Open Access Article
Open Access ArticleSurface plasmon resonance study of the interaction of N-methyl mesoporphyrin IX with G-quadruplex DNA†
M.
Perenon
,
H.
Bonnet
,
T.
Lavergne
 ,
J.
Dejeu
,
J.
Dejeu
 * and
E.
Defrancq
* and
E.
Defrancq

Univ. Grenoble Alpes, CNRS, DCM UMR-5250, F-38000 Grenoble, France. E-mail: jerome.dejeu@univ-grenoble-alpes.fr; Tel: +33 4 56 52 08 13
First published on 6th February 2020
Abstract
Surface plasmon resonance (SPR) was used to investigate the interaction between N-methyl mesoporphyrin IX (NMM) and different G-quadruplex (G4) topologies. The study was associated with circular dichroism analysis (CD) to assess the topology of the G4s when they interacted with NMM. We demonstrate the high selectivity of NMM for the parallel G4 structure with a dissociation constant at least ten times lower than those of other G4 topologies. We also confirm the ability of NMM to shift the G4 conformation from both the hybrid and antiparallel topologies toward the parallel structure.
Introduction
The double-helical structure of DNA in which two antiparallel strands are held together through canonical A/T and G/C base pairing was resolved over half a century ago. However, beyond double helical-based structures, the past decade has brought accumulating evidence of the existence of four-stranded nucleic acid structures, including G-quadruplexes and i-motifs.1,2 Especially, G-quadruplex DNA (G4-DNA) structures are formed from G-rich sequences through stacking of tetrads of Hoogsteen hydrogen-bonded guanines, and connected by loops.3 Sequencing and bioinformatics analyses of the human genome indicate that it contains as much as 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sequences having the potential to form stable G-quadruplex structures.4 In particular, they are found in the telomeric region where their stabilization has been shown to inhibit the activity of telomerase, an enzyme which is over-expressed in 80% of cancer cells, thus evidencing their potential as anticancer drug targets. However, G4 formation is not limited to the telomeric region: they are also over-represented in the promoter regions of a number of genes, including proto-oncogenes c-Myc, c-Kit, bcl-2 and KRAS. Thereby, a large number of data are now in agreement with a biologically relevant regulatory role for G-quadruplexes.5
000 sequences having the potential to form stable G-quadruplex structures.4 In particular, they are found in the telomeric region where their stabilization has been shown to inhibit the activity of telomerase, an enzyme which is over-expressed in 80% of cancer cells, thus evidencing their potential as anticancer drug targets. However, G4 formation is not limited to the telomeric region: they are also over-represented in the promoter regions of a number of genes, including proto-oncogenes c-Myc, c-Kit, bcl-2 and KRAS. Thereby, a large number of data are now in agreement with a biologically relevant regulatory role for G-quadruplexes.5
In this context, the G4 formation at the end of chromosomes (i.e. telomeres) and within the promoter region of oncogenes has been found worthy of acute attention as those structures are now considered as novel anticancer drug targets.6 Indeed, several studies have shown that the inhibition of the telomerase activity and transcriptional repression of oncogenes could be achieved using G4 stabilizing binding ligands.7
A major characteristic of G-quadruplex nucleic acid structures is their intrinsic polymorphic nature: depending on the length, sequence, medium and cations concentration, intramolecular G-quadruplexes show distinct structural topologies in which the strands are in parallel or antiparallel conformations, with the co-existence of different types of loops (lateral, diagonal or propeller) of variable lengths. This structural polymorphism greatly complicates the studies of structure–activity relationships of G4-interacting ligands.
