Open Access Article
Open Access ArticleCucurbiturils in nucleic acids research
Ekaterina Y.
Chernikova
 *a and
Daria V.
Berdnikova
*a and
Daria V.
Berdnikova
 *b
*b
aLaboratory of Photoactive Supramolecular Systems, A. N. Nesmeyanov Institute of Organoelement Compounds of Russian Academy of Sciences, Vavilova St. 28, Moscow, Russia. E-mail: chernikova@ineos.ac.ru
bDepartment Chemie-Biologie, Organische Chemie II, Universität Siegen, Adolf-Reichwein-Str. 2, 57076 Siegen, Germany. E-mail: berdnikova@chemie-bio.uni-siegen.de
First published on 10th November 2020
Abstract
During the past ten years, the importance of cucurbiturils (CB[n]) as macrocyclic hosts in supramolecular assemblies with various types of natural and synthetic nucleic acids (NAs) has increased explosively. As a component of such systems, CB[n] macrocycles can play a wide spectrum of roles from drug and gene delivery vehicles to catalysts/inhibitors of biochemical reactions and even building blocks for NA-based materials. The aim of this highlight article is to describe the development of the CB[n] applications in nucleic acids research and to outline the current situation and perspectives of this fascinating synergistic combination of supramolecular chemistry of CB[n] and NAs.
1. Introduction
Nucleic acids (NAs) such as deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) are central molecular scaffolds in almost all living organisms.1 Apart from their fundamental role as carriers of genetic information provided by DNA and viral RNA, nucleic acids are also responsible for intracellular recognition, transport and catalysis that is accomplished by various types of RNA. Along with extensive investigations of the properties and functions of naturally occurring nucleic acids, synthetic and chemically modified oligonucleotides are also attracting significant attention for the development of innovative diagnostic and therapeutic approaches in biology, medicine and materials science.2–5One of the recently emerged strategies towards the regulation of the functions of biomacromolecules is the supramolecular approach, which allows to manipulate the molecular counterparts in a supramolecular assembly and thereby govern the properties of the whole system.6 In general, the main supramolecular interaction occurring in NA-containing systems is the noncovalent association of small molecules and proteins to the binding sites of NA hosts. Introduction of additional host molecules that can be coupled to the supramolecular host–guest interplay of NAs and their ligands provides a range of flexible opportunities to tune the biological activity of both NAs and NA binders. Along these lines, cucurbiturils (CB[n]) acquired increasing popularity due to their good water solubility, outstanding binding properties towards cationic molecules, and good biocompatibility7,8 that makes them especially suitable for applications in the biomedical field. The recognition capabilities of CB[n] as host molecules have been thoroughly investigated and summarized in numerous review articles devoted to the synthesis and binding properties of CB[n]9 and their applications in stimuli responsive systems,10 drug encapsulation and delivery,11–14 fabrication of functional microcapsules,15 molecular recognition.16
During the last few years, the applications of macrocyclic hosts, such as cyclodextrins, cucurbiturils and calixarenes, in functional supramolecular assemblies with biomolecules have been summarized in several review articles.17–21 Thus, biomedical applications of the host–guest systems in non-viral gene delivery were collected by X. Ma and Y. Zhao.17 J. Jayawickramarajah and co-authors summarized recent advances in synthesis and investigation of covalent conjugates of DNA with macrocyclic hosts.18 Some studies on the modification of NAs scaffold through the non-covalent and covalent binding with the macrocyclic molecules were briefly discussed by T. G. Zhan and K. D. Zhang in a book chapter.21 The construction of protein assemblies by host–guest interactions with CB[n] has been comprehensively reviewed in 2017.19,20 However, the particular development and areas of application of CB[n] in nucleic acids research have not been reviewed, so far. To fill this gap, we provide an overview on the research progress (mainly from 2010 to present) in this exciting field on the borderline between biochemistry and supramolecular chemistry. We highlight the design principles and properties of supramolecular assemblies combining NA–ligand and CB[n]–ligand interactions and their potential applications in a range of bio-related fields, such as drug and gene delivery, DNA photocleavage, fluorescent sensing and imaging, electrochemical sensing, regulation of enzymatic processes, NA epigenetics, fabrication of biocompatible hydrogels and polymeric materials. The highlight is divided into sections according to the type and structure of nucleic acids. Some sections are additionally divided into sub-sections considering the particular areas of application. First, the systems involving naturally occurring double- and single-stranded DNA sequences are outlined. Further, the assemblies including non-canonical G-quadruplex DNA structures as well as synthetic and modified NAs are described. Finally, the RNA-based systems are discussed. In a separate section, we highlighted the assemblies comprising nucleic acids and both cucurbituril and cyclodextrin macrocyclic hosts.
2. Double-stranded DNA
2.1. Manipulation of the polyamine-assisted processes on DNA
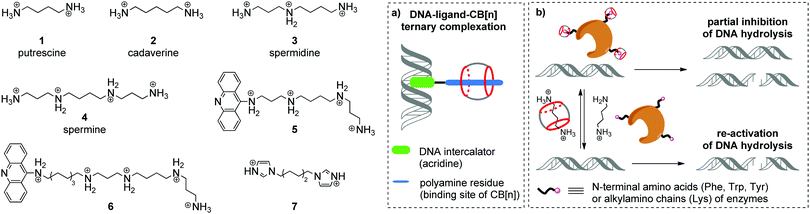
Aliphatic polyamines such as putrescine 1, cadaverine 2, spermidine 3 and spermine 4 (Scheme 1) are an important class of biogenic molecules involved in numerous physiological functions of eukaryotic and prokaryotic cells. Polyamines are essentially involved in the growth, proliferation and differentiation of cells and regulate DNA stability and gene expression.22,23 Under physiological conditions, polyamines get protonated and form corresponding polycations that are attracted to the negatively charged phosphate backbone of DNA and are accommodated in the minor groove due to electrostatic interactions and hydrogen bonding.24 These supramolecular interactions change the morphology of DNA and may stimulate B-to-Z conversion of the DNA double helix. Additionally, binding of polyamines assists the modulation of enzyme-catalyzed DNA relaxation and cleavage processes.25–27 At the same time, polyamines possess strong affinity towards CB[n] and readily form host–guest assemblies.16 The binding constants of polyamine complexes with CB[7] and CB[6] are ca. 1–2 orders and ca. 3–4 orders of magnitude higher, respectively, compared to those for polyamine–DNA assemblies. Along these lines, host–guest interactions with CB[n] represent a powerful tool to control the polyamine-induced transformations of DNA in biological systems by supramolecular complexation.In 2000, K. Kim, E. Nakamura and co-workers provided the first example of ternary supramolecular assemblies between small molecules, cucurbit[6]uril and DNA.28 For this purpose, bifunctional conjugates 5 and 6 comprising a spermine residue tethered to an acridine unit were obtained (Scheme 1). Threading of the spermine fragment into the CB[6] cavity, on one hand, and intercalation of the acridine residue into calf thymus DNA (ct DNA), on the other hand, resulted in formation of a stable ternary complex (Scheme 1a) that was identified by gel electrophoresis and an ethidium bromide displacement assay.28 Additionally, it was shown that complexation with CB[6] reduced the inhibiting activity of free spermine 4 towards the BanII enzyme in the nuclease-induced hydrolysis of pBR322 plasmid DNA.
Further studies on the enzyme-catalyzed reactions of DNA in the presence of CB[6]–polyamine complexes showed that CB[6] can work as a supramolecular switch that enhances or reduces the activity of spermidine 3 and spermine 4 in the corresponding processes.29 Thus, the 3–CB[6] complex was more efficient than free spermidine 3 in accelerating the TopoI-catalyzed relaxation of the supercoiled pBR322 plasmid DNA into its open form (topoisomerization). On the other hand, formation of the 4–CB[6] pseudorotaxane reduced the activity of spermine 4 in the restriction endonuclease BanII-catalysed hydrolysis of pBR322 plasmid DNA. Additionally, it was found that both pseudorotaxanes 3–CB[6] and 4–CB[6] had just a negligible effect on the DNA morphology and on the ratio of supercoiled and open circular forms of DNA, which is in sharp contrast with the behavior of free polyamines.
In 2015, W. Nau and co-workers also utilized the high affinity of CB[n] towards biologically active polyamines to control the enzymatic hydrolysis of DNA (Scheme 1b).30 It was shown that CB[6] and CB[7] macrocycles can inhibit the endonuclease-induced restriction of plasmid and linear forms of pGL3-Basic DNA by interaction with corresponding enzymes. The experimental data collected for various types of endonucleases (KpnI, SacI, XapI) and different DNA forms confirmed the generality of the inhibition effect. A possible mechanism of the enzyme de-activation by CB[n] includes formation of stable complexes between CB[n] and N-terminal amino acids (Phe, Trp, Tyr) or alkylamino chains (Lys) of enzymes. Competitive interactions of CB[n] species with polyamines 1–4 allowed to re-activate the endonucleases towards catalysis of the DNA hydrolysis, thus representing a new approach towards the regulation of biocatalytic processes by the reversible non-covalent interaction with CB[n].
An interesting example of a CB[6]-based host–guest system that mimics the action of a nuclease in a hydrolytic cleavage of DNA was reported by P. Yang and co-workers.31 Thus, a [2]pseudorotaxane comprising CB[6] and 1,6-bis(imidazole-1-yl)hexane 7 (Scheme 1) induced a pronounced DNA strand cleavage in pBR322 plasmid DNA, as shown by agarose gel electrophoresis, whereas compound 7 alone had almost no cleaving activity. According to the proposed mechanism, the hydrolytic cleavage occurred due to a cooperative effect of both CB[6] and 7 within a supramolecular complex. The imidazolium protonated the phosphate residue of a DNA strand. At the same time, the CB[6] carbonyl oxygen (Lewis base) deprotonated a water molecule and promoted its addition to the phosphorus atom, thus inducing phosphate ester cleavage and DNA strand cleavage. However, no follow-up studies were performed to assess the scope and limits of this approach.
As mentioned above, apart from their activity in the enzyme-catalyzed reactions of DNA, polyamines show an excellent ability to stabilize the left-handed helical structure of Z-DNA through linking the neighboring helixes and reducing the net negative charge repulsion between phosphate residues.27 Along with the right-handed duplex DNA forms (A-DNA, B-DNA), the left-handed Z-DNA is also biologically active and is involved in chromatin recombination and remodeling, gene regulation and nucleosome positioning.32 In 2018, X. Zhou and co-workers described an interesting approach to manipulate the B/Z-DNA transition by changing the concentration of free spermine through its complexation with CB[7].33 In this system, CB[7] extracted spermine 4 from Z-DNA by threading onto the central butanediamine moiety of the former, thus inducing the morphological transition of Z-DNA to B-DNA. Subsequent addition of 1-aminoadamantane led to the dissociation of the spermine–CB[7] complex and resulted in the reconversion to Z-DNA.
