 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Translation-coupled RNA replication and parasitic replicators in membrane-free compartments†
Ryo
Mizuuchi
 *ab and
Norikazu
Ichihashi
*ab and
Norikazu
Ichihashi
 acd
acd
aKomaba Institute for Science, The University of Tokyo, Meguro, Tokyo 153-8902, Japan. E-mail: mizuuchi@bio.c.u-tokyo.ac.jp
bJST, PRESTO, Kawaguchi, Saitama 332-0012, Japan
cDepartment of Life Science, Graduate School of Arts and Science, The University of Tokyo, Meguro, Tokyo 153-8902, Japan
dUniversal Biology Institute, The University of Tokyo, Meguro, Tokyo 153-8902, Japan
First published on 7th October 2020
Abstract
We report RNA self-replication through the translation of its encoded protein within membrane-free compartments generated by liquid–liquid phase separation. The aqueous droplets support RNA self-replication by concentrating a genomic RNA and translation proteins, facilitating the uptake of small substrates, and preventing the replication of parasitic RNAs through compartmentalization.
Bottom-up construction of an artificial cell is a viable approach to dissect the roles and mechanisms of cellular functions and provide novel cell-like reactors that can be readily manipulated.1–5 To better mimic cell behaviour and create complex biological systems, a key step in artificial cell construction is the compartmentalization of gene expression machinery, a central part of living cells that converts genetic information into proteins to facilitate cellular processes. Typically performed in artificial cells with phospholipid membranes, previous research integrated cell-free gene expression systems with diverse cellular phenomena including quorum sensing,6 cell deformation,7,8 energy generation,9 and replication of DNA and RNA.10,11 A phospholipid membrane, however, strictly limits the transport of molecules to and from compartments, which restricts the applicability of artificial cells. An alternative way to create artificial cells is to use membrane-free compartments, such as cell-like hydrogels and droplets based on liquid–liquid phase separation (LLPS). Although the expression of reporter genes was achieved in these systems,12–17 the demonstration of complex translation-activated biological processes has been challenging. Because membrane-free compartments are generally highly permeable, designable to sequester specific biomolecules, and molecularly crowded like living cells, they could provide unique regulation towards gene-expression and integrated biological reactions.3,5
LLPS provides membrane-less cell-like structures that are ubiquitously seen in natural cells as biomolecular condensates.18 The liquid-like state enables molecular exchange with environments and the selective sequestration of biomolecules, while maintaining the mobility of sequestered biomolecules.19,20 The development of such cell models could help to elucidate the roles of cellular condensates as well as to establish dynamic artificial cells. Because LLPS occurs by simple physical processes, it would also provide insights into primitive compartmentalization by phase separation at the origins of life, especially before the advent of membranes.21,22 To date, enzymatic and catalytic reactions, including the expression of fluorescent reporters, were demonstrated in two types of LLPS-based protocell models, coacervates12,15,23–25 and aqueous two-phase systems (ATPS).13,26–28 The integration of gene-expression with downstream biological processes in these systems, if achieved, would expand the versatility of LLPS-based droplets as artificial cells.
Here, we employ cell-free translation within an ATPS and demonstrate the self-replication of a genomic RNA by its encoded protein. The replication of genetic molecules using self-encoded proteins is essential in cells, allowing autonomous proliferation as well as Darwinian evolution.29,30 We further show additional roles of ATPS in facilitating the sustained replication of the genomic RNA: the prevention of parasitic RNA replication and the facile uptake of building blocks.
To demonstrate the translation-coupled RNA replication (TcRR) in ATPS, we combined a TcRR system based on previous studies29,31 with an ATPS of polyethylene glycol (PEG, 20 kDa, 15 wt%) and dextran (DEX, 9–11 kDa, 1.5 wt%) (Fig. 1A). The TcRR system consists of an artificial single-stranded genomic RNA (2041 nt) that encodes the core subunit of Qβ replicase and a reconstituted E.coli translation system based on the PURE (protein synthesis using recombinant elements) system,32 (Tables S1 and S2, ESI†). In the TcRR system, the core subunit translated from the genomic RNA forms a functional replicase by associating with EF-Tu and EF-Ts (elongation factors) in the translation system, and the active RNA polymerase replicates the genomic RNA. Vigorous mixing of the TcRR system with DEX and PEG generated DEX-rich phase droplets dispersed in a continuous PEG-rich phase (Fig. 1B). We confirmed that the genomic RNA was preferentially localized in DEX-rich phase droplets (Fig. 1B), consistent with previous studies that examined RNA localization in DEX/PEG ATPS.26,33 We then centrifuged the ATPS to obtain PEG- and DEX-rich phases separately, and investigated the localization of the translation proteins by subjecting both phases to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1C). The selective appearance of the bands in the DEX-rich phase confirmed that most translation proteins partitioned into the DEX-rich phase, which was previously conjectured,13 but not examined directly. These results showed that most components for the TcRR reaction were segregated in the ATPS droplets.