Most of the ligands developed so far are composed of an aromatic core, which can stack over the terminal tetrads, and side chains bearing positive charges for improving the interactions with the phosphodiester DNA backbone as well as for increasing water solubility.8,9 Among them, the porphyrin derivatives such as TMPyP4 have been extensively studied due to their relatively large aromatic framework.10 However, one major limitation of the use of positively charged porphyrin derivatives is the relatively lack of selectivity versus duplex. Therefore a number of metal complexes homologues of TMPYP4 have been developed. They showed a higher quadruplex versus duplex selectivity because the metal in the central cavity of the porphyrin core may preclude intercalation between the base pairs of duplex-DNA.11–15
N-Methyl mesoporphyrin IX (NMM, Fig. 1B) is an anionic porphyrin which was also known to bind to G-rich sequences. It was reported to exhibit a high fluorescence in presence of G4-DNA but not with duplex.16 It was also shown to be selective for G-DNA versus ssDNA, dsDNA, RNA–DNA hybrid, triplex and Holliday junctions thus emphasizing its interest for selective detection of G4-DNA.17 In this context, Yatsunyk and Coll. have studied by using circular dichroism (CD), UV-vis and fluorescence spectroscopies, and gel electrophoresis, the interactions of NMM with various G4-DNA structures in order to investigate the possible binding selectivity for the different G-quadruplex topologies (i.e. parallel, antiparallel or hybrid conformations). A good selectivity for G4-DNAversus duplex DNA was found and more interestingly NMM does not interact with an antiparallel G4-DNA (Tel 22) but with parallel G4-DNA.18 NMM also induces a conformational change from a hybrid to a parallel G4 structure. Yet it was noted that NMM showed quite similar binding constants for parallel and hybrid G4 structure with binding constants of 1 × 105 M−1 and 3 × 105 M−1, respectively. It should be also noticed that NMM binding with other quadruplex structures (e.g. antiparallel) has been reported but with a lower affinity.19
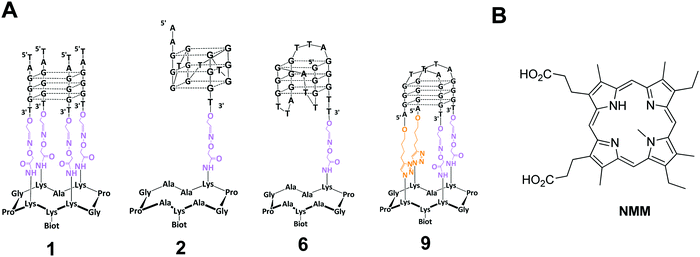 | ||
| Fig. 1 (A) Structure of G-quadruplex anchored of the cyclopeptide scaffold used in the present study and of NMM ligand. | ||
In this context, our objective was to further study the interaction of NMM with various G-quadruplex DNA and in particular to afford a quantitative dimension to the already observed phenomena, including the thermodynamic binding constants as well as kinetic parameters of the binding. These data are essential for the understanding of DNA–ligand interactions. Various biophysical techniques, including FRET melting, UV/vis spectrophotometry, circular dichroism (CD), NMR, and surface plasmon resonance (SPR) have been developed for studying G-quadruplex DNA/ligand interactions.20 In particular, SPR technique displays a number of advantages, including the following: (i) no need for special radioactive or fluorescent labeling of the molecules, (ii) time efficiency, (iii) use of very low quantity of materials and finally (iv) give access to association and dissociation rate.
In the present paper, we report on the study of the interactions of NMM with different G4 topologies 1–11 (parallel, hybrid, antiparallel, see Fig. 1 and Experimental section) by using SPR analysis. We demonstrate that NMM interacts more preferentially with parallel G-quadruplex DNA with a dissociation constant KD in the order of 100 nM whereas for hybrid topology a KD value of around 5–10 μM was obtained and almost no interaction was observed with antiparallel G-quadruplex topology. We also noticed that NMM is able to isomerize a hybrid topology to a parallel one. The results so obtained from SPR analysis confirm the high selectivity of the binding of NMM for parallel G-quadruplex DNA as previously reported in the literature.16 We demonstrate that the binding constants are different depending on the G4 conformation and that NMM interact with the anti-parallel conformation, as weakly, and can isomerize it into hybrid conformation.