2.2. Non-viral gene delivery
Gene therapy is a powerful method that allows to deliver therapeutic gene materials into target cells for the treatment of genetic disorders, cardiovascular and neurodegenerative diseases, and cancer without using drugs or surgery.34 There are two directions of gene therapy development that include applications of viral and non-viral (synthetic) gene vectors. Despite the excellent ability to overcome cellular barriers and immune defense mechanisms, viral vectors often possess genotoxicity, immunogenicity, as well as high costs and large-scale production problems. In contrast, non-viral analogues (cationic polymers, biopolymers or nanoparticles) are more friendly and can form tightly packed complexes with various NAs thereby protecting the gene material from destruction and facilitating its penetration across the cell membrane. Nevertheless, non-viral vectors often exhibit reduced transfection efficiency compared to the viral ones that significantly hinders their application as therapeutic agents in clinical practice.35Cationic dendrimers comprising polyamine branches are among the most prominent molecules for the design of non-viral gene delivery systems.35b,c Because of the spherical structure and multiple binding sites, dendrimers can efficiently condense various types of NA molecules into stable assemblies and, therefore, promote the cellular uptake. To overcome some fundamental problems related to cytotoxicity and biocompatibility of gene carriers, supramolecular nanoparticles constructed by host–guest interactions have been developed.36
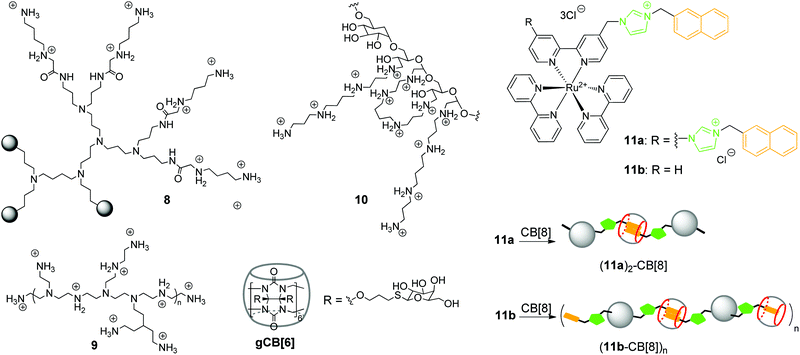
In 2002, K. Kim and co-workers obtained a noncovalent complex of CB[6] with poly(propylenimine) (PPI) dendrimer modified by diaminobutane-terminated groups 8 (Scheme 2).37
Later in 2018, R. Wang and co-workers also utilized the CB[n]-based strategy to reduce the inherent cytotoxicity of the PEI gene vector 9 with a large molecular weight (25 kDa, branched) (Scheme 2).38 For this propose, the supramolecular nanoparticles 9–pDNA and 9–CB[7]–pDNA with an average size of 251 nm and 174 nm, respectively, were prepared. An MTT assay in the absence and presence of CB[7] on three different human cell lines (HEK293, 293T, A549) revealed that CB[7] significantly reduced the cytotoxicity of PEI 9 in a dose-dependent manner. Apoptosis analysis with 293T cells with 9–pDNA and 9–CB[7]–pDNA showed that complexation with CB[7] markedly inhibited the rate of PEI-induced apoptosis. Additionally, it was found that CB[7] significantly alleviated the hemolytic activity of PEI. Confocal fluorescence microscopy and flow cytometry studies of the PEI–Cy5 conjugates in 293T cells clearly showed that complexation with CB[7] had just a minor effect on the cellular uptake. The efficiency of gene delivery of 9–pDNA moderately increased in the presence of CB[7] that was attributed to the improved biocompatibility of PEI upon encapsulation by CB[7].
Another work of K. Kim and co-workers describes a target-sensitive delivery system for the transfer of the genetic material into hepatocytes based on the galactose-linked CB[6] (Scheme 2).39 In this system, dextran functionalized with spermine side chains 10 was responsible for the biodegradability as well as effective condensation of pDNA. Self-assembly of the galactosylated CB[6] (gCB[6]) with the conjugate 10 was provided by formation of the strong inclusion complexes between gCB[6] and spermine residues on the dextran backbone (Scheme 2). It was found that the amount of gCB[6] bound to the polyplex significantly influenced the efficiency of pDNA condensation by reducing the amount of available spermine binding sites for the electrostatic attraction to pDNA. For the same reason, gCB[6] can decrease to some extent the affinity of the polyplex towards a negatively charged cell surface. By systematic alternation of several parameters, including the molar ratios of 10/gCB[6] and spermine nitrogen/pDNA phosphate, the optimized DNA-encapsulating delivery systems were identified. The delivery of the loaded pDNA was tested on the human hepatoma cells HepG2 bearing the galactose-specific asialoglycoprotein receptors (ASGPR). A control competition assay with free galactose and HeLa cells that did not contain ASGPR receptors confirmed that 10–(gCB[6])25 polyplex with the molar ratio of 25 exhibited target-specific interaction with HepG2 cell surface and enhanced gene transfection efficiency.
In addition to commonly used dendrimer–DNA complexes, several studies were focused on the development of novel types of vector delivery systems. Particularly, Y. Liu and co-workers reported a multifunctional supramolecular assembly consisting of the bis-naphthalene-derivatized Ru(bpy)3 complexes 11a and 11b as guests and CB[8] as a host (Scheme 2).40 The encapsulation of the naphthalene units in the CB[8] cavity yielded the water-soluble linear supramolecular dimer (11a)2–CB[8] or polypseudorotaxane (11b–CB[8])n with high molecular weight. As a gene delivery system, the polypseudorotaxane assembly combined several advantages including not only the efficient DNA condensation, but also inhibition of the DNA-cleaving ability of the HindIII (Haemophilus influenzae derivative) enzyme. Additionally, fluorescent properties of the polypseudorotaxane allowed to follow the transfection of the supercoiled pBR322 DNA into cells by confocal fluorescence microscopy.
2.3. Delivery and release of DNA-targeting drugs
The supramolecular design and fabrication of drug carrier systems operating by host–guest interactions with high delivery capacity and selectivity is a promising field of modern medicine and pharmacology. During the last decade, significant progress in the application of CB[n] in drug delivery systems has been achieved that is reflected in several comprehensive review articles.11–14 These reviews highlight the important issues regarding the enhancement of bioavailability and bioactivity of drugs by complexation with CB[n], such as improvement of solubility and stability, reduction of toxicity, protection against aggregation, stimulus-triggered release of included guest and other aspects. However, in view of the DNA-targeting systems, it is necessary to study not only the problems that arise on the route of a drug to a target, but also to understand mechanisms governing the drug release from the molecular carrier with subsequent association with DNA. To the best of our knowledge, up to date, there are only two publications available on the CB[n]-assisted delivery of clinically used drugs to DNA.41,42In 2016, P. Hazra and K. Gavvala with co-workers demonstrated a concept of the stimuli responsive translocation of guest-drug molecules from the CB[7] carrier to DNA.41 This approach was tested for two drugs, cryptolepine (12) (antimalarial and antineoplastic agent) and proflavine (13) (antiseptic drug), whose therapeutical activity arises from their interaction with DNA (Fig. 1). The authors provided some insights into modulation of photophysical properties of the CB[7]-bound drugs and applied these findings for the monitoring of the translocation process. Cryptolepine (12) forms a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion complex with CB[7] (Ka = 4.2 × 104 M−1) and can intercalate between the DNA base pairs (Ka = 2.1 × 104 M−1).41 The similar values of the association constants prevent full dissociation of the 12–CB[7] complex in the presence of DNA. However, introduction of acetylcholine chloride that had higher affinity towards CB[7] (Ka = 7.0 × 105 M−1) resulted in the displacement of cryptolepine molecules from the cavity and their subsequent intercalation into DNA. A similar principle has been applied to the displacement of the DNA binder proflavine (13) from the CB[7] cavity, although quantitative data for the evaluation of the equilibrium processes for the binary systems 13–CB[7] and 13–DNA were not obtained.42 It should be mentioned, however, that for both systems no experimental data confirming applicability of this concept in living cells were provided.
1 inclusion complex with CB[7] (Ka = 4.2 × 104 M−1) and can intercalate between the DNA base pairs (Ka = 2.1 × 104 M−1).41 The similar values of the association constants prevent full dissociation of the 12–CB[7] complex in the presence of DNA. However, introduction of acetylcholine chloride that had higher affinity towards CB[7] (Ka = 7.0 × 105 M−1) resulted in the displacement of cryptolepine molecules from the cavity and their subsequent intercalation into DNA. A similar principle has been applied to the displacement of the DNA binder proflavine (13) from the CB[7] cavity, although quantitative data for the evaluation of the equilibrium processes for the binary systems 13–CB[7] and 13–DNA were not obtained.42 It should be mentioned, however, that for both systems no experimental data confirming applicability of this concept in living cells were provided.
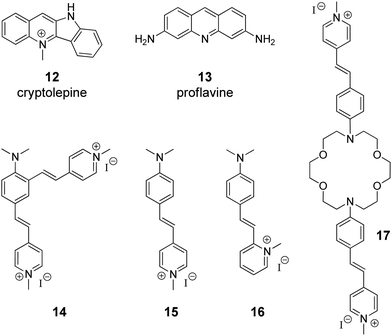 | ||
| Fig. 1 Structures of the DNA-targeting drugs cryptolepine (12) and proflavine (13) and structures of styryl(pyridinium) derivatives 14–17 used for CB[7]-assisted manipulation of the DNA-binding mode. | ||
2.4. CB-Induced changing of the ligand–DNA binding mode
Governing of the DNA-binding mode of small molecules in supramolecular assemblies is an attractive way to control ligand–DNA interactions without additional covalent modification of the binder. Along these lines, several three-component systems were developed, in which cucurbituril acted as a supramolecular control unit of the ligand–DNA binding mode. The first example of this concept for a CB-containing system was provided by S. Sun and co-workers in 2014 on fused bis(styrylpyridinium) dye 14 with two cationic binding sites (Fig. 1).43 Binding studies with ct DNA allowed to suggest that at high ligand-to-DNA ratios compound 14 aggregated on the DNA strand, whereas upon increasing the DNA concentration the aggregation switched to intercalation. Complexation with CB[7] resulted in the formation of the ternary system with one arm of dye 14 intercalated into DNA and the second one encapsulated in the macrocycle cavity. Such a supramolecular arrangement presumably enhanced the intercalative binding mode of 14. Complementary studies on the corresponding monochromophoric dye 15 (minor groove binder) showed that CB[7] extracted ligand 15 from the DNA binding site instead of forming a ternary assembly and, therefore, had no influence on the DNA binding mode of 15. In contrast to compound 15, it was proposed that the related dye 16 switches its DNA binding mode from groove binding to intercalation upon addition of CB[7] (Fig. 1).44 However, no unambiguous evidence for the change of the binding mode in this case was provided.Very recently, another contribution on CB-induced binding mode modulation was made by the Fedorova group.45 In this work, a three-component assembly including ct DNA, CB[7] and a bis(styryl)dye 17 was described (Fig. 1). In the presence of ct DNA, bis(styryl) dye 17 readily formed aggregates that were accommodated in the DNA minor groove. The addition of CB[7] disassembled the aggregates by encapsulation of one styrylpyridinium fragment of 17 whereas the second chromophore remained in the minor groove. In other words, complexation with CB[7] switched the system from aggregation towards the monomer groove binding that was additionally accompanied by increase of the fluorescence intensity of the bound ligand, thus allowing easy monitoring of the system rearrangement.
As can be seen from these examples, complexation with CB[n] mainly changes the ligand–DNA association from a rather heterogeneous unspecific binding (aggregation) to a more ordered one (partial groove binding). We envisage the future development of this approach as a fine switching of binding modes of a single ligand molecule, such as from minor groove binding to intercalation or from minor groove binding to major groove binding and vice versa.
