement of the absorption spectrum towards the red is also pursued via different chemical strategies, including the use of organometallic complexes,33,34 or enlarging the conjugation pattern of the sensitizers.35 More recently, the exploitation of non-linear optics, two-photon absorption (TPA)36–39 in particular, has been suggested.
Although various agents have been proposed for PDT and LAC, nowadays, the quasi-totality of clinically approved agents are based on porphyrins,40,41 phthalocyanines,42 or chlorins.43–46 These classes of molecules display red-shifted absorption, which may be further modulated by the inclusion of metal in the coordination sphere,42,47,48 relatively good ISC quantum yields as well as versatile and economic synthetic viability. On the other hand, their efficacy is somewhat limited by solubility issues49 and by aggregation phenomena50 that severely limit their bioavailability and lead to excited-state deactivation channels hampering the triplet population.
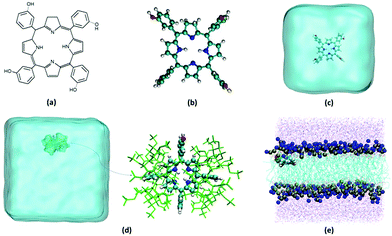
One of the most efficient PDT drugs on the market nowadays, yet still plagued by aggregations,51 is the so-called temoporfin (5,10,15,20-tetrakis(3-hydroxyphenyl)chlorin – mTHPC, Fig. 1a and b). Marketed in the EU under the brand name Foscan,52 mTHPC is a synthetic tetrapyrrole, a partially hydrogenated porphyrin derivative, known as chlorin (Fig. 1a). Temoporfin is a second-generation photosensitizer, which has higher skin penetration, deeper light penetration, and lower biotoxicity, and moreover, it requires shorter treatment time as well as a lower light dosage to achieve the desired PDT response compared with other clinically approved photosensitizers, such as hematoporphyrin derivatives and photofirin.52 Indeed, mTHPC possesses appropriate photophysical properties: excitation to the first excited singlet state by red light (630–680 nm),53 and efficient ISC to a longer-lived excited triplet state, which has recently been rationalized by molecular modeling.54 The latter triplet state may produce cytotoxic species by either a type I or type II reaction typical of photosensitizer agents used in photodynamic therapy.55
As mentioned, the most serious drawback of mTHPC, and many other organic sensitizers, is their hydrophobic nature, which severely limits their transportation in the bloodstream and causes aggregation-induced quenching (ACQ) in biological environments,56 hence resulting in weak emissions and inadequate generation of reactive oxygen species (ROS).57 To overcome the mTHPC aggregation issue, specific drug delivery strategies are currently being pursued,58 in particular, the use of molecular hosts based on beta-cyclodextrin (β-CD) has been proposed.59,60 The incorporation of mTHPC into beta-cyclodextrin61 (Fig. 1d) and its release in model lipid membranes62,63 (Fig. 1e) and cells64,65 have been experimentally assessed by monitoring its fluorescence quenching. Most notably, the encapsulation of mTHPC in trimethyl-β-cyclodextrin (TM-β-CD) also produces a strong enhancement of the fluorescence quantum yield that is almost totally quenched in solution, hence allowing for the possibility to efficiently follow drug delivery by optical spectroscopy.
A clear rationalization of the different aspects of PDT at the atomistic and electronic levels represents a challenge, since it requires multiscale modelling that is able to deal with both the treatment of excited states and the sampling of complex environments. In this respect, the development of PDT drug delivery systems represents a most paradigmatic case, since it requires the proper exploration of the conformational space of complex systems in complex environments, and at the same time, the need to provide a balanced description of physical and chemical phenomena taking place in both the ground and excited state manifolds.66 In particular, the complex systems involved in drug delivery and their interactions with biological structures requires the use of dynamic simulations using force field-based descriptions, also due to the key temporal spans that need to be covered. However, the sensitivity of excited state properties to even the slightest structural modifications also requires careful parameterization of the force fields.67–69
In this study, we report the optical and photophysical properties of mTHPC employing molecular simulations in different environments: (a) in vacuo, (b) in solution, (c) in solution encapsulated within TM-β-cyclodextrin (mTHPC:TM-β-CD complex), and (d) in interaction with a lipid bilayer mimicking a biological membrane. Herein, we provide a clear rationalization of the optical properties of the most relevant PDT drug; the coupling between its critical structural parameters, the degrees of freedom of different molecular environments, such as drug delivery systems or cellular compartments, and the effects of heterogeneous environments on its photophysical properties. The interactions of mTHPC with a model biological membrane will also help rationalize its efficiency as a PDT agent.
Methodology
Computational approach
Structural and optical properties of mTHPC were investigated with quantum mechanical/molecular dynamics (QM/MD) and classical MD simulations in different environments, namely in vacuum, in water, in complex with the TM-β-CD, and in contact with a model lipid bilayer composed of 1-palmitolyl-2-oleyl-sn-glycero-3-phosphocholine (POPC).Classical molecular dynamics and force field parameterization
Classical MD simulations were conducted using the NAMD program package.70 All simulations were carried out in the constant pressure and temperature ensemble (NPT)71 at 1 atm and 300.0 K, a time step of 2.0 fs was used to integrate Newton's equations of motion. A production run of 100 ns was performed after equilibration and thermalization, temperature and pressure were controlled by Langevin thermostat72 and Nosé–Hoover Langevin barostat,73,74 respectively. Periodic boundary conditions (PBC) were used and the Particle Mesh Ewald (PME) expansion75 was applied to calculate long-range electrostatics contributions with a 9 Å cutoff distance.The force field for mTHPC was parametrized with Generalized Amber Force Field (GAFF),76 while point charges were obtained by fitting to the restricted electrostatic potential (RESP) using the Antechamber program.77 GLYCAM0678 was used for the parametrization of TM-β-CD. mTHPC was solvated by a cubic water box of 50.0 × 50.0 × 5 0.0 Å3, while the mTHPC:TM-β-CD complex was prepared considering a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry (Fig. 1d) as suggested by previous work61 and solvated in a water box (90.0 × 85.0 × 85.0 Å3).