Materials and methods
Ligands and analytes preparations
All aqueous solutions were prepared with ultrapure water (Purelab UHQ Elga). The chemical products, NaCl, KCl, Tween 20, HEPES were purchased from Sigma-Aldrich and NMM from Santa Cruz Biotechnology (reference SC-396879). The different sequences that were used for the different systems are depicted in Table 1. Conjugates 1, 6 and 9 were synthetized according to our procedure already reported.21–23 The synthesis of conjugate 2 is depicted in the ESI.†| System | Sequence (5′–3′) | Origin | Topologya |
|---|---|---|---|
| a The listed topologies correspond to the dominant structure observed in presence of potassium rich buffers. We have also controlled the topology of all the studied systems in our conditions. | |||
| 1 | TAG·GGT | Telomeric sequence | Parallel22 |
| 2 | AAG·GGT·GGG·TGG·GTG·GGT | CEB25-L111(T) mini-satellite | Parallel24 |
| 3 | AAG·GGT·GGG·TGT·AAG·TGT·GGG·TGG·GTG·X | CEB25-WT mini-satellite | Parallel24 |
| 4 | AGG·GAG·GGC·GCT·GGG·AGG·AGG·GTT·AX | cKit | Parallel25 |
| 5 | GGA·GGG·TGG·GGA·GGG·TGG·GGA·AX | cMyc | Parallel26 |
| 6 | (TTA·GGG)3·GGG·TT3′ | Telomeric sequence | Hybrid23 |
| 7 | TTA·GGG·TTA·GGG·TTA·GGG·TTA·GGG·TTX | wtTel26 | Hybrid27 |
| 8 | TAGGG(TTAGGG)3TTX | wtTel23 | Hybrid28 |
| 9 | AGG·GTT·AGG·GT | Telomeric sequence | Antiparallel21 |
| 10 | (AGG-GCT)3·AGG·GTT·X | 22-CTA | Antiparallel29 |
| 11 | GGT·TGG·TGT·GGT·TGG·TTX | Thrombin binding aptamer (TBA) | Antiparallel30 |
SPR experiments
HS–(CH2)11–EG6–Biotin was procured from Prochimia. All other chemical products were purchased from Sigma-Aldrich. Cleaning procedure of the gold sensor chips included UV–ozone treatment during 10 min followed by rinsing with MilliQ water and ethanol. The cleaned gold surfaces were then functionalized according to the following procedure. Firstly, mixed self-assembled monolayers (SAMs) were formed at room temperature by dipping overnight gold sensors in the thiol mixture: 80% HS–(CH2)11–EG4–OH and 20% HS–(CH2)11–EG6–biotin (1 mM total thiol concentration in EtOH). After overnight adsorption, gold sensors were rinsed with ethanol and dried under nitrogen. The surface is then inserted in the BIAcore T200 device. All measurements were performed at 25 °C, using a running buffer (R.B.) composed of HEPES buffered saline: 10 mM HEPES pH 7.4, 50 mM NaCl, 100 mM KCl and 0.05% v/v surfactant P20. Streptavidin (100 ng mL−1) was injected (10 μL min−1) on the biotinylated SAM until saturation of the surface (around 2400 R.U.). The different systems 1–11 were injected at 2 μL min−1 on streptavidin-coated SAM surfaces until surface saturation, leading to a similar level of surface concentration around 4 pmol cm−2.Binding experiments were conducted by injection at 30 μL min−1 of NMM dissolved in R.B. at five different concentrations using a single cycle kinetic method (SCK). This method, developed by Karlsson et al., consists in sequential injections of an analyte at increasing concentrations without regeneration steps between each injection.31,32 The five concentrations were injected one after the other with no regeneration step even if dissociation is not complete. With the SCK method, the dissociation kinetic parameter is determined after the last concentration i.e. at the end of the experiment. No regeneration step was performed after the analyte injection but the sensor chip was changed for each experiment. A streptavidin surface, prepared as described below, was used as reference. Curves obtained on the reference surface were deduced from the curves recorded on the recognition one, allowing elimination of refractive index changes due to buffer effects. The binding rate constants of G-quadruplex DNA/ligands interactions were calculated by a non-linear analysis of the association and dissociation curves using the SPR kinetic evaluation software BIAcore T200 Software. The data were fitted using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 model. The association rate constants, kon1, and the dissociation rate constants, koff1 as well as the theoretical maximal response Rmax1 of the interaction were calculated. Finally, the equilibrium dissociation constants were obtained from the binding rate constants as KD1 = koff1/kon1 The reported values are the mean of 10 independent experiments, and the errors provided are standard deviations from the mean.
1 model. The association rate constants, kon1, and the dissociation rate constants, koff1 as well as the theoretical maximal response Rmax1 of the interaction were calculated. Finally, the equilibrium dissociation constants were obtained from the binding rate constants as KD1 = koff1/kon1 The reported values are the mean of 10 independent experiments, and the errors provided are standard deviations from the mean.
Circular dichroism studies
Circular dichroism studies were performed on a Jasco J-810 spectropolarimeter using 1 cm length quartz cuvette. Spectra were recorded at 25 °C with wavelengths range from 220 to 330 nm and were an average of three scans with a 0.5 s response time, a 1 nm data pitch, a 4 nm bandwidth and a 200 nm min−1 scanning speed. Samples of 2.5 μM G4 were annealed at 90 °C with NMM (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio) and cooled slowly overnight.33 In parallel, 2.5 μM G4 samples were annealed at 90 °C alone, cooled overnight, after which porphyrins were added and sample was incubated at 25 °C. The CD spectra were recorded at 1 min, 4 h, 24 h and 48 h after NMM addition. All experiments were performed in the same buffer as for the SPR experiments.