2.5. Supramolecular DNA sensing
Among various approaches towards real-time detection and monitoring of nucleic acids and investigation of their functions inside living cells, fluorescence spectroscopy offers many advantages as a versatile, highly reliable and sensitive method for both in vitro and in vivo assays.46 In general, fluorescent organic ligands may be used as stains for biomolecules, if their emission properties, i.e. emission intensity or emission color, change significantly upon the direct interaction of a ligand with an analyte.47–50 However, due to the large size of CB[n]–ligand complexes, their insertion into the structure of biomolecules is sterically limited. Therefore, CB[n]-based fluorescent sensing is mainly based on competitive host–guest approaches, such as indicator competition assay (ICA) and indicator displacement assay (IDA).51Application of complexes of tricyclic basic dyes acridine orange (18), methylene blue (19), and pyronine Y (20) (Scheme 3) with CB[8] for discrimination of DNA from RNA in solution and HeLa cells was reported by S. Sun co-workers.52 All dyes 18–20 formed supramolecular dimers inside the CB[8] cavity that resulted in significant quenching of their fluorescence in solution in comparison to the free dyes. In the presence of NA, the host–guest complexes dissociated and liberated the dyes that in turn bound to the NA leading to staining. Thus, the acridine orange complex (18)2–CB[8] interacted without selectivity with both ct DNA and S. cerevisiae RNA as indicated by the increase of the fluorescence. No significant effect was observed for the methylene blue complex (19)2–CB[8] in the presence of NAs. At the same time, the fluorescence of the pyronine Y complex (20)2–CB[8] increased only upon addition of DNA, whereas no response was obtained in the presence of RNA. Confocal fluorescent microscopy studies of 18, 20, (18)2–CB[8] and (20)2–CB[8] showed that 18 non-specifically stained the interior of HeLa cells, whereas 20 could not pass across the membrane and was accumulated in the extracellular matrix. It should be noted, however, that the selectivity of the fluorescence response of the dye–CB[8] complexes in the presence of NA in solution and HeLa cells was governed exclusively by the properties of the free dyes. Complexation with CB[8] had no effect on the selectivity and efficiency of staining and was helpful only to suppress autofluorescence of the free dyes and, therefore, to increase a signal-to-noise ratio.
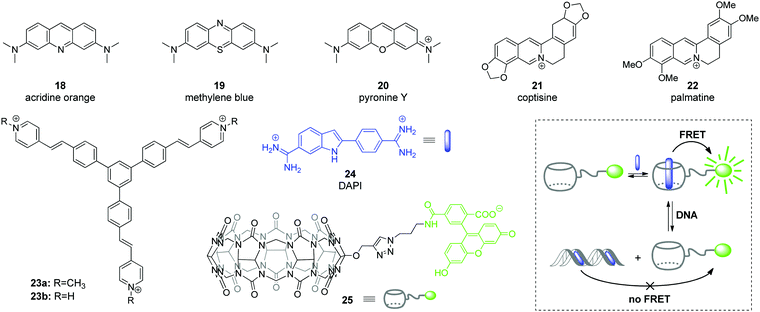 | ||
| Scheme 3 Fluorescent DNA markers 18–23 used for supramolecular DNA sensing as well as structures of the components 24, 25 and operation mode of a ratiometric host–guest FRET sensor for DNA detection. | ||
Abasic DNA sites (apurinic/apyrimidinic (AP) sites) are mutagenic and toxic DNA lesions resulting from the loss of nucleobases.53 AP sites are formed in course of enzymatic repair of damaged DNA or by the influence of external factors. Considering the relevance of AP sites to carcinogenesis, these lesions represent an important analytical and therapeutic target.53,54 In 2018, Y. Shao and co-workers described a supramolecular multicolor DNA sensor for the selective recognition of abasic DNA sites based on the ICA concept with CB[7].55 Two natural alkaloids, namely coptisine (21) and palmatine (22) have been used as fluorescent indicators for AP DNA sites (Scheme 3). Both alkaloids form 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion complexes with CB[7] that is accompanied by pronounced increase of their fluorescence. The direct interaction of coptisine (21) with DNA sequences containing an abasic site (AP-DNAs) results in the increase of the fluorescence intensity without a wavelength shift, irrespective of the opposite base type (purine or pyrimidine). Complexation with CB[7] shifts the intrinsic fluorescence of 21 from 548 nm to 520 nm, thus providing an opportunity to distinguish between CB[7]-bound and AP-DNA-bound ligand by emission color. Additionally, encapsulation of coptisine (21) by CB[7] makes the binding of the guest competitive and it was shown that the AP-DNA competition of 21 with CB[7] occurs to TXT-C/-T other than TXT-A/-G sites. The simultaneous presence of TXT-G, TXT-A, mismatched DNA and fully matched DNA does not interfere with the selective recognition of the TXT-C/-T sites by 21–CB[7] complex with a limit of detection of about 300 nM (S/N = 3). This strategy has been successively expanded to the recognition of the transversed abasic DNA sequences with adenine flanking the AP site and palmatine (22) as indicator. Thus, the addition of AXA-A/-C/-G/-T DNAs to the 22–CB[7] complex causes a red shift of the emission maximum of 18 nm irrespective of the opposite nucleotide structure. At the same time, addition of the fully matched DNA does not alter the emission wavelength, which allows to detect the presence of abasic sites. Overall, these results represent a promising CB-based ICA strategy for the multicolor AP-DNA detection. However, no in vitro studies were performed in this work.
1 inclusion complexes with CB[7] that is accompanied by pronounced increase of their fluorescence. The direct interaction of coptisine (21) with DNA sequences containing an abasic site (AP-DNAs) results in the increase of the fluorescence intensity without a wavelength shift, irrespective of the opposite base type (purine or pyrimidine). Complexation with CB[7] shifts the intrinsic fluorescence of 21 from 548 nm to 520 nm, thus providing an opportunity to distinguish between CB[7]-bound and AP-DNA-bound ligand by emission color. Additionally, encapsulation of coptisine (21) by CB[7] makes the binding of the guest competitive and it was shown that the AP-DNA competition of 21 with CB[7] occurs to TXT-C/-T other than TXT-A/-G sites. The simultaneous presence of TXT-G, TXT-A, mismatched DNA and fully matched DNA does not interfere with the selective recognition of the TXT-C/-T sites by 21–CB[7] complex with a limit of detection of about 300 nM (S/N = 3). This strategy has been successively expanded to the recognition of the transversed abasic DNA sequences with adenine flanking the AP site and palmatine (22) as indicator. Thus, the addition of AXA-A/-C/-G/-T DNAs to the 22–CB[7] complex causes a red shift of the emission maximum of 18 nm irrespective of the opposite nucleotide structure. At the same time, addition of the fully matched DNA does not alter the emission wavelength, which allows to detect the presence of abasic sites. Overall, these results represent a promising CB-based ICA strategy for the multicolor AP-DNA detection. However, no in vitro studies were performed in this work.
The ability of CB[8] to accommodate two guest molecules provided an efficient way to construct two-dimensional frameworks with an aggregation-induced emission (AIE) effect.56 To achieve this, branched guest molecules 23 comprising three styrylpyridinium arms attached to a phenyl core were designed (Scheme 3). The supramolecular components assembled through head-to-tail stacking between the 4-styrylpyridinium fragments inside the CB[8] cavity yielding stable 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest complexes. A highly ordered two-dimensional structure of 23–CB[8] has been confirmed both in solution and as a monolayer by various methods including NMR spectroscopy, dynamic light scattering and microscopic measurements. The formation of the rigid assembly 23–CB[8] was accompanied by a 25-fold enhancement of the fluorescence intensity and a red-shift of the emission maximum from 520 nm to 600 nm. Confocal fluorescence imaging demonstrated that complex 23–CB[8] can easily penetrate the membrane of HeLa cells, presumably through endocytosis or phagocytosis. The color of the intracellular emission is highly sensitive to the wavelength of the excitation light. Thus, the probe 23–CB[8] reveals orange fluorescence in the cytoplasmic region upon excitation with blue light (BP 460–495 nm, U-FUN) whereas the UV light excitation (BP 340–390 nm, U-FUW) simultaneously lights up the green emission in the cell nuclei along with orange emission in the cytoplasmic region. A possible reason for the green staining of the nuclei arises from a dynamic self-assembly process of 23–CB[8] or competition process between 23–CB[8] and DNA. Overall, supramolecular AIE probe 23–CB[8] can be considered as a new class of fluorescent probes for the intracellular DNA imaging.
1 host–guest complexes. A highly ordered two-dimensional structure of 23–CB[8] has been confirmed both in solution and as a monolayer by various methods including NMR spectroscopy, dynamic light scattering and microscopic measurements. The formation of the rigid assembly 23–CB[8] was accompanied by a 25-fold enhancement of the fluorescence intensity and a red-shift of the emission maximum from 520 nm to 600 nm. Confocal fluorescence imaging demonstrated that complex 23–CB[8] can easily penetrate the membrane of HeLa cells, presumably through endocytosis or phagocytosis. The color of the intracellular emission is highly sensitive to the wavelength of the excitation light. Thus, the probe 23–CB[8] reveals orange fluorescence in the cytoplasmic region upon excitation with blue light (BP 460–495 nm, U-FUN) whereas the UV light excitation (BP 340–390 nm, U-FUW) simultaneously lights up the green emission in the cell nuclei along with orange emission in the cytoplasmic region. A possible reason for the green staining of the nuclei arises from a dynamic self-assembly process of 23–CB[8] or competition process between 23–CB[8] and DNA. Overall, supramolecular AIE probe 23–CB[8] can be considered as a new class of fluorescent probes for the intracellular DNA imaging.
In 2019, W. Nau and co-workers applied the IDA concept to design a ratiometric host–guest DNA sensor based on Förster resonance energy transfer (FRET) process.57 In this system, the CB[7] host was labeled with an acceptor fluorophore carboxyfluorescein to give conjugate 25 (Scheme 3). The carboxyfluorescein residue was equatorially attached at the outer side of the macrocycle, so that the interior of conjugate 25 remained available for guest molecules. A well-known blue-fluorescent DNA stain 4′,6-diamidino-2-phenylindole (DAPI) 24 was chosen as a FRET acceptor guest. The host–guest complexation ensured the close proximity of the donor and acceptor dyes and prompted the highly efficient (>99%) intermolecular FRET process in the 24–25 system. The host–guest FRET pair was applied for the quantitative sensing of salmon sperm DNA. Upon gradual addition of double-stranded DNA (dsDNA) to the solution containing the pre-assembled FRET pair (0.5 μM 24 and 1 μM 25) a pronounced decrease of carboxyfluorescein fluorescence at 520 nm (FRET acceptor) along with enhancement of DAPI fluorescence at 450 nm (FRET donor) was observed. This clearly indicated the relocation of the guest 24 from the macrocyclic cavity of 25 to the DNA binding pocket. Notably, the ratio of the fluorescence intensities at two emission wavelengths (λem = 420 nm and λem = 520 nm) depended linearly on the DNA concentration thus allowing the quantitative detection of dsDNA from a picomolar range up to 20 mg mL−1, with a lower detection limit of 60 ng mL−1. Notably, the reliable DNA detection range provided by the CB[7]-based ratiometric chemosensor exceeded that of the commercial DNA staining dye SYBR Green I.
Overall, these inspiring examples of the CB-assisted DNA sensing demonstrate a potential of the supramolecular approach for increasing sensitivity and selectivity of DNA detection both in vitro and in vivo. We are surprised, however, that the CB-based strategies have never been applied for the sensing of quadruplex DNA and other non-canonical nucleic acid structures.
2.6. Photoresponsive systems
Viologen (4,4′-bipyridinium, V2+) 26 and its derivatives are well-known cationic redox active guests that form stable inclusion complexes with cucurbiturils.58 Viologens are extensively utilized as good electron acceptors in the electron transfer (ET) reactions in multi-component systems. Moreover, the viologen radical cation (V+˙) readily reduces oxygen in aqueous solution and generates reactive oxygen species (ROS), such as the superoxide anion-radical (O2−˙), singlet oxygen (1O2), and the hydroxyl radical (˙OH).59 In view of these properties, X. Peng and co-workers used the combination of the methylviologen residue with aromatic photosensitizers and complexation with CB[8] to construct several systems for DNA photocleavage.60–62 Thus, a stable ternary 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex was obtained by association of the donor–acceptor dyad comprising methylviologen 26 and phenothiazine 27 with CB[8] (Scheme 4).60 Upon light excitation, phenothiazine 27 transfers an electron to methylviologen inside the CB[8] cavity generating the corresponding radical ions 27+˙ and V+˙. The phenothiazine radical cation is subsequently ejected from CB[8] due to predominant formation of a stable viologen radical dimer (V+˙)2–CB[8] inside the cavity. Such encapsulation of the radical dimer significantly inhibits the charge recombination between 27+˙ and V+˙, thus extending the lifetime of V+˙ and favoring the generation of ROS. Given to this effect, the ternary assembly 26–27–CB[8] exhibits pronounced photocleavage of super-coiled pBR322 plasmid DNA upon irradiation with white light.