Next, we performed the TcRR reaction at 37 °C for 2 h in various conditions and determined the respective level of RNA replication. In the standard TcRR condition optimized without an ATPS (Table S2 ESI,† except with 16 mM magnesium acetate (Mg(OAc)2)),29,31 the replication of the genomic RNA was detectable only in the absence of the ATPS (Fig. 2A). We optimized the reaction condition with the ATPS and found that changing the concentration of Mg(OAc)2 from 16 mM to 8 mM significantly improved the TcRR reaction in the ATPS (Fig. S1, ESI†); similar DEX-rich phase droplets appeared to form in both Mg(OAc)2 conditions (8 mM in Fig. 1 and 16 mM in Fig. S2, ESI†). In 8 mM Mg(OAc)2, the genomic RNA replicated 4-fold in the ATPS, whereas the replication was negligible in the absence of one or both of DEX or PEG (Fig. 2B). We noticed that the TcRR reaction was detectable only if we mixed the translation system with DEX before the addition of PEG, perhaps because DEX keeps translation proteins soluble and functional despite the high concentration of PEG; PEG is known to inhibit cell-free translation probably due to protein precipitation.34,35 The size of DEX-rich phase droplets remained in a similar range after the incubation, although the number of smaller droplets diminished (Fig. S3, ESI†). Furthermore, in contrast to the bulk-optimized condition, the concentration of translation proteins in the DEX-rich phase droplets of the ATPS (Fig. 1C) allowed the dilution of the set of translation proteins (Table S1, ESI†) up to 4-fold without diminishing RNA replication (Fig. 2C).
 | ||
| Fig. 2 The TcRR reaction within the ATPS. The TcRR system with 3 nM genomic RNA was incubated at 37 °C for 2 h in (A) 16 mM or (B) 8 mM Mg(OAc)2 in the presence or absence of the ATPS, or in a continuous 1.5 wt% DEX 9–11 kDa or 15 wt% PEG 20 kDa phase. The RNA concentration was measured by quantitative RT-PCR. The dashed lines represent 1-fold replication (RNA concentration after the reaction divided by initial RNA concentration). (c) The TcRR reaction with diluted translation proteins (Table S1, ESI†) in the absence or presence of the ATPS (16 or 8 mM Mg(OAc)2, respectively). All error bars indicate standard errors (N = 3). | ||
One of the roles of compartments in prebiotic evolution is to allow sustainable replication of genetic replicators by limiting the propagation of parasitic ones.29,30,36 To examine whether the ATPS could shelter the genomic RNA from parasitic RNAs despite the lack of membranes, we prepared two types of droplets, which contained either the genomic RNA or a 510 nt parasitic RNA (Parasite-γ11537). The parasitic RNA lacks the replicase subunit gene, but it can replicate by exploiting the replicase expressed from the genomic RNA (Fig. 3A). We first encapsulated the Cy5-labeled genomic RNA and the Cy3-labeled parasitic RNA in distinct ATPS droplets (Fig. S4A, ESI†). By gently mixing the two ATPS, we examined the inter-droplet diffusion of the two RNAs. Although some droplets showed both fluorescence signals, the distribution of the fluorescence-labelled RNAs was not uniform across the droplet population, and one type of RNA dominated in most of the droplets (Fig. 3B). The uneven RNA distribution among the droplets persisted after 2 h incubation at 37 °C (Fig. 3C and Fig. S4B, ESI†), indicating that the localization of RNAs was maintained.