10 ratio) and cooled slowly overnight.33 In parallel, 2.5 μM G4 samples were annealed at 90 °C alone, cooled overnight, after which porphyrins were added and sample was incubated at 25 °C. The CD spectra were recorded at 1 min, 4 h, 24 h and 48 h after NMM addition. All experiments were performed in the same buffer as for the SPR experiments.
Results and discussions
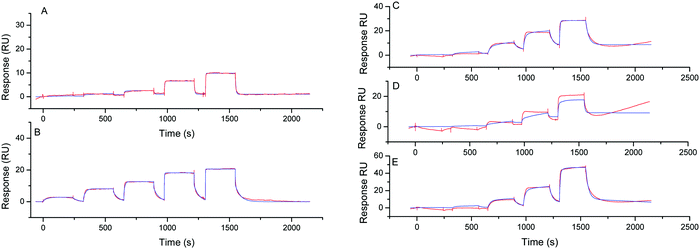
SAMs SPR biochips were used to study the interactions of NMM with the different G4 topologies (i. e. parallel, antiparallel and hybrid) in 10 mM HEPES pH 7.4, 50 mM NaCl, 100 mM KCl solution. We have recently reported that the kinetic parameters and the affinity were not influenced by the ionic strength of the buffer when cationic porphyrin were incubated with G-quadruplexes immobilized on those sensors.34 Analysis were performed by using SCK method which consists in the injection of the NMM analyte at five different concentrations after immobilization of the G-quadruplexes on the SPR chip.31,32The affinity of NMM for different parallel G-quadruplex topologies was first investigated. Different G-quadruplex DNA sequences were investigated: constrained system 1 which represents an intermolecular like parallel G4-DNA, system 2 with the mutated CEB-L111-(T) sequence anchored on the same cyclopeptide platform used for system 1 and systems 3–5 with CEB, c-Kit1 and c-Myc sequences, respectively. All these systems 1–5 form parallel G4 structures as confirmed by CD analyses which show a positive peak at 263 nm and a negative peak at 240 nm that are characteristic for parallel G4 topology (see the ESI,† Fig. S8).35 G4-Systems 1–5 were anchored on the SPR chip through biotin–streptavidin interactions.
The SPR signal responses related to NMM specific interaction with the G4-DNA monolayer (Fig. 2) were obtained after subtraction of the signals recorded on the reference flow-cell as well as the running buffer injection by applying a double referencing procedure.36
After rinsing with the running buffer (R.B.), the SPR signals returned to the baseline meaning that the binding was completely reversible. The sensorgrams were fitted to afford the kinetic constants (association, kon and dissociation, koff) to calculate the dissociation constant KD (KD = kon/koff). This last value was also obtained by fitting the steady state response versus the injected analyte concentration by using the Langmuir isotherm according to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry (Table 2 and Fig. S9, ESI†). The values presented in the Table 1 were the mean of 10 independent experiments.
1 binding stoichiometry (Table 2 and Fig. S9, ESI†). The values presented in the Table 1 were the mean of 10 independent experiments.
| System | Kinetic | Langmuir isotherm | ||
|---|---|---|---|---|
| k on (M−1 s−1) | k off (s−1) | K D (nM) | K D (nM) | |
| nd: not determined as the sensorgram could not be fitted decently to obtained the kinetic data of the interaction due to a too weak interaction.a Parallel conformation.b Hybrid conformation.c Antiparallel conformation. | ||||
| 1 | 3.0 × 105 | 0.20 | 669 ± 80 | 330 ± 8 |
| 2 | 2.3 × 106 | 0.18 | 100 ± 52 | 138 ± 53 |
| 3 | 6.0 × 105 | 0.04 | 65 ± 14 | 62 ± 10 |
| 4 | 4.5 × 105 | 0.08 | 170 ± 8 | 242 ± 57 |
| 5 | 2.9 × 105 | 0.05 | 180 ± 50 | 136 ± 16 |
| 6 | 2.1 × 104 | 0.24 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 ± 2120 500 ± 2120 |
4000 ± 2000 |
| 7 | 2.0 × 104 | 0.28 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 890 ± 3250 890 ± 3250 |
7200 ± 2200 |
| 8 | 3.9 × 104 | 0.26 | 6630 ± 867 | 1500 ± 700 |
| 9 | nd | nd | nd | >100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 10 | nd | nd | nd | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 550 ± 7300 550 ± 7300 |
| 11 | nd | nd | nd | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070 ± 8500 070 ± 8500 |
The dissociation constant, KD, was in the same order of magnitude whatever the method used for the calculation (Langmuir isotherm or provided from kinetic data).37 For all the parallel G-quadruplexes a KD value close to 100 nM was found excepted for system 1 for which an higher KD value was obtained (330–660 nM). This slight difference could be explained by a steric hindrance due to the cyclopeptide scaffold which could hamper NMM analyte to access to the inferior tetrad of 1. This phenomenon was less pronounced when comparing systems 2 and 3 as the DNA sequence is anchored on the cyclopeptide scaffold via a single linker chain resulting in minimal steric hindrance. We could notice that the kinetic association constant was high (kon ∼ 105–106 M−1 s−1) regardless of the sequence forming the parallel G4-DNA.