1 complex was obtained by association of the donor–acceptor dyad comprising methylviologen 26 and phenothiazine 27 with CB[8] (Scheme 4).60 Upon light excitation, phenothiazine 27 transfers an electron to methylviologen inside the CB[8] cavity generating the corresponding radical ions 27+˙ and V+˙. The phenothiazine radical cation is subsequently ejected from CB[8] due to predominant formation of a stable viologen radical dimer (V+˙)2–CB[8] inside the cavity. Such encapsulation of the radical dimer significantly inhibits the charge recombination between 27+˙ and V+˙, thus extending the lifetime of V+˙ and favoring the generation of ROS. Given to this effect, the ternary assembly 26–27–CB[8] exhibits pronounced photocleavage of super-coiled pBR322 plasmid DNA upon irradiation with white light.
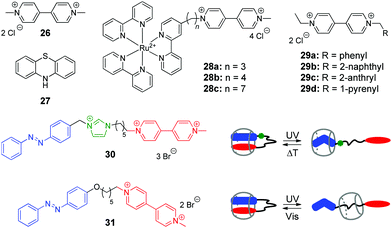 | ||
| Scheme 4 Structures of the photoresponsive redox guests 26–31 applied in the CB-based systems for DNA photocleavage. | ||
Two more systems developed by the Peng group represent covalent conjugates of viologens with Ru(bpy)32+ complexes 28a–28c61 and aromatic hydrocarbons 29a–29d (Scheme 4).62 Apart from light-induced generation of ROS by the viologen radical, covalent attachment of Ru(bpy)32+ in 28a–28c allows to recruit an additional mechanism of the DNA damage by the generation of the oxidized Ru3+ through the photoinduced intramolecular ET from Ru2+ to the viologen residue. Consequently, Ru3+ oxidizes guanine nucleobases and photocleave DNA without generating ROS. The increase of the alkyl spacer length in 28a–28c allows to improve the photocleavage efficiency by extending the lifetime of the charge transfer state. Encapsulation of the viologen moiety by CB[8] in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex further inhibits the intramolecular electron back transfer in the conjugates 28a–28c resulting in a relatively long-living charge transfer oxidation state (t ∼ 2 ms), which improves the photocleavage efficiency.61
1 complex further inhibits the intramolecular electron back transfer in the conjugates 28a–28c resulting in a relatively long-living charge transfer oxidation state (t ∼ 2 ms), which improves the photocleavage efficiency.61
Conjugates of viologen with aromatic hydrocarbons 29a–29d (Scheme 4) show a different behaviour.62 Thus, 2-anthryl 29c and 1-pyrenyl 29d derivatives bind to ct DNA through a mixed binding mode and both of them induce pronounced cleavage of plasmid pBR322 DNA upon irradiation with a xenon lamp. At the same time, phenyl 29a and 2-naphthyl 29b derivatives do not interact with DNA. However, upon complexation with CB[8] yielding 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes, all compounds 29a–29d exhibit efficient DNA photocleavage. As discussed above, the reason of this effect is encapsulation-induced inhibition of the intramolecular backwards ET in the conjugates that extends the lifetime of the charge separated excited state. Overall, X. Peng and colleagues clearly demonstrated the potential application of CB-based host–guest chemistry for DNA photocleavage.
1 complexes, all compounds 29a–29d exhibit efficient DNA photocleavage. As discussed above, the reason of this effect is encapsulation-induced inhibition of the intramolecular backwards ET in the conjugates that extends the lifetime of the charge separated excited state. Overall, X. Peng and colleagues clearly demonstrated the potential application of CB-based host–guest chemistry for DNA photocleavage.
In 2014, Y. Liu and co-workers synthesized and investigated host–guest complexes of photoresponsive azobenzene–viologen conjugates 30 and 31 (Scheme 4).63 Initially, upon association with CB[8], the conjugate molecules adopt a folded conformation with both E-azobenzene and viologen moieties immersed in the macrocycle cavity forming a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion complex. Irradiation of 30 and 31 at 365 nm results in the E–Z isomerization of the azobenzene fragment followed by the ejection of the viologen residue from the cavity and formation of pseudorotaxanes with different positioning of CB[8]. Thermal or photochemical backwards Z–E-isomerization of azobenzene restores the initial loop-shaped structures of the E-30–CB[8] and E-31–CB[8]. Most notably, the E-configured complexes show a remarkable DNA condensation effect on pBR322 plasmid DNA. In contrast, the pseudorotaxane structures Z-30–CB[8] and Z-31–CB[8] possess no DNA condensation ability but rather display significant DNA cleaving properties upon UV light irradiation. This can be explained by liberation of the viologen residue from the cavity upon isomerization of azobenzene and consequent UV-induced generation of ROS by viologen radical in solution. Overall, host–guest interactions of conjugates 30 and 31 with CB[8] represent an example of the photoswitching between DNA-condensing and DNA-cleaving properties.
1 inclusion complex. Irradiation of 30 and 31 at 365 nm results in the E–Z isomerization of the azobenzene fragment followed by the ejection of the viologen residue from the cavity and formation of pseudorotaxanes with different positioning of CB[8]. Thermal or photochemical backwards Z–E-isomerization of azobenzene restores the initial loop-shaped structures of the E-30–CB[8] and E-31–CB[8]. Most notably, the E-configured complexes show a remarkable DNA condensation effect on pBR322 plasmid DNA. In contrast, the pseudorotaxane structures Z-30–CB[8] and Z-31–CB[8] possess no DNA condensation ability but rather display significant DNA cleaving properties upon UV light irradiation. This can be explained by liberation of the viologen residue from the cavity upon isomerization of azobenzene and consequent UV-induced generation of ROS by viologen radical in solution. Overall, host–guest interactions of conjugates 30 and 31 with CB[8] represent an example of the photoswitching between DNA-condensing and DNA-cleaving properties.
A conceptually different application of a photoreaction in the host–guest system involving CB[7], DNA and hydroxypropyl-β-cyclodextrin (HP-β-CD) has been provided by our groups in 2015 and is described in detail in Section 2.7.
2.7. Functional systems comprising a cucurbituril/cyclodextrin pair
Cyclodextrins (CD) represent another important class of macrocyclic host molecules that are widely applied in biochemistry, pharmacy and medicine due to their low toxicity, biocompatibility and chemical stability.64,65 In contrast to cucurbiturils with their affinity towards cationic guests, cyclodextrins tend to encapsulate preferentially neutral hydrophobic guests. Hence, simultaneous presence of both macrocycles in one supramolecular system allows to enhance its functionality due to the interplay of orthogonal binding preferences of the hosts. In this section, we describe the examples of assemblies including both CD and CB[n] macrocycles and NAs.In 2015, our groups have developed a supramolecular five-component cascade that allowed to control ligand–DNA interactions in real time by light.66 Such a DNA-binding/displacement system has been realized based on a fine balance between different host–guest interactions of the precursor 32 and the photoinduced intercalator 33 with HP-β-CD, dsDNA and CB[7] (Scheme 5). Starting from the encapsulation of precursor 32 by HP-β-CD to provide the delivery of the substrate in aqueous solution, the cascade continues with photoinduced in situ formation of the intercalator 33, its release from the cyclodextrin host and subsequent association with DNA. The final step of the cascade is the removal of 33 from the DNA binding site by CB[7].
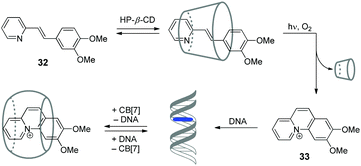 | ||
| Scheme 5 Association and redistribution equilibria of the precursor 32 and photocyclization product 33 in the presence of HP-β-CD, CB[7] and DNA. | ||
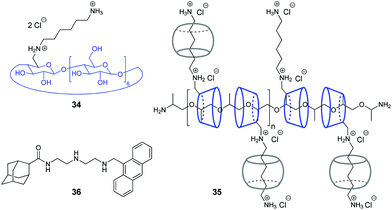
Y. Liu and co-workers have shown that assemblies comprising a CB/CD pair can efficiently provide the condensation of DNA in a controllable way.67,68 Condensation of NAs represents one of the key steps in gene therapy that allows to prevent rapid degradation of oligonucleotides by nucleases.69 Thus, in 2007, the authors constructed a pseudopolyrotaxane 35 by threading poly(propylene glycol) diamine (PPG4000) through the modified β-cyclodextrins 34 and subsequent assembly with different amounts of CB[6] to give the complexes with various content of the CB[6]-bound alkylammonium pendants (0, 20, 40, 70, 100%) (Fig. 2).67 It was found, that the resulting supramolecular assemblies can efficiently provide condensation of pEGFP-C2 plasmid DNA consisting of circular supercoiled DNA (form I) and relaxed circular DNA (form II) into nanoparticles. Depending on the percentage of CB[6], the DNA condensation efficiency of pseudopolyrotaxanes 35 varied nonlinearly reaching the highest value when 70% of the cationic β-CDs in the supramolecular strand were associated with CB[6].
In 2016, the same group developed a more simple, yet efficient DNA condensation system including the CB[6]/β-CD pair.68 Thus, a ternary supramolecular complex was obtained by self-assembly of modified β-cyclodextrin 34, CB[6] and anthryl adamantane 36 (Fig. 2). Specifically, like in the case of pseudopolyrotaxane 35, CB[6] associated with the cationic alkylammonium tail of β-CD 34, whereas the adamantane residue of 36 formed the inclusion complex with the cyclodextrin cavity. It was found that the synergistic effect of the components in the ternary assembly resulted in pronounced condensation of pBR322 plasmid DNA into uniform spherical nanoparticles. At the same time, no obvious condensation effect was observed upon interaction of individual compounds or binary inclusion complexes with DNA, except for the 36–CB[6] complex that slightly induced DNA-condensation. In 2016, the Liu group also described a supramolecular assembly comprising a CB[6]/β-CD pair for the condensation and delivery of siRNA to cancer cells. This system is described in detail in Section 5.
Overall, the supramolecular systems highlighted above mainly include double-helix DNAs obtained from natural sources, such as calf thymus DNA, salmon testes DNA and various types of plasmid DNAs. However, the application of CB[n]-containing assemblies is by no means limited to the canonical double-stranded forms of DNA. Below we collected examples of the CB[n]-based supramolecular systems with G-quadruplex DNA, synthetic and modified DNA oligonucleotides and RNA.