2 stoichiometry (Fig. 1d) as suggested by previous work61 and solvated in a water box (90.0 × 85.0 × 85.0 Å3).
Lipid14 General Amber Force Field79 was used to represent the POPC unit, while the lipid bilayer was set up with the CHARMM-GUI Membrane interface.80,81 100 POPC units were used in each membrane leaflet, the bilayer was fully hydrated with 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 551 water molecules and was neutralized by 104 K+ and Cl− ions, the total simulation box size is 81.0 × 81.0 × 138.0 Å3. POPC is the most widely used lipid model to simulate biological membranes. Indeed, a single lipid membrane model is an oversimplification of a biological environment, since the structural properties of the membrane may also be affected by the presence of other fatty acids or cholesterol. However, the use of a single lipid, greatly reduces the degrees of freedom, while still providing a realistic amphiphilic model that discriminates between the polar head and the lipid core region.82–84
551 water molecules and was neutralized by 104 K+ and Cl− ions, the total simulation box size is 81.0 × 81.0 × 138.0 Å3. POPC is the most widely used lipid model to simulate biological membranes. Indeed, a single lipid membrane model is an oversimplification of a biological environment, since the structural properties of the membrane may also be affected by the presence of other fatty acids or cholesterol. However, the use of a single lipid, greatly reduces the degrees of freedom, while still providing a realistic amphiphilic model that discriminates between the polar head and the lipid core region.82–84
The TIP3P water model85 was consistently used in all the simulated systems. MD simulations were visualized and analyzed via the VMD code;86 the most relevant non-covalent interactions (NCI) throughout the MD trajectories were analyzed and depicted with NCIPlot code.87 The study of the gradient of the electronic density clearly differentiates NCIs in terms of steric clashes, dispersion, and hydrogen bonding.
Quantum mechanics molecular dynamics
Hybrid quantum mechanics/molecular mechanics molecular dynamic simulations88 (QM/MM MD) were performed to sample the ground state conformational space by the Terachem/Amber89 and NAMD/Terachem interface.90 mTHPC was included in the QM partition and described by the B3LYP91 density functional and 6-31G basis set. Water and TM-β-CD were treated at the MM level of theory, with the force field parameterization previously described. For details of the QM/MM simulations, see ESI,† (Table S1).Electronic structure calculations
Single point energy calculations and geometry optimizations were performed using the Gaussian 09 program package92 with Density Functional Theory (DFT) at B3LYP91 and 6-31G(d) basis set level of theory. The potential energy surface scan to parameterize the mTHPC force field and the conformational analysis were performed with the same level of theory, the energies along the scan have also been estimated using the second order Moller–Plesset perturbation theory (MP2).To simulate the absorption spectra, vertical electronic excitations from the ground state were obtained via time-dependent DFT93 (TD-DFT). The choice of the exchange–correlation functional and basis set were further justified by a benchmark on the calculation of the vertical excitation energies from the ground state equilibrium geometry reported in ESI.† While the influence of the basis set on the excitation energies appears to be negligible, the functional has a more pronounced effect. More specifically, ωB97X-D and M06-2X provide the correct separation of the Qx and Qy bands, in the red wavelengths domain, while B3LYP only yields an unresolved broad band. Hence, ωB97XD/6-31G(d) clearly represents the best compromise between computational cost and accuracy, and was chosen to further take into account the dynamical and vibrational effects. To this end, all UV-vis absorption spectra were generated from the convolution of vertical excitations from 100 snapshots randomly extracted from the MD trajectories. The convolution of the vertical excitation energies and oscillator strengths was performed using Gaussian functions of full width at half-length (FWHL) of 0.15 eV.
Experimental procedures
Materials
The photosensitizer, mTHPC, was kindly provided by Biolitec research GmbH (Jena, Germany) with purity >99%. TM-β-CD (purity >98%, M.W. 1430 Da) was purchased from AraChem (Tilburg, the Netherlands). β-CDs in the powder were weighed and then dissolved in Dulbecco's phosphate-buffered saline (DPBS) (pH 7.4) at 4 °C at the final concentration of 10 mM using molar weight provided by the supplier. mTHPC stock solution (2 mM) was prepared in absolute ethanol (99.6%) and was kept at 4 °C in the dark. mTHPC concentration in the solution was estimated by means of a spectrophotometric method using molar extinction coefficient of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 at 650 nm in ethanol.94
000 M−1 cm−1 at 650 nm in ethanol.94
Inclusion complexes between β-CDs and mTHPC were formed using the co-precipitation method.95 Briefly, the stock ethanol solution of mTHPC (2 mM) was added to TM-β-CDs dissolved in DPBS (pH 7.4) at 4 °C and 1 mM. The final content of ethanol in the mTHPC/β-CD solutions did not exceed 0.5%. The solution was thoroughly mixed for 15 min under magnetic stirring.
Spectroscopic measurements
Absorption spectra were recorded using a Lambda 35 spectrophotometer (PerkinElmer, USA) using 1 cm optical path quartz cuvettes. All spectroscopic measurements were carried out in triplicate at room temperature (23–25 °C). The optical density of all samples did not exceed 0.3 a.u.Results and discussion
In the following section, we report the structural and optical properties of mTHPC in molecular environments of increasing complexity. Using molecular simulations, the interaction of temoporfin with water, TM-β-CD and a lipid bilayer is modelled, and subsequently compared with the corresponding experimental data, when available. For this purpose, different levels of theory are employed, the evolution of critical structural parameters along with photophysical properties are assessed, and the effect of the molecular environment is comparatively discussed.mTHPC is composed of four functionalized phenols that adorn the core chlorin ring. The phenol rings constitute one of the leading factors potentially inducing specific intermolecular interactions with the environment, such as hydrogen bonds, while the chlorin core will be the most crucial unit dictating the optical properties of the chromophore. In addition, the inclusion of mTHPC in the TM-β-CD complex will also determine the orientation of the peripheral substituents and will, in turn, modify their flexibility. Hence, a proper parameterization of the force field to catch all these subtle phenomena, and in particular, the rotational flexibility of the phenol substituents with respect to the chlorin core is crucial.