The KD values obtained from SPR analysis were found lower than those obtained by Yatsunyk and Coll. for NMM binding with Tel22 sequence by using UV and CD titration assays (KD ∼ 3 μM).18 Nevertheless the SPR analysis confirmed the high-affinity interaction of NMM for parallel G4-DNA.
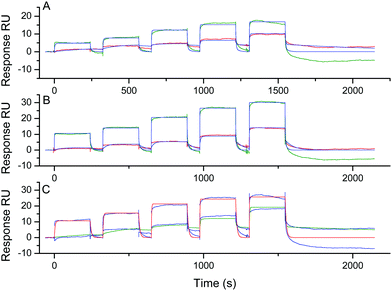
To evaluate the selectivity of NMM, we next investigated its interaction with systems 6–11 showing other G-quadruplex DNA topologies. The sequences in systems 6–8 are known to fold into hybrid topology and the sequences for systems 9–11 are reported to fold into antiparallel topology. The topology for each system was confirmed by CD analysis: a positive peak at 290 nm and negative one at 240 nm (Fig. S10, ESI†) were observed for systems 6–8 and a positive peak at 290 nm and a negative one at 260 nm (Fig. S11, ESI†) were observed for 9–11, which are characteristic of hybrid and antiparallel topology, respectively. Again, the different systems were anchored on SPR chips through biotin–streptavidin interactions and the same procedure as below were used to calculate the affinity constants. The sensorgrams obtained for the hybrid topology (systems 6–8) are presented in Fig. 3.
The dissociation constant, were found to be much higher than for the parallel systems 1–5 with KD values of around 5–10 μM as calculated by both methods (Fig. S12, ESI,† and Table 2). These values are concordant with the affinity for hybrid conformation reported in the literature by using singular value decomposition analysis of CD and UV-vis titration.18 We could observe that the association with the hybrid systems is more difficult than with the parallel systems with an association rate (kon) decreasing from 105 to 104 M−1 s−1. On the other hand, the dissociation is slightly faster with the hybrid systems with a dissociation rate (koff) increasing from the range of 0.04–0.08 s−1 for parallel systems 3–5 to 0.24–0.28 s−1 for hybrid systems 6–8 (Table 2). These results suggest a functional role for the loops in the recognition process: their presence above the tetrad for the hybrid systems may disturb the stacking of NMM on the G-tetrad (vide infra).
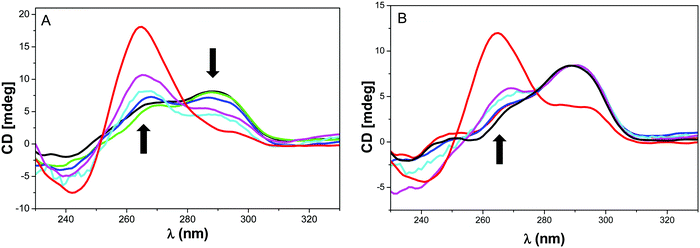
It was reported that NMM could induce the rearrangement of Tel2218 and bimolecular telomere sequence (dTAGGGUTAGGGT) in diluted K+ buffer from an hybrid to a parallel topology.38 To assess the effect of NMM on the two hybrid systems 7–8, the ligand was incubated with the folded hybrid-type G-quadruplexes and CD profiles were collected against time (Fig. 4). As a control experiment, the folding of the G4 forming sequences 7 and 8 were performed in presence of NMM that shows the predominant formation of parallel topologies (in red in Fig. 4).