We then performed the TcRR reaction by encapsulating the genomic and parasitic RNAs together in the same ATPS or by gently mixing two ATPS, each containing one type of RNA (Fig. 3D). When we pre-mixed the genomic and parasitic RNAs in the same ATPS (#1), the genomic RNA barely replicated due to the high replication of the parasitic RNA. However, when mixing two ATPS that separately contained the genomic and parasitic RNAs in a 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (which equalized the concentration of the two RNAs in the mixture) (#2), the genomic RNA replicated at the same level as the control reaction performed with an ATPS in the absence of the parasitic RNA (#3), whereas the replication of the parasitic RNA was inhibited compared to the reaction performed with the pre-mixed genomic and parasitic RNAs (#1). We also performed the reaction with a mixed genomic and parasitic RNA system in the absence of ATPS (#4), and found that the replication of the genomic RNA was more efficient with the ATPS droplets, with a significant suppression of replication of the parasitic RNA (#2). These results suggested that the ATPS could support the RNA self-replication by compartmentalizing the genomic and parasitic RNAs in distinct droplets.
1 ratio (which equalized the concentration of the two RNAs in the mixture) (#2), the genomic RNA replicated at the same level as the control reaction performed with an ATPS in the absence of the parasitic RNA (#3), whereas the replication of the parasitic RNA was inhibited compared to the reaction performed with the pre-mixed genomic and parasitic RNAs (#1). We also performed the reaction with a mixed genomic and parasitic RNA system in the absence of ATPS (#4), and found that the replication of the genomic RNA was more efficient with the ATPS droplets, with a significant suppression of replication of the parasitic RNA (#2). These results suggested that the ATPS could support the RNA self-replication by compartmentalizing the genomic and parasitic RNAs in distinct droplets.
In the evolution of genomic RNAs, parasitic RNAs likely appear through mutations in compartments where a genomic RNA already exists.30,36,37 We therefore also examined whether the ATPS could prevent the replication of a small amount of parasitic RNA (de novo synthesized or contaminated) without manual segregation as performed in Fig. 3D. As described in the Supplementary Results section (ESI†), we found that the ATPS prevented such parasitic RNA (∼220 nt) replication (Fig. S5, ESI†). Moreover, in the TcRR reaction without the addition of known parasitic RNAs, a genomic RNA (R3038) replicated slightly better in the ATPS (#5, Fig. 3E) than in bulk (#6) by preventing the appearance of parasitic RNAs, even with the inhibitory ATPS condition (Fig. 2A and B). These results emphasize the effectiveness of the membrane-free compartments to support RNA self-replication by limiting parasitic RNA replication.
While the ATPS appeared to prevent inter-droplet RNA diffusion (Fig. 3), LLPS-based droplets are generally permeable to small molecules.19,20 We therefore wondered whether the TcRR reaction could continue by externally supplying small substrates. We initiated the TcRR reaction in the ATPS with 62.5 μM cytidine triphosphate (CTP, used in RNA polymerization), and then added 625 μM CTP to the ATPS after 2 h incubation and continued the reaction. The extra CTP added was as low as 0.03 volume of the ATPS so that the composition of the ATPS was basically unchanged. While in the absence of additional CTP a genomic RNA replicated only up to the resource limitation (corresponding to approximately 120 nM genomic RNA), the TcRR reaction with added CTP exceeded this limitation and used up nearly the entire of the added CTP by 5 h (Fig. 4), suggesting that the ATPS droplets were fully permeable to the extra genetic materials, which continued the TcRR reaction.