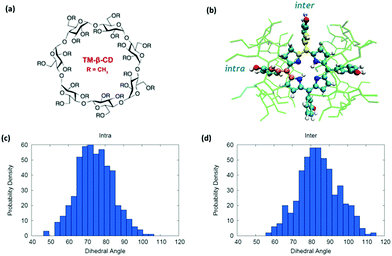
Dihedral angles defining the rotation of the peripheral phenolic groups of mTHPC are depicted in Fig. 2. While full rotations are not expected to take place due to clashes between the phenyl hydrogens and the porphyrin core; phenolic moieties are expected to span a much wider range in gas phase and in solution. However, this may change considerably in the cyclodextrin complex due to much larger steric hindrance. As previously demonstrated, the mTHPC:TM-β-CD complex has a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry61 (Fig. 1d) and is held together by dispersion interactions between the two TM-β-CD that globally shelter the hydrophobic mTHPC from the water environment.
2 stoichiometry61 (Fig. 1d) and is held together by dispersion interactions between the two TM-β-CD that globally shelter the hydrophobic mTHPC from the water environment.
A gas-phase QM scan of Temoporfin's peripheral dihedral angle, performed both at DFT and MP2 level of theory (Fig. S2–S3, ESI†), confirms a large degree of freedom of the phenolic group as seen by the nearly flat potential energy curve in the ±60–120° region. The sharp increase in the potential energy around 0° and ±180° is due to steric repulsion with the core and is indicative of the emergence of two distinct conformers. Hence, to properly represent this situation a free rotation of the phenol ring was implemented in the classical force field for mTHPC by setting the force constant to zero, the results for the distribution of the dihedral angles in the different environments obtained with the classical force field match remarkably the ones of the QM or QM/MM MD. Note that the mTHPC FF was parametrized to better represent the PS in confined environments, hence, it is somewhat expected that its performance will be less adequate in vacuo and in solution where the rotational flexibility is larger.
In addition to the globally reduced flexibility, two different situations occur in the temoporfin-cyclodextrin complex (mTHPC:TM-β-CD complex), potentially presenting differences in steric hindrance due to the specific complexation pattern. As seen in Fig. 2b, two of the phenolic groups of Temoporfin are located inside the cyclodextrin pockets (intra), while the other two reside in the hydrophobic region between the two cyclodextrins (inter). In addition to the reduced mobility evidenced by the sharper distribution reported in Fig. 2, as compared to the water case (Fig. S10–S12, ESI†), subtle differences also emerge between the intra and inter distributions, where the intra-phenol appears to experience a slightly more impeded rotation than the inter (Fig. 2c and d). This is probably due to the highly constrained interior of the cyclodextrin core that is explored by the intra-phenol moeity. The reduced mobility of the phenol rings in the macromolecular complex as opposed to the gas phase or solution may also offer an additional reason for the enhancement of fluorescence observed in the host complex. The free rotation of the peripheral moieties would offer a further non-radiative decay pathway that will lead to the deactivation of the excited state. Indeed, the restricted intramolecular rotation has been recognized as a cause of the fluorescence enhancement in different organic luminophores.96,97 As it is the case for diverse fluorophores,98–100 the aggregation of chromophores, and hence the formation of dark excitonic states, could also be considered as a supplementary phenomenon leading to fluorescence quenching in solution.
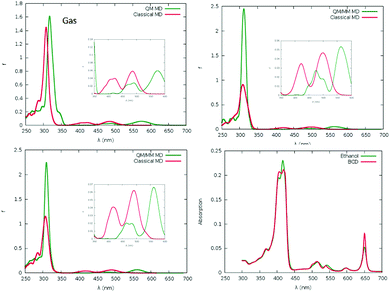
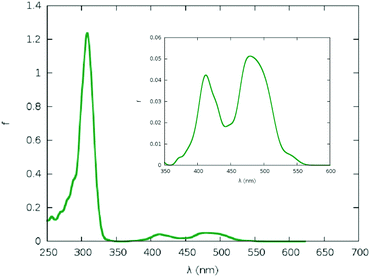
The experimental absorption spectra of mTHPC are reported in Fig. 3, and unsurprisingly present all the features of chlorin-based chromophores. In particular, the absorption bands in the 500–680 nm region, corresponding to the Q-bands, as well as the more intense Soret bands in the blue region. It is evident from the analysis of the QM based MD simulation of mTHPC in the gas phase (Fig. S6, ESI†), as well as the QM/MM MD simulation of mTHPC in solution (Fig. S10, ESI†) that two low-energy large-amplitude degrees of freedom dominate the conformational space: the out-of-plane deformation of the conjugated ring system and the rotation of the peripheral phenolic units. These degrees of freedom are critical in describing the global vibration of the drug and may have an influence on the observed optical properties. The out-of-plane normal modes may be responsible for the red-shift of the absorption spectra with respect to vertical transitions calculated from the ground state geometry only (Fig. 3 and ESI†) due to the significant destabilization of the ground state rather than the excited state, as is common for similar π-conjugated systems.42,44,101–103 Although the influence of the rotation of the peripheral substituents on the absorption spectral maxima is expected to be marginal, it may, nonetheless, induce a broadening of the absorption and emission bands, and more notably, may have a key role in inducing thermal quenching of the luminescence. While the possibility to take into account the out-of-plane deformation effects via a reasonable sampling of the conformational space is well established and has been thoroughly demonstrated in similar systems,36,42,66,101 the effects of the peripheral dihedral rotations require much deeper attention, due to their strong non-harmonic behaviour. In addition, the effect of the molecular surrounding, and in particular of the embedding in confined environments, such as cyclodextrin, should be properly taken into account.