3. G-Quadruplex DNA
G-Quadruplex DNA (G4-DNA) represents one of the non-canonical types of DNA that is formed in G-rich DNA sequences upon stacking of two or more guanine quartets.70 In mammalian cells, quadruplex DNA structures participate in several important processes, such as gene suppression, senescence and initiation of the cellular response to DNA damage.71 Due to the essential biological role of G4-DNA, control of its folding and functions by interaction with exogenous ligands offers new possibilities for biology and medicine. Along these lines, a CB-based approach has been developed that allows to manipulate folding of G4-DNA by supramolecular assembly.In 2017, X. Zhou and co-workers described the first example of the CB-assisted supramolecular control of G-quadruplex DNA formation.72 In this study, the quadruplex-forming oligonucleotide 5′-TTAGGGTTAGGGTTAGGGTTAGGG-3′ (H24) from the human telomere sequence was used. It was shown that azobenzene derivatives 37 and 38 induced folding of the single-stranded H24 oligonucleotide into a parallel G-quadruplex structure (Scheme 6). Addition of CB[7] resulted in the disassembly of the 37–DNA complexes and subsequent unfolding of the quadruplex due to the translocation of dye 37 to the CB[7] cavity. Introduction of a competitive CB-binder 1-aminoadamantane led to the displacement of ligand 37 from the CB[7] cavity that consequently resulted in the refolding of the G4 structure of the 37–DNA complex. Therefore, the supramolecular interplay between the DNA-binder, CB[7] and 1-aminoadamantane provided a general principle for the reversible supramolecular switching of the G-quadruplex structure. Notably, the folding/unfolding of the G4-DNA can be performed reversibly over several cycles by repetitive addition of CB[7] and 1-aminoadamantane. It should be noted that in contrast to the piperidine-substituted azobenzene 37, the trimethylammonium-substituted azobenzene 38 failed to form a stable complex with CB[7], thus making the supramolecular control of the G4 folding impossible. Additionally, the described supramolecular switching was applied to manipulate the enzymatic reaction between thrombin and fibrinogen with a specially designed thrombin inhibitor consisting of two thrombin-binding aptamers connected by H24 DNA sequence. Folding/unfolding of the linker into the quadruplex structure switched the activity of the inhibitor and, therefore, regulated the thrombin-catalysed conversion of fibrinogen to fibrin.
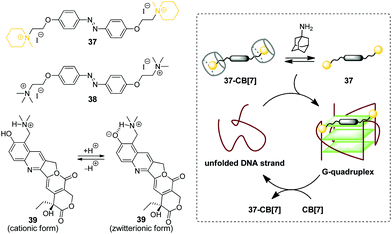 | ||
| Scheme 6 Governing the folding of G-quadruplex DNA by reversible association of DNA binders 37–39 with CB[7]. | ||
In 2018, P. Hazra and co-workers applied a similar concept for supramolecular G-quadruplex manipulation.73 Like in the previous study, the authors used human telomeric DNA H24 and CB[7] as a molecular container. The antitumor drug topotecan (39) was chosen as a mediator of the quadruplex folding (Scheme 6). Complexation of 39 with H24 DNA induced the formation of the G-quadruplex, whereas addition of CB[7] led to the translocation of the ligand to the CB[7] cavity and subsequent quadruplex unfolding. Notably, upon addition of CB[7], the fluorescence of the ligand changed from green to violet due to the transition of topotecan (39) from the zwitterionic to cationic form upon translocation from the DNA binding site to the CB[7] cavity because of the CB-induced pKa shift. Therefore, this system offered an advantage to follow the DNA structural transformations by fluorescence spectroscopy. Overall, these two studies provided a proof-of-principle for the supramolecular control of the G4-DNA structure by competitive interaction of the quadruplex-binding ligands with CB[7]. However, further development of this approach is required to reveal its potential for the practical applications.
4. Synthetic and modified DNA
In this section, we collected the systems comprising epigenetically modified DNAs as well as artificially synthesized sequences that do not have naturally occurring analogues. Some of these studies are focused on the biological functions of DNA, whereas in other ones DNA is utilized as a supramolecular building block without a reference to its biological role.4.1. CB-Assisted supramolecular sensing and imaging
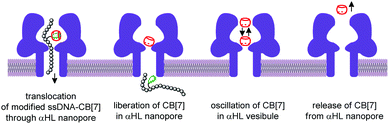
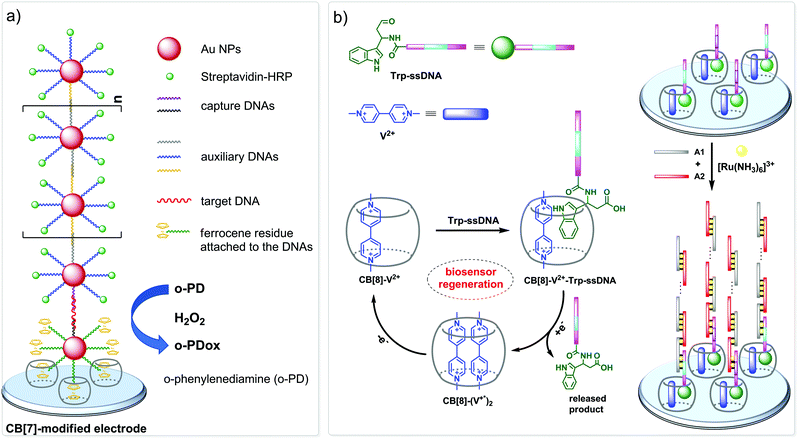
The nanopore sensing is a powerful analytical method that relies on monitoring of ionic current fluctuations produced by the interactions between the analyte and the binding sites of nanopores.74 The frequency of current signatures is used to identify the analyte and quantify its concentration. Since 2015, H.-C. Wu and co-workers are developing a versatile method for nanopore sensing of biologically related analytes based on DNA–CB[7] host–guest probes.75–83 In the method developed by H.-C. Wu and co-workers, introduction of the analyte leads to the release or in situ formation of the single-stranded DNA (ssDNA) bearing a ferrocene–CB[7] or adamantane–CB[7] complex, referred to as a DNA probe. Threading of the DNA probe through the α-hemolysin (αHL) nanopore results in the dissociation of the host–guest complex and liberation of CB[7] (Fig. 3). Subsequent trapping and oscillation of free CB[7] in the vestibule of αHL produces a unique pattern of the current signature events that endows the detection with very high confidence at the single molecule level. The application of this strategy allowed to detect a large range of analytes of different size and nature, such as vascular endothelial growth factor (VEGF), thrombin, cocaine,75 ssDNA, mono-/multivalent antibodies,76 microRNA,77 and cancer biomarkers,78,79 with high levels of sensitivity. Selective epigenetic modification of 5-methylcytosine, 5-hydroxymethylcytosine80 and 8-oxo-2′-deoxyguanosine81 by attachment of the ferrocene–CB[7]80 or adamantane–CB[7]81 complexes allowed to apply the CB-based αHL nanopore sensing to detect the presence of these nucleobases in single-stranded DNA oligonucleotides. Recently, the method was used for simultaneous monitoring of protease activities and local pH values.82 Finally, this approach has been applied to determine the binding constants of the host–guest interactions of CB[6] and CB[7] with small molecules and proteins.83 However, the resulting binding constants were not fully consistent with the previously reported data making this method suitable for qualitative analysis only.CB-Based assemblies with NAs are also useful for the construction of electrochemical sensing systems for biomolecules, which gain increasing popularity due to low costs, reliability, simplicity of exploitation and high sensitivity.84 Modification of biosensors by CB[n] anchoring offers new opportunities to generate stimulus-responsive surfaces in a reversible manner.85 Up to date, two examples of CB[n]-functionalized surfaces as analytical tools for prostate and breast cancer diagnostics have been reported. In 2016, F. Zhang and co-workers developed an electrochemical sensing platform on the basis of a CB[7]-modified electrode for the detection of breast cancer susceptibility gene (BRCA) DNA (Scheme 7a).86 The recognition of the targeted DNA sequences occurred in two steps: (i) strong host–guest complexation between CB[7] and a ferrocene residue attached to the DNA on the gold nanospheres (FcNS); (ii) homogenous hybridization of a targeted DNA with FcNS and horseradish peroxidase-labeled DNA/Au nanospheres (HRPNS) concatamers. When brought together, FcNS and HRPNS concatamers supplied the gain of the electrochemical signal. The sensitivity of the platform towards target BRCA DNA increased with decreasing number of the mismatched bases reaching the highest level of signal when DNA strands were fully complementary. The concentration range of the BRCA DNA detection was 1 × 10−7 M – 5 × 10−11 M with the detection limit of 25 pM (S/N = 3). Importantly, the responsiveness of the CB[7]–FcNS host–guest system to the pH changes allowed to achieve the moderate recyclability of the sensing platform (80% after 5 cycles).
Later on, host–guest recognition properties of CB[8] were used to construct a biosensor with a remarkable response towards microRNA-182-5p, a prostate cancer biomarker.87 Thus, two ssDNAs complementary to the target miRNA were labelled with tryptophane residues (Scheme 7b). A charge-transfer complex between CB[8] and methyl viologen (V2+) immobilized on the electrode surface was capable to trap the tryptophane-labelled ssDNAs Trp-ssDNA to form strong heteroternary complex CB[8]–V2+–ssDNA. At last, to trigger an electrochemical signal, the electrode with the preorganized heteroternary complex was modified by hybridization with ssDNA and introduction of electroactive species [Ru(NH3)6]3+. A redox stimulus led to a one-electron reduction of methyl viologen and expulsion of Trp-ssDNA outside the CB[8] cavity. The reversible assembly and release was possible over 10 cycles, demonstrating excellent stability and regeneration rate. Additionally, the screening of cancer-specific biomarkers showed that this biosensor exhibited high selectivity towards miRNA-182-5p over the interfering species miRNA-21, miRNA-141, and miRNA-155.
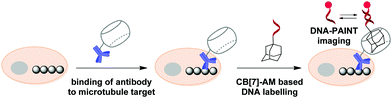
In 2018, S. Agasti and co-workers described an interesting approach towards biorthogonal fluorescent imaging in cells and tissues based on highly selective host–guest interaction between CB[7] and 1-aminoadamantane AM.88 In particular, the developed method was applied to realize high density DNA labelling for PAINT-based (Points Accumulation for Imaging in Nanoscale Topography) super-resolution imaging of fixed HeLa cells (Scheme 8). To achieve this, the methanol-fixed cells were treated with CB[7]-conjugated antibodies to target cellular microtubules. Subsequently, an AM-derivatized 11nt-ssDNA strand was incubated with CB[7]-tagged cells for specific DNA immobilization on microtubules through CB[7]–AM interactions. Finally, the complementary ATTO655-conjugated 11nt-DNA was incubated with the cells for the DNA-PAINT imaging. Single molecule blinking was recorded using a 642 nm excitation laser line. As a result, a significant increase in resolution as compared to the diffraction-limited image was observed by visualizing a dense microtubule region. Notably, the CB[7]–aminoadamantane system demonstrated exceptional serum stability and maintained high coupling efficiency even after incubation of the components at 37 °C for 16 h. Overall, the described CB[7] noncovalent label provided a synthetic alternative to the widely applied biotin–streptavidin system with some more advanced features, such as increased imaging resolution.
4.2. Supramolecular regulation and mimicking of biological processes
In 2017, a CB-based supramolecular approach towards reversible control of the enzymatic reactions at 5-formylcytosine (5fC) sites in DNA has been developed.89 The 5fC nucleotide 40 plays an important role in active DNA demethylation and acts as an epigenetic mark in mammals, thus representing an attractive chemical target for the intervention in biological processes. The adamantane derivative bearing a hydroxylamine group AD has been developed as a chemical tag providing quantitative and selective labelling of 5fC sites (Scheme 9). CB[7] selectively binds to the adamantyl residue of the modified 5fC-AD nucleotide 41, however this host–guest interaction does not disrupt the natural hydrogen bonding of nucleobases in DNA duplex. Based on these findings, complexation with CB[7] has been applied for the inhibition of several 5fC-targeting biochemical reactions, such as restriction endonuclease digestion, DNA polymerase elongation, and polymerase chain reaction. The reason for these effects is the steric bulk of CB[7] that prevents enzymes from binding to the DNA substrate to catalyse the corresponding processes. Introduction of a competitive binder 1-aminoadamantane AM (Scheme 9) allows to remove the macrocycle from the 5fC-AD site of 41 and re-activate the corresponding enzymatic reactions, thus providing a reversible supramolecular control. Overall, this study outlines a range of potential applications of CB-based systems in epigenetics.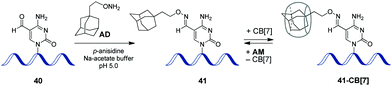 | ||
| Scheme 9 Epigenetic labelling of 5-formylcytosine (5fC) sites in DNA with adamantyl residue and complexation with CB[7]. | ||
The first example of a covalent cucurbituril–DNA conjugate has been provided in 2017 by J. Jayawickramarajah and L. Issacs with co-workers.90 Based on self-assembling DNA-small molecule chimeras, the authors constructed a synthetic transducer that selectively converted binding of adenosine triphosphate (ATP) (biological input) into displacement of a model inhibitor of carbonic anhydrase II (CA-II) 42 from the CB[7] cavity (functional output) (Scheme 10). To design the DNA chimeras, an ATP-binding DNA aptamer was split in two halves.