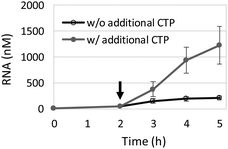 | ||
| Fig. 4 The TcRR reaction through the supply of additional CTP. The TcRR system with 10 nM of a genomic RNA was incubated in the ATPS at 37 °C for 5 h. The concentration of the translation system (Table S1, ESI†) was halved, and the initial CTP concentration was 62.5 μM. Water or 625 μM additional CTP was mixed with the ATPS (open and filled circles, respectively) at 2 h (indicated as the arrow), followed by further incubation. The RNA concentrations were measured by quantitative RT-PCR. The error bars indicate standard errors (N = 3). | ||
In this study, we integrated in vitro gene-expression with RNA replication within LLPS-based droplets. In a DEX/PEG ATPS, the genomic RNA and translation proteins were confined in DEX-rich phase droplets and functioned simultaneously to activate RNA self-replication by its encoded Qβ replicase protein (Fig. 2B). Two characteristics of LLPS-based droplets,19,20 concentration of internalized components and permeability to small molecule analytes, allowed us to reduce translation proteins without hindering the TcRR reaction (Fig. 2C) and to supply substrates externally to continue the reaction (Fig. 4). Such a persistent genome replication system functional with a lower input of translation proteins would be a step toward realizing a self-regenerating and long-lasting artificial cell.4
Moreover, the ATPS supported the RNA self-replication by sheltering a genomic RNA from various short parasitic RNAs (Fig. 3 and Fig. S5, ESI†). The primary mechanism for this effect was to segregate the parasitic RNAs into distinct droplets and prevent inter-droplet RNA diffusion. Such effective compartmentalization is a key feature of cell-like structures for the evolution of genetic entities and previously shown only in compartments with explicit boundaries.29,30,36,37 Therefore, future studies should examine whether the functional RNA is evolvable even in membrane-free compartments, an attractive question in the field of origins-of-life. More discussions can be found in the Supplementary Discussion (ESI†).
In conclusion, we demonstrated translation-integrated RNA replication within DEX/PEG ATPS droplets, where the replication machinery was concentrated, accessible to external building blocks, and protected from takeover by parasitic RNAs. These results validate LLPS-based droplets as artificial cells for implementing complex biological reactions as well as model systems to study a primitive type of compartmentalization.
We thank Hiroyuki Noji, Miho Yanagisawa, and Chiho Watanabe for useful discussion and Tony Z. Jia for helpful comments on the manuscript. This work was supported by JST, PRESTO grant number JPMJPR19KA, Japan.
Conflicts of interest
There are no conflicts to declare.Notes and references
- F. Caschera and V. Noireaux, Curr. Opin. Chem. Biol., 2014, 22, 85–91 CrossRef CAS.
- M. Li, X. Huang, T. Y. D. Tang and S. Mann, Curr. Opin. Chem. Biol., 2014, 22, 1–11 CrossRef CAS.
- E. Dubuc, P. A. Pieters, A. J. van der Linden, J. C. van Hest, W. T. Huck and T. F. de Greef, Curr. Opin. Biotechnol., 2019, 58, 72–80 CrossRef CAS.
- N. Yeh Martín, L. Valer and S. S. Mansy, Emerg. Top. Life Sci, 2019, 3, 597–607 CrossRef.
- N. Laohakunakorn, L. Grasemann, B. Lavickova, G. Michielin, A. Shahein, Z. Swank and S. J. Maerkl, Front. Bioeng. Biotechnol., 2020, 8, 213 CrossRef.
- G. Rampioni, F. D’Angelo, M. Messina, A. Zennaro, Y. Kuruma, D. Tofani, L. Leoni and P. Stano, Chem. Commun., 2018, 54, 2090–2093 RSC.
- E. Godino, J. N. López, D. Foschepoth, C. Cleij, A. Doerr, C. F. Castellà and C. Danelon, Nat. Commun., 2019, 10, 4696 CrossRef.
- T. Furusato, F. Horie, H. T. Matsubayashi, K. Amikura, Y. Kuruma and T. Ueda, ACS Synth. Biol., 2018, 7, 953–961 CrossRef CAS.
- S. Berhanu, T. Ueda and Y. Kuruma, Nat. Commun., 2019, 10, 1325 CrossRef.
- P. Van Nies, I. Westerlaken, D. Blanken, M. Salas, M. Mencía and C. Danelon, Nat. Commun., 2018, 9, 1583 CrossRef.
- H. Kita, T. Matsuura, T. Sunami, K. Hosoda, N. Ichihashi, K. Tsukada, I. Urabe and T. Yomo, ChemBioChem, 2008, 9, 2403–2410 CrossRef CAS.
- E. Sokolova, E. Spruijt, M. M. K. Hansen, E. Dubuc, J. Groen, V. Chokkalingam, A. Piruska, H. A. Heus and W. T. S. Huck, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11692–11697 CrossRef CAS.
- P. Torre, C. D. Keating and S. S. Mansy, Langmuir, 2014, 30, 5695–5699 CrossRef CAS.
- J. Thiele, Y. Ma, D. Foschepoth, M. M. K. Hansen, C. Steffen, H. A. Heus and W. T. S. Huck, Lab Chip, 2014, 14, 2651–2656 RSC.