For system 7, the CD signal at 264 nm was found to increase with a concomitant decrease of the signal at 295 nm which suggests a shift from hybrid to a conformation with substantial parallel component (Fig. 4A). This behaviour was less pronounced with system 8 that indicates a more difficult isomerization from hybrid to parallel topology for this sequence. This was confirmed by the control experiment for which a band at 295 nm is still present. The NMM-induced structural conversion was found to be slow and a minimum of 24 h is required to observe the equilibrium. This time scale is too long to consider a structural reorganization during SPR experiments. Thus the KD values obtained by SPR do reflect the inherent interaction of NMM with the hybrid G-quadruplex topology.
The affinity for antiparallel topologies was next studied by SPR by using systems 9–11 (Fig. S13, ESI†). System 9 is a constrained antiparallel G-quadruplex derived from the telomeric sequence,21 system 10 (22-CTA) is the a telomeric mutant sequence reported to form an antiparallel G428 and system 11 is the sequence of thrombin binding aptamer (TBA) which is known to adopt antiparallel structure (Fig. S11, ESI†).29 For constrained system 9, the SPR signals were barely detectable suggesting a very low affinity (>100 μM, Fig. S10, ESI†). For systems 10 and 11, a KD value can be only extracted from the fitting of the response with the Langmuir isotherm (Fig. 5). Indeed, the sensorgram could not be fitted to obtain the kinetic data of the interaction. Although the values were low (Table 2), this is in agreement with previously reported studies by fluorescence. Indeed, an enhancement of fluorescence was measured when NMM was added to antiparallel G-quadruplex DNA (basket or chair). This enhancement was weaker than for the parallel G4 but revealed an interaction between NMM and the antiparallel G4.
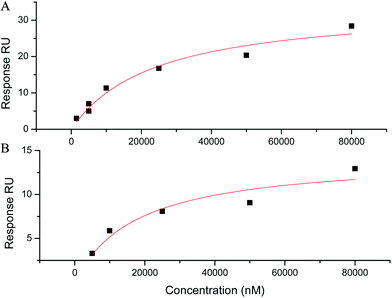 | ||
Fig. 5 Adsorption isotherm (square) and fitting curve (line) using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Langmuir interaction model for the interaction of NMM with antiparallel G-quadruplexes (A) system 11 and (B) system 11. 1 Langmuir interaction model for the interaction of NMM with antiparallel G-quadruplexes (A) system 11 and (B) system 11. | ||
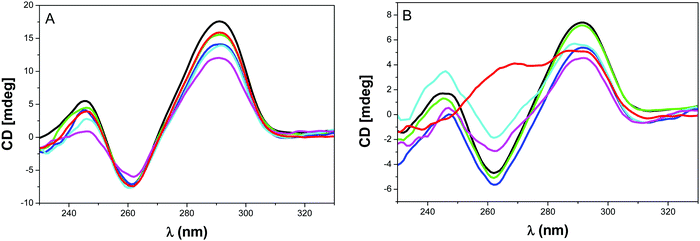
In order to investigate if any rearrangement of the antiparallel topology could occur during the recognition by NMM, CD analysis was performed by using the telomeric systems 9 and 10 (Fig. 6) and system 11 (Fig. S14, ESI†). As anticipated no change was observed upon addition and incubation of NMM to system 9 or by annealing the G4-DNA9 in presence of NMM because the constrain preclude any isomerization of the system (Fig. 6A). This confirms the results from SPR showing that NMM do not interact with this antiparallel topology. The same lack of structural reorganization was also obtained with TBA 11 as previously observed (Fig. S14, ESI†).18
The result with system 10 was rather unanticipated. Indeed, NMM did not modify the topology when it was added after the annealing step even after long incubation time. However when NMM is added during the annealing of G-quadruplex DNA 10, we could observe an increase in the CD signal at 264 nm (line red in Fig. 6B) that suggests a partial isomerization from antiparallel to parallel or hybrid structure. To the best of our knowledge, this is the first observation of a shift from antiparallel to another topology induced by NMM.