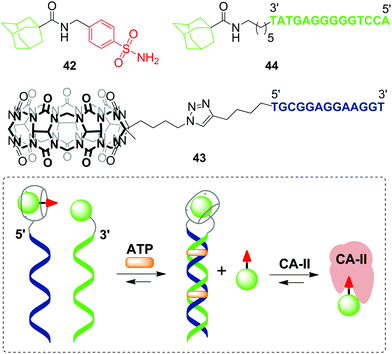 | ||
| Scheme 10 A synthetic transducer based on a split DNA aptamer that converts an ATP binding input into release of the CA-II inhibitor. | ||
Very recently, a supramolecular platform that mimics a cooperative function of the natural transcription factor pair has been developed based on the combination of DNA minor groove binders pyrrole–imidazole polyamides (PIP) and a host–guest pair CB[7]–adamantane (Fig. 4).91 Four types of DNA templates comprising a Widom 601 sequence and PIP-binding sites were constructed and reconstituted to form nucleosomes. Cooperative binding of the host 45 and guest conjugates 46–49 to DNA along with a CB[7]–adamantane complexation increased the DNA recognition length of PIP and allowed versatile binding modes. Additional modification of the host or guest conjugates with an epigenetic modulator (bromodomain inhibitor, not shown on Fig. 4) provided the system with the ability to enhance the level of histone acetylation in nucleosomes.
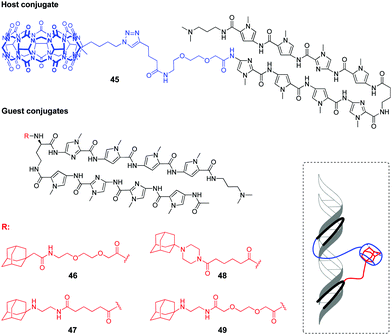 | ||
| Fig. 4 Structures of the host and guest conjugates mimicking a natural DNA transcription factor pair and a scheme of complexation. | ||
4.3. Molecular platforms for mechanistical studies of host–guest interactions
Very recently, O. Seitz and co-workers performed a systematic study to evaluate the distance limits of bivalent interactions with both experimental measurements and statistical mechanics analysis.92 As an experimental model, the authors used the host–guest interactions between Cy5-labelled CB[7]–DNA conjugates and Cy3-labelled adamantane–DNA conjugates. CB[7] or adamantane derivatives were attached to DNA duplexes with different spacing between the units (21, 42, 63, 84 or 105 nucleotides) forming the corresponding CB[7] arrays 50 (host arrays) or adamantane arrays (guest arrays) 51, 52 (Fig. 5). Upon the interaction of the host 50 and guest 51, 52 arrays, the occurrence of FRET in the Cy3–Cy5 dye pair revealed complexation through CB[7]–adamantane interactions and allowed to determine the dissociation constants. Combination of the experimental data and theoretical modelling led to the following generalizing conclusions on the bivalency limitations: (i) the distance between recognition modules, (ii) the flexibility of the scaffold and (iii) dissociation constant of the monovalent interaction. The usage of two adamantane guests with different binding affinity towards CB[7] (Fig. 5) allowed to deduce that low-affinity receptor–ligand interactions reduce the concentration threshold for switching from bivalent interactions to cross-linking. Overall, these results increase the understanding of the factors that are necessary to design bivalent interactive blocks for biological systems.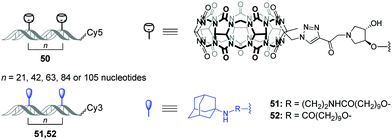 | ||
| Fig. 5 Structures of CB[7]–DNA and adamantane–DNA conjugates 50–52 applied for mechanistic studies of the bivalent non-covalent interactions. | ||
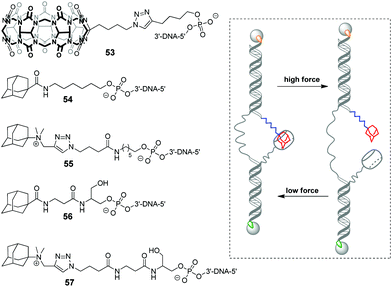
In 2019, a DNA-based platform to investigate the mechanical strength of the host–guest interactions in the CB[7]–adamantane system was reported.93 The platform comprised two dsDNA handles, which were tethered to CB[7] 53 and different adamantane derivatives 54–57, respectively (Scheme 11). The dsDNA handles were attached to two optically trapped beads through streptavidin–biotin and digoxigenin–antibody interactions and they were also linked to each other by a polythymidine (T50) spacer. A controllable increase of the distance between the beads allowed to detect a rupture event indicating the disassembly of the host–guest complex. It was found that a positively charged adamantane guest possessed higher mechanical stability (49 pN) than a neutral one (44 pN), which was consistent with the reported binding preferences of CB[7]. Surprisingly, it was additionally found that a hexyl group adjacent to the adamantane residue in 54 and 55 served as a chaperone assisting the formation of the CB[7]–adamantane complex.
4.4. Supramolecular nanomaterials comprising CB[n] and DNA
Although the majority of applications of CB[n] in NA research is focused on biological functions of DNA, there are a few examples of systems comprising DNA as a building block with programmable structure and hybridization. Thus, in 2015, a representative example of all-supramolecular double network hydrogel formed by combination of CB[8]- and DNA-based hydrogels was described.94 In general, double network hydrogels consist of two independent hydrogels with contrasting properties that interpenetrate each other providing the resulting material with combined physical properties of the individual components. In this particular case, the double network was constructed by “one pot” mixing of four building blocks, namely a Y-shaped DNA oligomer, a DNA linker, phenylalanine-derivatized carboxymethyl cellulose (CMC-phe) and CB[8] (Scheme 12a). The complementary sequences of the DNA Y-scaffold and DNA linker selectively hybridised to form the first crosslinked network. In parallel, CMC-phe and CB[8] also selectively self-assembled due to the host–guest interactions between phenylalanine and CB[8] yielding the second crosslinked network. Because of its structure, the double network hydrogel had increased mechanical properties and thermal stability. Dynamic supramolecular interactions between the building blocks provided the material with shear-thinning and thixotropic properties. Additionally, the hydrogel possessed a full biodegradability profile as every single network could be targetedly cleaved by the corresponding enzymes, such as nucleases and cellulases. Owing to these properties, this supramolecular hydrogel has a great potential as injectable soft material for in vivo applications.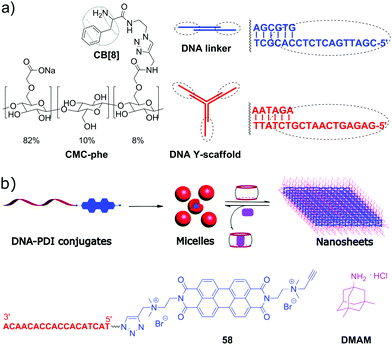 | ||
| Scheme 12 (a) Molecular building blocks self-assembling into a double network hydrogel based on the interpenetrating CB[8]- and DNA-based networks; (b) structure of the ssDNA–perylenebisdiimide conjugates, schematic representation of their self-assembly into spherical micelles and rearrangement into nanosheets upon addition of CB[10]. Adapted from ref. 95, copyright RSC. | ||
In 2019, S. Liu and co-workers realized a CB-based supramolecular tuning of the nanostructures formed by ssDNA 18-mer–perylenebisdiimide (PDI) conjugates 58 (Scheme 12b).95 Due to the strong π–π stacking between PDI units, conjugates 58 obtained by tailoring of a PDI moiety to a DNA oligonucleotide self-assembled in aqueous solution into spherical micelles with a diameter of about 24 nm, according to TEM data. Addition of CB[8] resulted in complete disassembly of the micelles due to encapsulation of the PDI fragments in CB[8]. If CB[10] was added instead of CB[8], the micelles rearranged into crystalline 2D nanosheets with sizes up to the micrometer scale (Scheme 12b). Such drastic morphological changes were driven by both π–π stacking of PDI units and encapsulation of multiple PDI units inside the CB[10] cavity. Introduction of the competitive guest 3,5-dimethyl-1-amino-adamantane DMAM with a stronger binding affinity towards CB[10] resulted in the disassembly of the nanosheets and recovery of the spherical micelles of 58. Additionally, controlled attachment of Au-nanoparticles to the 58–CB[10] nanosheets was demonstrated pointing to the potential applicability of these nanoarchitectures as addressable templates for loading functional molecules or drugs.
Very recently, the same group developed stimuli-responsive DNA-based micelles constructed of CB[7]-modified DNA oligonucleotides 59 and two types of hydrophobic guests such as dendrimer-appended ferrocene 60 and adamantane derivative 61 functionalized with alkyl tails (Scheme 13).96 Host–guest complexation between the CB[7] functionality and guest entities yielded supramolecular amphiphilic complexes 59–60 and 59–61 that further assembled into spherical micelles with a diameter of about 18 and 30 nm, respectively, as shown by TEM analysis. The micelles could encapsulate the hydrophobic dye Nile Red, on one hand, and anchor DNA-conjugated Au-nanoparticles through DNA hybridization, on the other hand, which proved that 60 or 61 formed the core whereas DNA strands were located at a periphery of the micelles. Additionally, in the case of 59–60, dual morphological responsiveness of the system was achieved. Thus, addition of competitive 1-aminoadamantane guest resulted in the irreversible micelle disassembly whereas the redox activity of the ferrocene residue in 60 provided the reversible transformation of 59–60 from spherical micelles into the disassociated state and back. It is expected that the ability of DNA-based micelles to load functional molecules make them well suitable for drug delivery with controllable release or for cancer treatment.