In Fig. 3, we also report the comparison between the absorption spectra calculated as the convolution of vertical transitions from a series of snapshots obtained by sampling mTHPC's conformational space with either a QM/MM approach, in which the whole chromophore is treated quantum mechanically, or a MM approach, in which the dynamics of the chromophore is described by a force field. The agreement between the different levels of the theory is good, confirming the validity of the chosen approach, the only difference being the slightly more red shifted Q-band obtained with the QM/MM. Interestingly, the Q-band region also appears more sensitive to the specific chemical environment, in particular for the absorption spectrum of the mTHPC:TM-β-CD. Indeed, in this case, the inversion of the intensity between the Qx and Qy Soret bands observed experimentally, constituting one of the spectroscopic signatures of complex formation, is clearly identified and nicely reproduced by the full classic and the hybrid sampling protocol.
The reasonable agreement between the calculated spectroscopic properties also constitutes a further validation of the novel force field, with an obvious improvement over the parametrization used in our previous work.61 Note that experimental results have been conducted in ethanol instead of water,104,105 because of the global low solubility of the sensitizers. In the simulations, we decided to use water as a solvent to mimic the most biologically relevant solvent. Moreover, the protic nature of ethanol solvent should lead to a similar solvatochromic effect as water.
Having established the robustness and precision of our force field parameterization that has been validated by means of different levels of theory and proven to reproduce both the structural parameters and the optical properties of mTHPC in different environments, we move to examine the interaction, between the chromophore and a lipid bilayer. Indeed, the mode of action as a PDT drug should involve the disruption of the cell membrane induced by the activated 1O2 following the absorption of light. The results of the MD simulation are reported in Fig. 4 in the form of the density profile along the membrane axis for the main functional group and pictorially using a representative snapshot. At the beginning of the simulation the photosensitizer is placed in bulk water and unsurprisingly, due to its hydrophobic nature, mTHPC develops favorable interactions with the lipid membrane with a persistence time exceeding hundreds of ns.
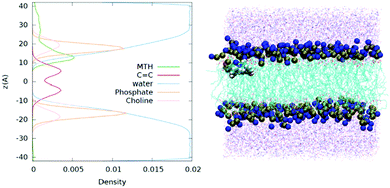 | ||
| Fig. 4 Density Profile along the membrane axis and a representative snapshot showing mTHPC embedded in a POPC lipid bilayer. | ||
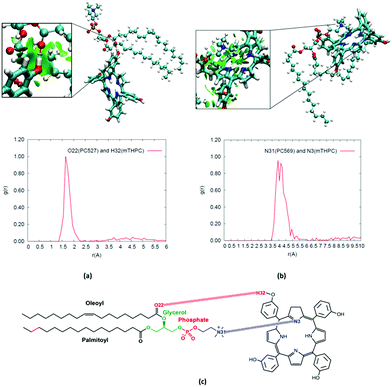
In the obtained most stable configuration, mTHPC is positioned around the polar head regions, and more precisely, at the interface between the phosphate and the fatty acid hydrophobic core. Indeed, mTHPC is more exposed to the lipid environment than to the water bulk. This behaviour is not totally unexpected and can be traced back to the concomitant presence of a hydrophobic core and phenol groups that may develop specific non-covalent interactions with the polar head regions. However, the position of mTHPC is most favorable for PDT purposes. Indeed, while O2 will permeate the membrane due to its hydrophobic nature as also revealed by MD simulations,106 it will also have a non-negligible concentration in the region occupied by the photosensitizer. In addition, our density profile also shows that mTHPC partially overlaps with the position of the lipid carbon–carbon double bond. This fact indicates that 1O2 will be produced in close proximity to its reactive target, hence minimizing the quenching probability. It has also been reported that photosensitizers, also porphyrin-based, may exert photopermeabilization of the lipid membrane via an alternative contact dependent mechanism, that involves electron abstraction from solvent or lipids.107,108 Although further studies, including the modelling of additional membrane components, such as cholesterol, should be needed to confirm this scenario, our density profile, and the partial overlap identified, claims in favour of the possibility of this photophysical route, concerning the chromophore positioning and the key geometrical factors.
To better characterize the factors influencing the stability of the persistent aggregate evidenced in the MD trajectory, we performed a detailed analysis of the specific interactions taking place between mTHPC and the lipid constituents. As reported in Fig. 5, in addition to dispersion interactions typical of hydrophobic compounds, the formation of specific interactions mainly involving hydrogen-bonding between the phenol groups and the negatively charged phosphates, contributes to locking the chromophore at the interface region.