Conclusion
In this paper, the interaction between N-methyl mesoporphyrin IX (NMM) and different G-quadruplex topologies (parallel, hybrid and antiparallel) were investigated by SPR and CD. The NMM affinity properties were influenced by the conformation of the G4. Indeed, NMM was confirmed to be selective for the parallel form and a dissociation constant KD around 100 nM was obtained. This is near 10 time less than for the hybrid topology (several micromolar) and 100 time less than the antiparallel (several ten micromolar). This difference of KD was due to an increase of the association kinetic rate and a decrease of the dissociation for NMM binding to the studied parallel structures. This could be explained by the absence of loops on the upper and lower tetrads of parallel topology systems that may facilitate the π stacking of NMM on those tetrads and therefore increases the association rate. This is in good agreement with the mechanism proposed for the interaction of NMM with parallel G-quadruplex DNA.39 The authors reported that the interaction involves an efficient π stacking of NMM with the terminal G-tetrad as well as the localisation of the N-methyl group into the G-quadruplex core aligned with the potassium cation. In case of hybrid or antiparallel structures, the steric hindrance of the terminal G-tetrad either by the lateral or by the diagonal loops may hamper the interactions37,40 and therefore necessitate a rearrangement of the topology to allow the access to the terminal tetrad (i.e. hybrid to parallel shift as reported18 or antiparallel to parallel/hybrid as observed in this study). This is emphasized by the very weak affinity observed with the constrained antiparallel system 9 for which no structural accommodation is possible.We also confirmed that NMM could shift the hybrid form to parallel after a long time of incubation (24 h). When the folding was performed in presence of NMM the parallel structure was readily formed. We also demonstrate for the first time that NMM could promote the isomerization of an antiparallel G-quadruplex structure (22 CTA) toward parallel or hybrid topology.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to Agence Nationale pour la Recherche (ANR G4-TopiPro ANR-16-CE11-0006-01), ARCANE and CBH-EUR-GS (ANR-17-EURE-0003). Dr M.-P. Teulade-Fichou, Dr D. Verga and Dr C. Beauvineau are acknowledged for fruitful discussions. The authors acknowledge also support from ICMG FR 2607 for providing synthesis and purification of oligonucleotides and SPR facilities.References
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182 CrossRef CAS PubMed.
- M. Zeraati, D. B. Langley, P. Schofield, A. L. Moye, R. Rouet, W. E. Hughes, T. M. Bryan, M. E. Dinger and D. Christ, Nat. Chem., 2018, 10, 631 CrossRef CAS PubMed.
- A. I. Karsisiotis, C. O’Kane and M. Webba da Silva, Methods, 2013, 64, 28–35 CrossRef CAS PubMed.
- C. K. Kwok, G. Marsico, A. B. Sahakyan, V. S. Chambers and S. Balasubramanian, Nat. Methods, 2016, 13, 841 CrossRef CAS PubMed.
- D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627 CrossRef CAS PubMed and references cited in.
- S. Neidle, Nat. Rev. Chem., 2017, 1, 1 CrossRef and references cited in.
- S. Muller and R. Rodriguez, Expert Rev. Clin. Pharmacol., 2014, 7, 663 CrossRef PubMed.
- A. R. Duarte, E. Cadoni, A. S. Ressurreicao, R. Moreira and A. Paulo, ChemMedChem, 2018, 13, 869 CrossRef CAS PubMed and references cited in.
- S. Asamitsu, T. Bando and H. Sugiyama, Chem. – Eur. J., 2019, 25, 417 CrossRef CAS PubMed.
- C. Romera, O. Bombarde, R. Bonnet, D. Gomez, P. Dumy, P. Calsou, J. F. Gwan, J. H. Lin, E. Defrancq and G. Pratviel, Biochimie, 2011, 93, 1310 CrossRef CAS PubMed.
- J. Dejeu, T. Lavergne, J. D. Nora, E. Defrancq and G. Pratviel, Inorg. Chim. Acta, 2016, 452, 98 CrossRef CAS.
- L. Sabater, M.-L. Nicolau-Travers, A. De Rache, E. Prado, J. Dejeu, O. Bombarde, J. Lacroix, P. Calsou, E. Defrancq, J.-L. Mergny, D. Gomez and G. Pratviel, J. Biol. Inorg. Chem., 2015, 20, 729 CrossRef CAS PubMed.
- A. J. Gaier and D. R. McMillin, Inorg. Chem., 2015, 54, 4504 CrossRef CAS PubMed.
- E. Boschi, S. Davis, S. Taylor, A. Butterworth, L. A. Chirayath, V. Purohit, L. K. Siegel, J. Buenaventura, A. H. Sheriff, R. Jin, R. Sheardy, L. A. Yatsunyk and M. Azam, J. Phys. Chem. B, 2016, 120, 12807 CrossRef CAS PubMed.
- L. Sabater, P. J. Fang, C. F. Chang, A. De Rache, E. Prado, J. Dejeu, A. Garofalo, J. H. Lin, J.-L. Mergny, E. Defrancq and G. Pratviel, Dalton Trans., 2015, 44, 3701 RSC.