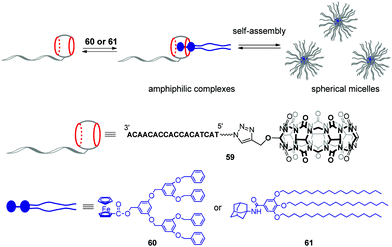 | ||
| Scheme 13 Formation of supramolecular CB[7]–DNA conjugates and their self-assembly into spherical micelles. | ||
5. RNA
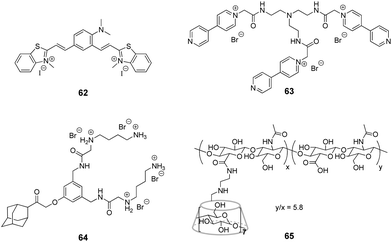
Although the application of CB[n] in systems including DNA is extensively developed, studies focused on the interactions with RNA in CB[n]-based supramolecular assemblies are still rather rare. To the best of our knowledge, the research on ligand–CB[n]–RNA systems is, so far, limited to five examples that are highlighted below. It should be noted, however, that some of the above described studies on ligand–CB[n]–DNA interactions in cells indirectly involved different types of cellular RNAs, whose interactions with the ligand or CB[n] were considered as undesirable.The examples of application of CB[n] in the RNA field include, first of all, the enhancement of the RNA staining by encapsulation of the fluorescent RNA binders in the macrocyclic cavity. Thus, in 2013, CB[7] was used as a transporting agent providing the delivery of fluorescent RNA marker 62 to living cells (Fig. 6).97 Studies on fixed HeLa cells showed that dye 62 selectively stained RNA in nucleoli and cytoplasm with only slight interference of the DNA staining in the nucleus. However, the application of 62 for RNA staining in living HeLa cells was hampered by the failure of the dye to cross the cell membrane. Nevertheless, the ability of CB[7] and its complexes for intracellular uptake through endocytosis as well as the propensity of CB[7] to form a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex (CB–ligand) with dye 62 allowed to overcome this obstacle. Thus, in contrast to the free dye 62, the 62–CB[7]2 complex penetrated into the cells and the cell nuclei where the ligand was released from CB[7] and selectively stained the ribosomal RNA (rRNA) in the nucleus and cytoplasm. The light-up effect of the dye fluorescence upon binding to rRNA was detected by confocal fluorescence microscopy. Importantly, the majority of the stained cells remained alive indicating low cytotoxicity of both CB[7] and the dye 62. Another example of fluorescent staining and discrimination of DNA from RNA in solution and HeLa cells with the complex of pyronine Y (20) with CB[8] has been described in Section 2.5.
1 complex (CB–ligand) with dye 62 allowed to overcome this obstacle. Thus, in contrast to the free dye 62, the 62–CB[7]2 complex penetrated into the cells and the cell nuclei where the ligand was released from CB[7] and selectively stained the ribosomal RNA (rRNA) in the nucleus and cytoplasm. The light-up effect of the dye fluorescence upon binding to rRNA was detected by confocal fluorescence microscopy. Importantly, the majority of the stained cells remained alive indicating low cytotoxicity of both CB[7] and the dye 62. Another example of fluorescent staining and discrimination of DNA from RNA in solution and HeLa cells with the complex of pyronine Y (20) with CB[8] has been described in Section 2.5.
In 2018, H.-C. Wu and colleagues developed a novel method for the selective detection of microRNAs based on triplex molecular beacons and CB[n]-assisted nanopore sensing.77 Apart from microRNA, this approach has been applied for the detection of a range of small molecules, DNA and proteins and is described in detail in Section 4.1.
Another area of application of CB[n]-based assemblies in the RNA field is the design of nanovehicles for the delivery of small interfering RNAs (siRNA). siRNA represents a class of short double-stranded non-coding RNA molecules that participate in the RNA interference process, which is an important mechanism used by an organism to defend against external invasion by inhibiting the expression of specific genes.98,99 The propensity of CB[n] to form stable inclusion complexes with charged guests was used to construct supramolecular polymeric nanostructures, which could electrostatically absorb siRNA and deliver it to cancer cells.
Very recently, J. Liu and co-workers reported supramolecular polymer nanocapsules resulting from the self-assembly of triviologen derivative 63 and CB[8] (Fig. 6).100 The high positive charge density allowed the nanocapsules to absorb survivin siRNA electrostatically and deliver it into cancer cells. The in vitro studies on MCF-7 breast cancer lines showed good biocompatibility of the system, efficient siRNA transfection and pronounced downregulation of the survivin protein expression. Overall, the association with nanocapsules significantly improved the intracellular uptake and reduced the enzymatic degradation of siRNA providing a promising supramolecular approach for gene therapy.
More complex supramolecular system for siRNA delivery comprising CB[6] and β-CD as macrocyclic hosts has been recently developed by Y. Liu and co-workers.101 Thus, a ternary assembly of the amphiphilic guest 64 with CB[6] and cyclodextrin-tethered hyaluronic acid 65 (Fig. 6) resulted in the formation of functional nanoparticles that are capable of targeted delivery of siRNA to PC-3 human prostate cancer cells. The CB[6]-induced pKa shift significantly increased the degree of protonation of the guest's polyamine groups at neutral pH, that played a key role in the electrostatic association of siRNA with the nanoparticles. The hyaluronic acid residues provided the targeting capability because they could be specifically recognized by hyaluronic acid receptors that are overexpressed on the surface of cancer cells. Notably, the transfection efficiency of the system exceeded that of conventional transfection reagents. Overall, these self-assembled nanoparticles provided an elegant example of a supramolecular non-viral vector for targeted delivery and release of siRNA in cancer cells.
6. Conclusions
Starting from the pioneering work by K. Kim and co-workers in 2000 up to now – mainly within the past ten years – the applications of CB[n]s in nucleic acids research have significantly developed and expanded. The studies highlighted in this review cover a wide range of areas, such as governing biological processes of DNA, changing DNA morphology, sensing of NA, gene and drug delivery, DNA photocleavage, supramolecular mimicking of biological processes as well as mechanistic studies of non-covalent interactions and design of biocompatible nanomaterials. All the described systems function without interference with the native structure/base pairing of NA and are based on (i) CB[n]-assisted tuning of the properties of NA-binding ligands, (ii) non-covalent interactions of CB[n] with epigenetically modified sites on NA strands or (iii) covalent attachment of CB[n] to a NA. Considering this, we suggest that the cornerstone of the successful development in this area is the absence of the direct interaction between the native structure of nucleic acids and CB[n].The uniqueness of CB[n] macrocycles in NA context is enhanced by their ability to cross the cell membrane and to penetrate into the cell nuclei in free state or together with encapsulated guests. This feature points out novel approaches not only for CB[n]-assisted drug delivery to NA but also for drug deactivation or/and overdose treatment. Considering this, we were very surprised to find only two reported examples of the CB[7]-based delivery of clinically used drugs to DNA (Section 2.3). Therefore, we assume that in the near future, further development of CB[n]-assisted drug delivery to nucleic acids could be extended to existing NA-targeting drugs, in particular, anticancer agents. Apart of drug delivery, the ability of CB[n] to cross the cell membrane opens up new perspectives in intracellular NA-sensing and gene transfection.
To sum up, we believe that the large potential of the application of CB[n] in the field of NA chemistry is far from being fully explored and realized. Thus, the majority of the reviewed examples are related to the canonical naturally occurring DNAs or synthetic/modified DNA oligonucleotides, whereas the systems including non-canonical naturally occurring DNA structures (G-quadruplex DNA, triplex DNA, etc.) and RNA are just barely explored. Along these lines, we expect the increasing interests of scientists in connecting supramolecular functionality of CB[n] with unique biological functions of the non-canonical therapeutically relevant DNA and RNA. Furthermore, we anticipate future involvement of CB[n] in the development of the RNA-targeting therapy that is currently emerging as a significant alternative or supplement to the existing treatment of viral and bacterial infections, cancer and several RNA-associated genetic diseases.102 Additionally, we are looking forward to the successful transfer of the elaborated functional systems involving CB[n] and NAs to in vitro and in vivo conditions for the cases when this has not been accomplished yet. Overall, we hope that this review will be helpful to order and summarize the achievements and provide a basis for new research ideas and discoveries in this exciting field.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Generous financial support from Deutsche Forschungsgemeinschaft (project BE 7202/1-1) and Russian Science Foundation (project 19-43-04127) is gratefully acknowledged. We thank Prof. Dr Heiko Ihmels (Universität Siegen, Germany) for the proof-reading of the manuscript and valuable advices.Notes and references
- Nucleic acids in chemistry and biology, ed. G. M. Blackburn, M. J. Gait, D. Loakes and D. M. Williams, RSC publishing, Cambridge, 3rd edn, 2006 Search PubMed.
- Z. Wu and L. Zhang, Biomater. Sci., 2019, 7, 4944 RSC.
- Z. Suo, J. Chen, X. Hou, Z. Hu, F. Xing and L. Feng, RSC Adv., 2019, 9, 16479 RSC.
- A. C. Hill and J. Hall, Mater. Chem. Front., 2020, 4, 1074 RSC.
- T. MacCulloch, A. Buchberger and N. Stephanopoulos, Org. Biomol. Chem., 2019, 17, 1668 RSC.
- M. J. Hannon, Chem. Soc. Rev., 2007, 36, 280 RSC.
- D. Shetty, J. K. Khedkar, K. M. Parkad and K. Kim, Chem. Soc. Rev., 2015, 44, 8747 RSC.
- W. Liu, S. K. Samanta, B. D. Smith and L. Isaacs, Chem. Soc. Rev., 2017, 46, 2391 RSC.
- K. I. Assaf and W. M. Nau, Chem. Soc. Rev., 2015, 44, 394 RSC.
- L. Isaacs, Acc. Chem. Res., 2014, 47, 2052 CrossRef CAS.
- D. H. Macartney, Isr. J. Chem., 2011, 51, 600 CrossRef CAS.
- K. I. Kuok, S. Li, I. W. Wyman and R. Wang, Ann. N. Y. Acad. Sci., 2017, 1398, 108 CrossRef CAS.
- D. Das, K. I. Assaf and W. M. Nau, Front. Chem., 2019, 7, 619 CrossRef CAS.
- S. Walker, R. Oun, F. J. McInnes and N. J. Wheate, Isr. J. Chem., 2011, 51, 616 CrossRef CAS.
- J. Liu, Y. Lan, Z. Yu, C. S. Y. Tan, R. M. Parker, C. Abell and O. A. Scherman, Acc. Chem. Res., 2017, 50, 208 CrossRef CAS.
- S. J. Barrow, S. Kasera, M. J. Rowland, J. Barrio and O. A. Scherman, Chem. Rev., 2015, 115, 12320 CrossRef CAS.
- X. Ma and Y. Zhao, Chem. Rev., 2015, 115, 7794 CrossRef CAS.
- X. Zhou, P. Pathak and J. Jayawickramarajah, Chem. Commun., 2018, 54, 11668 RSC.
- S. Dun, C. Ottmann, L.-G. Milroy and L. Brunsveld, J. Am. Chem. Soc., 2017, 139, 13960 CrossRef.
- C. Hou, Z. Huang, Y. Fang and J. Liu, Org. Biomol. Chem., 2017, 15, 4272 RSC.
- T. G. Zhan and K. D. Zhang, Artificial host molecules modifying biomacromolecules, Handbook of Macrocyclic Supramolecular Assembly, Springer, Singapore, 2019 Search PubMed.
- K. Igarashia and K. Kashiwagic, Int. J. Biochem. Cell Biol., 2019, 107, 104 CrossRef.
- D.-H. Bae, D. J. R. Lane, P. J. Jansson and D. R. Richardson, Biochim. Biophys. Acta, Gen. Subj., 2018, 1862, 2053 CrossRef CAS.
- A. Kabir, M. Hossain and G. S. Kumar, J. Chem. Thermodyn., 2013, 57, 445 CrossRef CAS.
- K. S. Srivenugopal, D. E. Wemmer and D. R. Morris, Nucleic Acids Res., 1987, 15, 2563 CrossRef CAS.
- H. Kirino, R. Kuwahara, N. Hamasaki and T. Oshima, J. Biochem., 1990, 107, 661 CrossRef CAS.
- L. Yang, S. Wang, T. Tian and X. Zhou, Curr. Med. Chem., 2012, 19, 557 CrossRef CAS.
- H. Isobe, N. Tomita, J. W. Lee, H.-J. Kim, K. Kim and E. Nakamura, Angew. Chem., Int. Ed., 2000, 39, 4257 CrossRef CAS.
- H. Isobe, S. Sato, J. W. Lee, H.-J. Kim, K. Kim and E. Nakamura, Chem. Commun., 2005, 1549 RSC.
- C. P. Carvalho, A. Norouzy, V. Ribeiro, W. M. Nau and U. Pischel, Org. Biomol. Chem., 2015, 13, 2866 RSC.