This interaction, as well as its persistence, is confirmed by the structure of the radial distribution function presented in Fig. 5a, for the involved phenolic hydroxyl and the phosphate oxygen groups. The sharp peak at around 1.6 Å is very indicative of a typical and persistent hydrogen bond. The same conclusions can be drawn from the NCI plots for some representative snapshots. The features of a hydrogen bond formation, represented by the concentrated green circle around the phosphate and phenol group, are indeed clearly evident in the zoom of Fig. 5a. Although the hydrogen bonding pattern was somewhat expected, other non-covalent interactions are also pertinent, namely, the cation–π interactions between the aromatic mTHPC core and the positively charged choline in the lipid head groups are clearly evidenced both by the radial distribution function and by the NCI plot (Fig. 5b). In addition, as evidenced in the ESI,† the more structured and viscous environment offered by the lipid bilayer induces a partial rigidification of mTHPC, and hence a more impeded rotation around the phenol rings. However, this situation is much weaker than the one observed in the case of the cyclodextrin complex, the enhancement of fluorescence observed when mTHPC is in the presence of biological membranes should, hence, be mainly ascribed to the absence of π-stacking (aggregation).62,63,109
The absorption spectrum calculated in the POPC interface is also reported in Fig. 6 and shows that most of the characteristic spectral features are maintained. Indeed, both the Q and the Soret bands are clearly distinguishable, and interestingly, in contrast with what was observed in the case of the mTHPC:TM-β-CD complex, no inversion between the Qx and Qy band is observed. However, the energy differences between the two bands are underestimated by our computational approach, as compared to experiment. This difference could mostly be ascribed to the neglect of purely quantum vibronic coupling through the explicit calculation of Franck–Condon parameters.110,111 However, such an approach, despite some interesting recent developments, is still far from being standardized.112
Conclusions
In this contribution, we report the detailed analysis of the structural and optical properties of a known photosensitizer, mTHPC, clinically used for PDT purposes in different complex environments, including a cyclodextrin-based encapsulating drug delivery agent, and a model lipid bilayer. Our results show that the confined environment of the cyclodextrin induces a strong reduction of the mobility of the peripheral phenol rings. Furthermore, clear differences in the mobility degree appear for intra and inter substituents. The strong reduction of the phenyl ring rotation is also correlating well with the observed enhancement of the fluorescence quantum yield upon encapsulation. Our results have also highlighted the formation of persistent aggregates between mTHPC and lipid membranes, with the sensitizer residing preferentially in the interface between the polar heads and the lipid core. This situation is favoured by interactions, such as the formation of hydrogen bonds and cation–π interactions with the polar heads that counterbalances the hydrophobic dispersion interactions developed with the lipid core. Interestingly, the position of the sensitizer represents one of the most ideal ones for PDT purposes, since it encompasses both the area of maximum oxygen concentration and a partial overlap with the position of the oxidizable lipid double-bond. In addition, the optical properties have been determined, including the sampling of the conformational space of the ground state, highlighting a rather marginal, but non-innocent, effect of the environment and the necessity to carefully parameterize the crucial force-field parameters.In the future, we plan to pursue the study of mTHPC interactions with cyclodextrins and lipid membranes in a two-fold perspective. From the one side, the release from the encapsulating complex and the subsequent insertion in the lipid membrane will be studied by using enhanced sampling techniques. In addition, the intersystem crossing, and hence the population of the triplet state manifold, for mTHPC in different environments will be studied including the possibility to perform QM/MM based non-adiabatic dynamics to quantitatively assess the quantum yield and timescales of the first event of the photosensitization chain.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Calculations were partially performed at the TUBITAK ULAKBIM High Performance and Grid Computing Center (TRUBA resources), the LPCT (http://www.lpct.univ-lorraine.fr) local computing resources as well as the computational resources at CCBG (http://www.ccbg.boun.edu.tr) funded by the Bogazici University Research Fund (BAP-SUP Project No. 8245). The authors thank biolitec research GmbH (Jena, Germany) for providing with mTHPC.References
- Y. Wang, J. Zhao, Z. Chen, F. Zhang, Q. Wang, W. Guo, K. Wang, H. Lin and F. Qu, Biomaterials, 2019, 217, 119282 CrossRef CAS PubMed.
- N. Philbrick, Smithsonian, 2016, 2795–2838 Search PubMed.
- A. Azizi, P. Shohrati, M. Goudarzi, S. Lawaf and A. Rahimi, Photodiagn. Photodyn. Ther., 2019, 27, 203–209 CrossRef CAS PubMed.
- J. Shen, Q. Liang, G. Su, Y. Zhang, Z. Wang, C. Baudouin and A. Labbé, Transl. Vis. Sci. Technol., 2019, 8, 1–10 Search PubMed.
- L. Zhang, Z. Ji, J. Zhang and S. Yang, Photodiagn. Photodyn. Ther., 2019, 28, 159–165 CrossRef CAS PubMed.
- M. Hamblin, Photochem. Photobiol., 2020, 96, 506–516 CrossRef CAS PubMed.
- P. Jarrett and R. Scragg, Photochem. Photobiol. Sci., 2017, 16, 283–290 RSC.
- T. Karppinen, L. Ylianttila, H. Kautiainen, T. Reunala and E. Snellman, Acta Derm. – Venereol., 2015, 95, 579–582 CrossRef PubMed.
- J. Zhang, C. Jiang, J. P. Figueiró Longo, R. B. Azevedo, H. Zhang and L. A. Muehlmann, Acta Pharm. Sin. B, 2018, 8, 137–146 CrossRef PubMed.
- R. L. Yanovsky, D. W. Bartenstein, G. S. Rogers, S. J. Isakoff and S. T. Chen, Photodermatol., Photoimmunol. Photomed., 2019, 35, 295–303 CrossRef PubMed.
- P. Majumdar, R. Nomula and J. Zhao, J. Mater. Chem. C, 2014, 2, 5982–5997 RSC.
- E. Buytaert, M. Dewaele and P. Agostinis, Biochim. Biophys. Acta, Rev. Cancer, 2007, 1776, 86–107 CrossRef CAS PubMed.
- Y.-Y. Wang, Y.-C. Liu, H. Sun and D.-S. Guo, Coord. Chem. Rev., 2019, 395, 46–62 CrossRef CAS.
- Z. Tan, J. Zhang, L. Lin and B. Li, in Optics in Health Care and Biomedical Optics VII, ed. Q. Luo, X. Li, Y. Gu and Y. Tang, 2016, vol. 10024, p. 100241C Search PubMed.
- J. Kou, D. Dou and L. Yang, Oncotarget, 2017, 8, 81591–81603 CrossRef PubMed.
- Q. Yu, E. M. Rodriguez, R. Naccache, P. Forgione, G. Lamoureux, F. Sanz-Rodriguez, D. Scheglmann and J. A. Capobianco, Chem. Commun., 2014, 50, 12150–12153 RSC.
- F. Yi, X. Zheng, F. Fang, J. Zhang, B. Zhou and X. Chen, Exp. Dermatol., 2019, 0–3 Search PubMed.
- H.-C. Huang, S. Mallidi, G. Obaid, B. Sears, S. Tangutoori and T. Hasan, Applications of Nanoscience in Photomedicine, Elsevier, 2015, pp. 487–510 Search PubMed.
- H. Liu, Y. Liu, L. Wang, X. Ruan, F. Wang, D. Xu, J. Zhang, X. Jia and D. Liu, Technol. Cancer Res. Treat., 2019, 18, 1–6 CrossRef PubMed.
- J. Wang, M. Zheng and Z. Xie, J. Colloid Interface Sci., 2019, 535, 84–91 CrossRef CAS PubMed.
