How strong are hydrogen bonds in the peptide model?†
Abstract
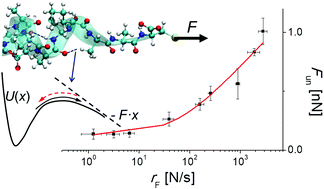
Detailed knowledge of intramolecular hydrogen bonds, including their nanomechanics, in a peptide secondary structure is crucial for understanding mechanisms of numerous biochemical processes. Single-molecule force spectroscopy has become a powerful tool to study directly the mechanical properties of single biopolymers and monitoring the hydrogen bonds. However, the interpretation of such experiments, due to their poor temporal resolution relative to the rate of intramolecular dynamics, requires the support of molecular simulations. In this work, we provide a methodology for determining the kinetic and energetic characteristics of hydrogen bonds in a template model of the protein secondary structure. Our approach, based on the steered molecular dynamics method, employs dynamic force spectroscopy calculations and uses two advanced theoretical models of force-induced unbinding. A systematic analysis of the simulated data with these models allowed for quantitative characterization of a single hydrogen bond in the α-helix of the AAKA(AEAAKA)5AC peptide model and detailed explanation of the mechanism of the α-helix unfolding. The methodology proposed here may be extended to other molecular structures stabilized by internal hydrogen bonds.


 Please wait while we load your content...
Please wait while we load your content...