- H. Arthanari, S. Basu, T. L. Kawano and P. H. Bolton, Nucleic Acids Res., 1998, 26, 3724 CrossRef CAS.
- J. Ren and J. B. Chaires, Biochemistry, 1999, 38, 16067 CrossRef CAS.
- J. M. Nicoludis, S. P. Barrett, J.-L. Mergny and L. A. Yatsunyk, Nucleic Acids Res., 2012, 40, 5432 CrossRef CAS PubMed.
- A. Kreig, J. Calvert, J. Sanoica, E. Cullum, R. Tipanna and S. Myong, Nucleic Acids Res., 2015, 43, 7961 CrossRef CAS PubMed.
- P. Murat, Y. Singh and E. Defrancq, Chem. Soc. Rev., 2011, 40, 5293 RSC.
- R. Bonnet, T. Lavergne, B. Gennaro, N. Spinelli and E. Defrancq, Chem. Commun., 2015, 51, 4850 RSC.
- P. Murat, D. Cressend, N. Spinelli, A. Van der Heyden, P. Labbé, P. Dumy and E. Defrancq, ChemBioChem, 2008, 9, 2588 CrossRef CAS PubMed.
- P. Murat, R. Bonnet, A. Van der Heyden, N. Spinelli, P. Labbé, D. Monchaud, M.-P. Teulade-Fichou, P. Dumy and E. Defrancq, Chem. – Eur. J., 2010, 16, 6106–6114 CrossRef CAS PubMed.
- S. Amrane, M. Adrian, B. Heddi, A. Serero, A. Nicolas, J.-L. Mergny and A. T. Phan, J. Am. Chem. Soc., 2012, 134, 5807 CrossRef CAS PubMed.
- V. Kuryavyi, A. T. Phan and D. J. Patel, Nucleic Acids Res., 2010, 38, 6757 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, H. Y. Gaw and D. J. Patel, Nat. Chem. Biol., 2005, 1, 167 CrossRef CAS PubMed.
- A. Ambrus, D. Chen, J. Dai, T. Bialis, R. A. Jones and D. Yang, Nucleic Acids Res., 2006, 34, 2723 CrossRef CAS PubMed.
- J. Dai, M. Carver and D. Yang, Biochimie, 2008, 90, 1172 CrossRef CAS PubMed.
- K. W. Lim, P. Alberti, A. Guédin, L. Lacroix, J.-F. Riou, N. J. Royle, J.-L. Mergny and A. T. Phan, Nucleic Acids Res., 2009, 37, 6239 CrossRef CAS.
- R. F. Macaya, P. Schultze, F. W. Smith, J. A. Roe and J. Feigon, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3745 CrossRef CAS.
- R. Karlsson, P. S. Katsamba, H. Nordin, E. Pol and D. G. Myszka, Anal. Biochem., 2006, 349, 136 CrossRef CAS PubMed.
- W. Palau and C. Di Primo, Talanta, 2013, 114, 211 CrossRef CAS PubMed.
- LC-MS analysis of NMM was carried out before and after 90 °C heating and showed that NMM is stable in these conditions (see ESI,† Fig. S5–S7).
- E. Prado, L. Bonnat, H. Bonnet, T. Lavergne, A. van der Heyden, G. Pratviel, J. Dejeu and E. Defrancq, Langmuir, 2018, 34, 13057 CrossRef CAS PubMed.
- S. Masiero, R. Trotta, S. Pieraccini, S. De Tito, R. Perone, A. Randazzo and G. P. Spada, Org. Biomol. Chem., 2010, 8, 2683 RSC.
- R. L. Rich and D. G. Myszka, Curr. Opin. Biotechnol., 2000, 11, 54 CrossRef CAS PubMed.
- A factor 5 was considered for a representative difference between the different affinity.
- S. Paramasivan, I. Rujan and P. H. Bolton, Methods, 2007, 43, 324 CrossRef CAS PubMed.
- J. M. Nicoludis, S. T. Miller, P. D. Jeffrey, S. P. Barrett, P. R. Rablen, T. J. Lawton and L. A. Yatsunyk, J. Am. Chem. Soc., 2012, 134, 20446 CrossRef CAS PubMed.
- Y. Wang and D. J. Patel, Structure, 1993, 1, 263 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis and characterization of CEB25-L111(T) G4 parallel conjugate 2, Langmuir isotherm (complexes 1–8), CD profile (complexes 2, 6–11), SCK sensorgrams (complexes 9–11). See DOI: 10.1039/c9cp06321h |
| This journal is © the Owner Societies 2020 |