- Y. Hu, J. F. Honek, B. C. Wilson and Q. B. Lu, J. Biophotonics, 2019, 12, 1–11 Search PubMed.
- Y. Ni, H. Zhang, C. Chai, B. Peng, A. Zhao, J. Zhang, L. Li, C. Zhang, B. Ma, H. Bai, K. L. Lim and W. Huang, Adv. Healthcare Mater., 2019, 8, 1–9 Search PubMed.
- T. J. Dougherty and S. L. Marcus, Eur. J. Cancer, 1992, 28, 1734–1742 CrossRef.
- J. T. Ferreira, J. Pina, C. A. F. Ribeiro, R. Fernandes, J. P. C. Tomé, M. S. Rodríguez-Morgade and T. Torres, Chem. – Eur. J., 2020, 26, 1789–1799 CrossRef CAS PubMed.
- H. Abrahamse and M. R. Hamblin, Biochem. J., 2016, 473, 347–364 CrossRef CAS PubMed.
- M. Kang, C. Zhou, S. Wu, B. Yu, Z. Zhang, N. Song, M. M. S. Lee, W. Xu, F.-J. Xu, D. Wang, L. Wang and B. Z. Tang, J. Am. Chem. Soc., 2019, 141, 16781–16789 CrossRef CAS PubMed.
- Q. Li, Y. Li, T. Min, J. Gong, L. Du, D. L. Phillips, J. Liu, J. W. Y. Lam, H. H. Y. Sung, I. D. Williams, R. T. K. Kwok, C. L. Ho, K. Li, J. Wang and B. Z. Tang, Angew. Chem., Int. Ed., 2020, 59, 9470–9477 CrossRef CAS PubMed.
- H. Chen, J. Tian, W. He and Z. Guo, J. Am. Chem. Soc., 2015, 137, 1539–1547 CrossRef CAS PubMed.
- B. Xia, B. Wang, J. Shi, Y. Zhang, Q. Zhang, Z. Chen and J. Li, Acta Biomater., 2017, 51, 197–208 CrossRef CAS PubMed.
- S. Bonnet, B. Limburg, J. D. Meeldijk, R. J. M. K. Gebbink and J. A. Killian, J. Am. Chem. Soc., 2011, 133, 252–261 CrossRef CAS PubMed.
- S. H. C. Askes, G. U. Reddy, R. Wyrwa, S. Bonnet and A. Schiller, J. Am. Chem. Soc., 2017, 139, 15292–15295 CrossRef CAS PubMed.
- A. Francés-Monerris, I. Tuñón and A. Monari, J. Phys. Chem. Lett., 2019, 10, 6750–6754 CrossRef PubMed.
- Y. Zhao, Z. Zhang, Z. Lu, H. Wang and Y. Tang, ACS Appl. Mater. Interfaces, 2019, 11, 38467–38474 CrossRef CAS PubMed.
- J. D. Knoll and C. Turro, Coord. Chem. Rev., 2015, 282–283, 110–126 CrossRef CAS PubMed.
- L. Li, B. Bae, T. H. Tran, K. H. Yoon, K. Na and K. M. Huh, Carbohydr. Polym., 2011, 86, 708–715 CrossRef CAS.
- O. Sengul, E. B. Boydas, M. Pastore, W. Sharmouk, P. C. Gros, S. Catak and A. Monari, Theor. Chem. Acc., 2017, 136, 67 Search PubMed.
- J. D. Bhawalkar, N. D. Kumar, C. F. Zhao and P. N. Prasad, J. Clin. Laser Med. Surg., 1997, 15, 201–204 CrossRef CAS PubMed.
- Y. Shen, A. J. Shuhendler, D. Ye, J. J. Xu and H. Y. Chen, Chem. Soc. Rev., 2016, 45, 6725–6741 RSC.
- F. Bolze, S. Jenni, A. Sour and V. Heitz, Chem. Commun., 2017, 53, 12857–12877 RSC.
- J. W. Hofman, M. G. Carstens, F. V. Zeeland, C. Helwig, F. M. Flesch, W. E. Hennink and C. F. V. Nostrum, Pharm. Res., 2008, 25, 2065–2073 CrossRef CAS PubMed.
- M. García-Díaz, D. Sánchez-García, J. Soriano, M. L. Sagristà, M. Mora, Á. Villanueva, J. C. Stockert, M. Cañete and S. Nonell, MedChemComm, 2011, 2, 616–619 RSC.
- B. Koca, E. Hamuryudan, S. Catak, A. Erdogmus, A. Monari and V. Aviyente, J. Phys. Chem. C, 2019, 123, 24417–24425 CrossRef CAS.
- Y. Li, Y. Yu, L. Kang and Y. Lu, Int. J. Clin. Exp. Med., 2014, 7, 4867–4876 Search PubMed.
- H. Gattuso, A. Monari and M. Marazzi, RSC Adv., 2017, 7, 10992–10999 RSC.
- A. V. Kachynski, A. Pliss, A. N. Kuzmin, T. Y. Ohulchanskyy, A. Baev, J. Qu and P. N. Prasad, Nat. Photonics, 2014, 8, 1–7 CrossRef.
- M. Marazzi, H. Gattuso, A. Giussani, H. Zhang, M. Navarrete-Miguel, C. Chipot, W. Cai, D. Roca-Sanjuán, F. Dehez and A. Monari, J. Phys. Chem. Lett., 2019, 10, 7133–7140 CrossRef CAS PubMed.
- Z. Zhou, J. Liu, J. Huang, T. W. Rees, Y. Wang, H. Wang, X. Li, H. Chao and P. J. Stang, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 20296–20302 CrossRef CAS PubMed.
- Z. Zhang, W. Sang, L. Xie and Y. Dai, Coord. Chem. Rev., 2019, 399, 213022 CrossRef CAS.
- X. Lu, W. Zhu, T. Chen, Q. Peng, C. Yu and M. Yang, Chem. Phys. Lett., 2019, 735, 136737 CrossRef CAS.