- T. Y. Dora tang, D. Van Swaay, A. DeMello, J. L. Ross Anderson and S. Mann, Chem. Commun., 2015, 51, 11429–11432 RSC.
- X. Zhou, H. Wu, M. Cui, S. N. Lai and B. Zheng, Chem. Sci., 2018, 9, 4275–4279 RSC.
- L. Aufinger and F. C. Simmel, Angew. Chem., Int. Ed., 2018, 57, 17245–17248 CrossRef CAS.
- S. Alberti, A. Gladfelter and T. Mittag, Cell, 2019, 176, 419–434 CrossRef CAS.
- C. D. Crowe and C. D. Keating, Interface Focus, 2018, 8, 820180032 CrossRef.
- N. Martin, ChemBioChem, 2019, 20, 2553–2568 CrossRef CAS.
- R. R. Poudyal, F. Pir Cakmak, C. D. Keating and P. C. Bevilacqua, Biochemistry, 2018, 57, 2509–2519 CrossRef CAS.
- T. Yoshizawa, R. S. Nozawa, T. Z. Jia, T. Saio and E. Mori, Biophys. Rev., 2020, 12, 519–539 CrossRef CAS.
- J. Crosby, T. Treadwell, M. Hammerton, K. Vasilakis, M. P. Crump, D. S. Williams and S. Mann, Chem. Commun., 2012, 48, 11832–11834 RSC.
- B. Drobot, J. M. Iglesias-Artola, K. Le Vay, V. Mayr, M. Kar, M. Kreysing, H. Mutschler and T. Y. D. Tang, Nat. Commun., 2018, 9, 3643 CrossRef.
- R. R. Poudyal, R. M. Guth-Metzler, A. J. Veenis, E. A. Frankel, C. D. Keating and P. C. Bevilacqua, Nat. Commun., 2019, 10, 1–13 CrossRef.
- C. A. Strulson, R. C. Molden, C. D. Keating and P. C. Bevilacqua, Nat. Chem., 2012, 4, 941–946 CrossRef CAS.
- D. N. Cacace and C. D. Keating, J. Mater. Chem. B, 2013, 1, 1794–1803 RSC.
- B. W. Davis, W. M. Aumiller, N. Hashemian, S. An, A. Armaou and C. D. Keating, Biophys. J., 2015, 109, 2182–2194 CrossRef CAS.
- N. Ichihashi, K. Usui, Y. Kazuta, T. Sunami, T. Matsuura and T. Yomo, Nat. Commun., 2013, 4, 2494 CrossRef.
- R. Mizuuchi and N. Ichihashi, Nat. Ecol. Evol., 2018, 2, 1654–1660 CrossRef.
- R. Mizuuchi, N. Ichihashi and T. Yomo, ChemBioChem, 2016, 17, 1229–1232 CrossRef CAS.
- Y. Shimizu, A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa and T. Ueda, Nat. Biotechnol., 2001, 19, 751–755 CrossRef CAS.
- T. Z. Jia, C. Hentrich and J. W. Szostak, Orig. Life Evol. Biosph., 2014, 44, 1–12 CrossRef CAS.
- C. K. Bakke, L. M. Jungbauer and S. Cavagnero, Protein Expr. Purif., 2006, 45, 381–392 CrossRef CAS.
- X. Ge, D. Luo and J. Xu, PLoS One, 2011, 6, e28707 CrossRef CAS.
- S. Matsumura, Á. Kun, M. Ryckelynck, F. Coldren, A. Szilágyi, F. Jossinet, C. Rick, P. Nghe, E. Szathmáry and A. D. Griffiths, Science, 2016, 354, 1293–1296 CrossRef CAS.
- T. Furubayashi, K. Ueda, Y. Bansho, M. Daisuke, N. Shota, R. Mizuuchi and N. Ichihashi, eLife, 2020, 9, e56038 CrossRef.
- R. Mizuuchi, N. Ichihashi, K. Usui, Y. Kazuta and T. Yomo, ACS Synth. Biol., 2015, 4, 292–298 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, supplementary results, supplementary discussion, Fig. S1–S5, Tables S1–S3. See DOI: 10.1039/d0cc06606k |
| This journal is © The Royal Society of Chemistry 2020 |