- Z. Liu, Y. Xue, M. Wu, G. Yang, M. Lan and W. Zhang, Biomacromolecules, 2019, 20, 4563–4573, DOI:acs.biomac.9b01368.
- J. Y. Chen, N. K. Mak, C. M. N. Yow, M. C. Fung, L. C. Chiu, W. N. Leung and N. H. Cheung, Photochem. Photobiol., 2000, 72, 541 CrossRef CAS PubMed.
- M. O. Senge and J. C. Brandt, Photochem. Photobiol., 2011, 87, 1240–1296 CrossRef CAS PubMed.
- W. M. Sharman, C. M. Allen and J. E. V. Lier, Drug Discovery Today, 1999, 4, 507–517 CrossRef CAS PubMed.
- M. De Vetta, O. Baig, D. Steen, J. J. Nogueira and L. González, Molecules, 2018, 23, 2932 CrossRef PubMed.
- A. D. Quartarolo, D. Pérusse, F. Dumoulin, N. Russo and E. Sicilia, J. Porphyrins Phthalocyanines, 2013, 17, 980–988 CrossRef CAS.
- F. Bächle, N. Siemens and T. Ziegler, Eur. J. Inorg. Chem., 2019, 7089–7116 CrossRef.
- X. He, B. Situ, M. Gao, S. Guan, B. He, X. Ge, S. Li, M. Tao, H. Zou, B. Z. Tang and L. Zheng, Small, 2019, 1905080, 1–8 Search PubMed.
- I. Yakavets, M. Millard, V. Zorin, H. P. Lassalle and L. Bezdetnaya, J. Controlled Release, 2019, 304, 268–287 CrossRef CAS PubMed.
- I. Yakavets, H. P. Lassalle, D. Scheglmann, A. Wiehe, V. Zorin and L. Bezdetnaya, Nanomaterials, 2018, 8, 1–15 CrossRef PubMed.
- I. Yakavets, M. Millard, L. Lamy, A. Francois, D. Scheglmann, A. Wiehe, H. P. Lassalle, V. Zorin and L. Bezdetnaya, Cancers, 2019, 11, 1–16 CrossRef PubMed.
- I. Yakavets, H. P. Lassalle, I. Yankovsky, F. Ingrosso, A. Monari, L. Bezdetnaya and V. Zorin, J. Photochem. Photobiol., A, 2018, 367, 13–21 CrossRef CAS.
- I. Yakavets, I. Yankovsky, L. Bezdetnaya and V. Zorin, Dyes Pigm., 2017, 137, 299–306 CrossRef CAS.
- V. Reshetov, D. Kachatkou, T. Shmigol, V. Zorin, M. A. D’Hallewin, F. Guillemin and L. Bezdetnaya, Photochem. Photobiol. Sci., 2011, 10, 911–919 RSC.
- I. Yankovsky, E. Bastien, I. Yakavets, I. Khludeyev, H. P. Lassalle, S. Gräfe, L. Bezdetnaya and V. Zorin, Eur. J. Pharm. Sci., 2016, 91, 172–182 CrossRef CAS PubMed.
- I. Yakavets, I. Yankovsky, M. Millard, L. Lamy, H. P. Lassalle, A. Wiehe, V. Zorin and L. Bezdetnaya, Int. J. Pharm., 2017, 529, 568–575 CrossRef CAS PubMed.
- X. Assfeld, A. Monari, M. Marazzi and H. Gattuso, Front. Chem., 2018, 6, 86 CrossRef PubMed.
- J. Cerezo, F. J. Avila Ferrer, G. Prampolini and F. Santoro, J. Chem. Theory Comput., 2015, 11, 5810–5825 CrossRef CAS PubMed.
- V. Barone, I. Cacelli, N. De Mitri, D. Licari, S. Monti and G. Prampolini, Phys. Chem. Chem. Phys., 2013, 15, 3736–3751 RSC.
- G. Prampolini, F. Ingrosso, A. Segalina, S. Caramori, P. Foggi and M. Pastore, J. Chem. Theory Comput., 2019, 15, 529–545 CrossRef CAS PubMed.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- S. E. Feller, Y. Zhang, R. W. Pastor and B. R. Brooks, J. Chem. Phys., 1995, 103, 4613–4621 CrossRef CAS.
- R. L. Davidchack, R. Handel and M. V. Tretyakov, J. Chem. Phys., 2009, 130, 234101 CrossRef PubMed.
- W. G. Hoover, Phys. Rev. A: At., Mol., Opt. Phys., 1986, 34, 2499–2500 CrossRef PubMed.
- P. Procacci, S. Marsili, A. Barducci, G. F. Signorini and R. Chelli, J. Chem. Phys., 2006, 125, 164101 CrossRef PubMed.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- J. Wang, W. Wang, P. A. Kollman and D. A. Case, J. Mol. Graphics Modell., 2006, 25, 247–260 CrossRef CAS PubMed.
- A. Uhe, S. Kozuch and S. Shaik, J. Comput. Chem., 2010, 1–8 Search PubMed.
- C. J. Dickson, B. D. Madej, Å. A. Skjevik, R. M. Betz, K. Teigen, I. R. Gould and R. C. Walker, J. Chem. Theory Comput., 2014, 10, 865–879 CrossRef CAS PubMed.
- E. L. Wu, X. Cheng, S. Jo, H. Rui, K. C. Song, E. M. Dávila-Contreras, Y. Qi, J. Lee, V. Monje-Galvan, R. M. Venable, J. B. Klauda and W. Im, J. Comput. Chem., 2014, 35, 1997–2004 CrossRef CAS PubMed.
- S. Jo, J. B. Lim, J. B. Klauda and W. Im, Biophys. J., 2009, 97, 50–58 CrossRef CAS PubMed.
- N. Ivanova and A. Ivanova, J. Comput. Chem., 2018, 39, 387–396 CrossRef CAS PubMed.
- K. Pluhackova, S. A. Kirsch, J. Han, L. Sun, Z. Jiang, T. Unruh and R. A. Böckmann, J. Phys. Chem. B, 2016, 120, 3888–3903 CrossRef CAS PubMed.
- S. J. Marrink, V. Corradi, P. C. T. Souza, H. I. Ingólfsson, D. P. Tieleman and M. S. P. Sansom, Chem. Rev., 2019, 119, 6184–6226 CrossRef CAS PubMed.
- P. Marck and L. Nilsson, J. Phys. Chem. A, 2001, 105, 9954–9960 CrossRef.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14(33–38), 27–28 Search PubMed.
- J. Contreras-García, E. R. Johnson, S. Keinan, R. Chaudret, J. Piquemal, D. N. Beratan and W. Yang, J. Chem. Theory Comput., 2011, 7, 625–632 CrossRef PubMed.
- J. Sgrignani and A. Magistrato, ACS Catal., 2015, 5, 3864–3875 CrossRef CAS.
- C. M. Isborn, A. W. Götz, M. A. Clark, R. C. Walker and T. J. Martínez, J. Chem. Theory Comput., 2012, 8, 5092–5106 CrossRef CAS PubMed.
- M. C. R. Melo, R. C. Bernardi, T. Rudack, M. Scheurer, C. Riplinger, J. C. Phillips, J. D. C. Maia, G. B. Rocha, J. V. Ribeiro, J. E. Stone, F. Neese, K. Schulten and Z. Luthey-Schulten, Nat. Methods, 2018, 15, 351–354 CrossRef CAS PubMed.
- A. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- W. C. Frisch, M. J. Trucks, G. W. Schlegel, H. B. Scuseria, G. E. Robb, M. A. Cheeseman, J. R. Scalmani, G. Barone, V. Mennucci, B. Petersson, G. A. Nakatsuji, H. Caricato, M. Li, X. Hratchian, H. P. Izmaylov, A. F. Bloino, J. Zheng and G. Sonnenb, 2009.
- C. M. Isborn, N. Luehr, I. S. Ufimtsev and T. J. Martínez, J. Chem. Theory Comput., 2011, 7, 1814–1823 CrossRef CAS PubMed.
- R. Bonnett, P. Charlesworth, B. D. Djelal, S. Foley, D. J. Mcgarvey and T. G. Truscott, In Vivo, 1999, 325–328 CAS.
- E. M. M. D. Valle, Process Biochem., 2004, 39, 1033–1046 CrossRef.
- Q. Zeng, Z. Li, Y. Dong, C. Di, A. Qin, Y. Hong, L. Ji, Z. Zhu, C. K. W. Jim, G. Yu, Q. Li, Z. Li, Y. Liu, J. Qin and B. Z. Tang, Chem. Commun., 2007, 70–72 RSC.
- H. Yang, X. Zhou, T. Hui, Y. Han, X. Jiang and J. Yan, RSC Adv., 2019, 9, 12078–12084 RSC.
- T. Etienne, L. Chbibi, C. Michaux, E. A. Perpète, X. Assfeld and A. Monari, Dyes Pigm., 2014, 101, 203–211 CrossRef CAS.
- M. Kasha, Radiat. Res., 1963, 20, 55 CrossRef CAS PubMed.
- N. J. Hestand and F. C. Spano, Acc. Chem. Res., 2017, 50, 341–350 CrossRef CAS PubMed.
- T. Etienne, T. Very, E. A. Perpète, A. Monari and X. Assfeld, J. Phys. Chem. B, 2013, 117, 4973–4980 CrossRef CAS PubMed.
- H. Gattuso, M. Marazzi, F. Dehez and A. Monari, Phys. Chem. Chem. Phys., 2017, 19, 23187–23193 RSC.
- A. Segalina, X. Assfeld, A. Monari and M. Pastore, J. Phys. Chem. C, 2019, 123, 6427–6437 CrossRef CAS.
- M. Millard, I. Yakavetsa, M. Piffoux, A. Brun, F. Gazeau, J. M. Guigner, J. Jasniewski, H. P. Lassalle, C. Wilhelm and L. Bezdetnaya, Drug Delivery, 2018, 25, 1790–1801 CrossRef CAS PubMed.
- C. Lange, C. Lehmann, M. Mahler and P. J. Bednarski, Comparison of cellular death pathways after mTHPC-mediated photodynamic therapy (PDT) in five human cancer cell lines, 2019, vol. 11 Search PubMed.
- R. M. Cordeiro, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 438–444 CrossRef CAS PubMed.
- T. T. Tasso, J. C. Schlothauer, H. C. Junqueira, T. A. Matias, K. Araki, É. Liandra-Salvador, F. C. T. Antonio, P. Homem-De-Mello and M. S. Baptista, J. Am. Chem. Soc., 2019, 141, 15547–15556 CrossRef CAS PubMed.
- I. O. L. Bacellar, M. C. Oliveira, L. S. Dantas, E. B. Costa, H. C. Junqueira, W. K. Martins, A. M. Durantini, G. Cosa, P. Di Mascio, M. Wainwright, R. Miotto, R. M. Cordeiro, S. Miyamoto and M. S. Baptista, J. Am. Chem. Soc., 2018, 140, 9606–9615 CrossRef CAS PubMed.
- M. De Vetta, L. González and J. J. Nogueira, ChemistryOpen, 2018, 7, 475–483 CrossRef CAS PubMed.
- F. Santoro, R. Improta, A. Lami, J. Bloino and V. Barone, J. Chem. Phys., 2007, 126, 084509 CrossRef PubMed.
- R. Improta, V. Barone and F. Santoro, Angew. Chem., Int. Ed., 2007, 46, 405–408 CrossRef CAS PubMed.
- J. Cerezo, D. Aranda, F. J. Avila Ferrer, G. Prampolini and F. Santoro, J. Chem. Theory Comput., 2020, 16, 1215–1231 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Dihedral distributions, RMSD plots, free rotation plot, experimental UV/VIS spectral data for mTHPC in water, TM-β-CD and POPC, Cartesian coordinates for mTHPC. See DOI: 10.1039/d0cp02055a |
| This journal is © the Owner Societies 2020 |