- F.-J. Huo, C.-X. Yin and P. Yang, Bioorg. Med. Chem. Lett., 2007, 17, 932 CrossRef CAS.
- A. Rich and S. Zhang, Nat. Rev. Genet., 2003, 4, 566 CrossRef CAS.
- S.-R. Wang, J.-Q. Wang, G.-H. Xu, L. Wei, B.-S. Fu, L.-Y. Wu, Y.-Y. Song, X.-R. Yang, C. Li, S.-M. Liu and X. Zhou, Adv. Sci., 2018, 5, 1800231 CrossRef.
- M. R. Cring and V. C. Sheffield, Gene Ther., 2020 DOI:10.1038/s41434-020-00197-8.
- (a) X. Guo and L. Huang, Acc. Chem. Res., 2012, 45, 971 CrossRef CAS; (b) Pharmaceutical applications of dendrimers, ed. A. Chauhan and H. Kulhari, Elsevier, Amsterdam, 1st edn, 2020 Search PubMed; (c) M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259 CrossRef CAS.
- X. Ma and Y. Zhao, Chem. Rev., 2015, 115, 7794 CrossRef CAS.
- Y.-B. Lim, T. Kim, J. W. Lee, S.-M. Kim, H.-J. Kim, K. Kim and J.-S. Park, Bioconjugate Chem., 2002, 13, 1181 CrossRef CAS.
- Q. Huang, S. Li, Y.-F. Ding, H. Yin, L.-H. Wang and R. Wang, Biomater. Sci., 2018, 6, 1031 RSC.
- S. K. Kim, K. M. Park, K. Singha, J. Kim, Y. Ahn, K. Kim and W. J. Kim, Chem. Commun., 2010, 46, 692 RSC.
- W. Zhang, H.-Y. Zhang, Y.-H. Zhang and Y. Liu, Chem. Commun., 2015, 51, 16127 RSC.
- R. K. Koninti, S. Sappati, S. Satpathi, K. Gavvala and P. Hazra, ChemPhysChem, 2016, 17, 506 CrossRef CAS.
- K. Gavvala and S. Satpathi, J. Lumin., 2016, 171, 234 CrossRef CAS.
- S. Sun, Y. Yuan, Z. Li, S. Zhang, H. Zhang and X. Peng, New J. Chem., 2014, 38, 3600 RSC.
- A. Manna and S. Chakravorti, Spectrochim. Acta, Part A, 2015, 150, 120 CrossRef CAS.
- E. Y. Chernikova, A. Y. Ruleva, V. B. Tsvetkov, Y. V. Fedorov, V. V. Novikov, T. M. Aliyeu, A. A. Pavlov, N. E. Shepel and O. A. Fedorova, Org. Biomol. Chem., 2020, 18, 755 RSC.
- N. N. Alder, Fluorescence spectroscopy and its applications in analysing biomolecular processes, in Biomolecular and Bioanalytical Techniques:Theory, Methodology and Applications, ed. V. Ramesh, John Wiley & Sons, Chichester, 1st edn, 2019 Search PubMed.
- H. K. Saeed, S. Sreedharan and J. A. Thomas, Chem. Commun., 2020, 56, 1464 RSC.
- W. Chyan and R. T. Raines, ACS Chem. Biol., 2018, 13, 1810 CrossRef CAS.
- S. J. Smith, C. R. Nemr and S. O. Kelley, J. Am. Chem. Soc., 2017, 139, 1020 CrossRef CAS.
- W. J. Peveler and W. R. Algar, ACS Chem. Biol., 2018, 13, 1752 CrossRef CAS.
- S. Sinn and F. Biedermann, Isr. J. Chem., 2018, 58, 357 CrossRef CAS.
- F. Li, Y. Xu, H. Li, C. Wang, A. Lu and S. Sun, New J. Chem., 2014, 38, 1396 RSC.
- N. Kitsera, M. Rodriguez-Alvarez, S. Emmert, T. Carell and A. Khobta, Nucleic Acids Res., 2019, 47, 8537 CrossRef CAS.
- J. L. Illuzzi and D. M. Wilson III, Curr. Med. Chem., 2012, 19, 3922 CrossRef CAS.
- Y. Zhou, L. Gao, X. Tong, Q. Li, Y. Fei, Y. Yu, T. Ye, X.-S. Zhou and Y. Shao, Anal. Chem., 2018, 90, 13183 CrossRef CAS.
- H. Liu, Z. Zhang, Y. Zhao, Y. Zhou, B. Xue, Y. Han, Y. Wang, X. Mu, S. Zang, X. Zhou and Z. Li, J. Mater. Chem. B, 2019, 7, 1435 RSC.
- S. Zhang, K. I. Assaf, C. Huang, A. Hennig and W. M. Nau, Chem. Commun., 2019, 55, 671 RSC.
- W. Ong, M. Gómez-Kaifer and A. E. Kaifer, Org. Lett., 2002, 4, 1791 CrossRef CAS.
- J. S. Bus, S. D. Aust and J. E. Gibson, Biochem. Biophys. Res. Commun., 1974, 58, 749 CrossRef CAS.
- S. Sun, W. Gao, F. Liu, J. Fan and X. Peng, J. Mater. Chem., 2010, 20, 5888 RSC.
- S. Sun, Y. He, Z. Yang, Y. Pang, F. Liu, J. Fan, L. Sun and X. Peng, Dalton Trans., 2010, 39, 4411 RSC.
- T. Zhang, S. Sun, F. Liu, Y. Pang, J. Fan and X. Peng, Phys. Chem. Chem. Phys., 2011, 13, 9789 RSC.
- H.-B. Cheng, Y.-M. Zhang, C. Xu and Y. Liu, Sci. Rep., 2014, 4, 4210 CrossRef.
- K. Uekama, F. Hirayama and H. Arima, Pharmaceutical applications of cyclodextrins and their derivatives, in Cyclodextrins and Their Complexes: Chemistry, Analytical Methods, Applications, Wiley-VCH, Weinheim, 2006 Search PubMed.
- A. I. Day and J. G. Collins, in Supramolecular Chemistry: From Molecules to Nanomaterials, ed. P. Gale and J. Steed, Wiley-VCH, Weinheim, 2012 Search PubMed.
- D. V. Berdnikova, T. M. Aliyeu, T. Paululat, Y. V. Fedorov, O. A. Fedorova and H. Ihmels, Chem. Commun., 2015, 51, 4906 RSC.
- C.-F. Ke, S. Hou, H.-Y. Zhang, Y. Liu, K. Yang and X.-Z. Feng, Chem. Commun., 2007, 3374 RSC.
- X.-J. Zhang, Y.-M. Zhang, Z. Wang, Y. Chen and Y. Liu, ChemistrySelect, 2016, 4, 685 CrossRef.
- A. Estévez-Torres and D. Baigl, Soft Matter, 2011, 7, 6746 RSC.
- C. K. Kwok and C. J. Merrick, Trends Biotechnol., 2017, 35, 997 CrossRef CAS.
- D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627 CrossRef CAS.
- T. Tian, Y. Song, L. Wei, J. Wang, B. Fu, Z. He, X.-R. Yang, F. Wu, G. Xu, S.-M. Liu, C. Li, S. Wang and X. Zhou, Nucleic Acids Res., 2017, 45, 2283 CAS.
- S. Satpathi, R. K. Singh, A. Mukherjee and P. Hazra, Phys. Chem. Chem. Phys., 2018, 20, 7808 RSC.
- L. Q. Gu, O. Braha, S. Conlan, S. Cheley and H. Bayley, Nature, 1999, 398, 686 CrossRef CAS.
- T. Li, L. Liu, Y. Li, J. Xie and H.-C. Wu, Angew. Chem., Int. Ed., 2015, 54, 7568 CrossRef CAS.
- B. Guo, Y. Sheng, K. Zhou, Q. Liu, L. Liu and H.-C. Wu, Angew. Chem., Int. Ed., 2018, 57, 3602 CrossRef CAS.
- X. Wu, B. Guo, Y. Sheng, Y. Zhang, J. Wang, S. Peng, L. Liu and H.-C. Wu, Chem. Commun., 2018, 54, 7673 RSC.
- Z. Zhang, T. Li, Y. Sheng, L. Liu and H.-C. Wu, Small, 2019, 15, 1804078 CrossRef.
- L. Liu, T. Li, S. Zhang, P. Song, B. Guo, Y. Zhao and H.-C. Wu, Angew. Chem., Int. Ed., 2018, 57, 11882 CrossRef CAS.
- T. Zeng, L. Liu, T. Li, Y. Li, J. Gao, Y. Zhao and H.-C. Wu, Chem. Sci., 2015, 6, 5628 RSC.
- L. Liu, Y. Li, T. Li, J. Xie, C. Chen, Q. Liu, S. Zhang and H.-C. Wu, Anal. Chem., 2016, 88, 1073 CrossRef CAS.
- L. Liu, Y. You, K. Zhou, B. Guo, Z. Cao, Y. Zhao and H.-C. Wu, Angew. Chem., Int. Ed., 2019, 58, 14929 CrossRef CAS.
- Y. You, K. Zhou, B. Guo, Q. Liu, Z. Cao, L. Liu and H.-C. Wu, ACS Sens., 2019, 4, 774 CrossRef CAS.
- S. Singh, A. Deep, G. Mohanta and V. K. Meena, in Developments in the electrochemical bionanosensors for the predictive diagnosis of prostate and breast cancer, ed. P. Chandra, Springer Nature Singapore Pte Ltd, 2017 Search PubMed.
- M. Wiemann and P. Jonkheijm, Isr. J. Chem., 2018, 58, 314 CrossRef CAS.
- S. Yang, M. You, L. Yang, F. Zhang, Q. Wang and P. He, J. Electroanal. Chem., 2016, 783, 161 CrossRef CAS.
- Y. Chang, Y. Zhuo, Y. Chai and R. Yuan, Anal. Chem., 2017, 89, 8266 CrossRef CAS.
- R. Sasmal, N. D. Saha, M. Pahwa, S. Rao, D. Joshi, M. S. Inamdar, V. Sheeba and S. S. Agasti, Anal. Chem., 2018, 90, 11305 CrossRef CAS.
- S.-R. Wang, Y.-Y. Song, L. Wei, C.-X. Liu, B.-S. Fu, J.-Q. Wang, X.-R. Yang, Y.-N. Liu, S.-M. Liu, T. Tian and X. Zhou, J. Am. Chem. Soc., 2017, 139, 16903 CrossRef CAS.
- X. Zhou, X. Su, P. Pathak, R. Vik, B. Vinciguerra, L. Isaacs and J. Jayawickramarajah, J. Am. Chem. Soc., 2017, 139, 13916 CrossRef CAS.
- Z. Yu, M. Ai, S. K. Samanta, F. Hashiya, J. Taniguchi, S. Asamitsu, S. Ikeda, K. Hashiya, T. Bando, G. N. Pandian, L. Isaacs and H. Sugiyama, Chem. Commun., 2020, 56, 2296 RSC.
- N. Dubel, S. Liese, F. Scherz and O. Seitz, Angew. Chem., Int. Ed., 2019, 58, 907 CrossRef CAS.
- S. Pandey, D. V. D. W. Kankanamalage, X. Zhou, C. Hu, M. E. Hoque, L. Isaacs, J. Jayawickramarajah and H. Mao, J. Am. Chem. Soc., 2019, 141, 18385 CrossRef CAS.
- C. Li, M. J. Rowland, Y. Shao, T. Cao, C. Chen, H. Jia, X. Zhou, Z. Yang, O. A. Scherman and D. Liu, Adv. Mater., 2015, 27, 3298 CrossRef CAS.
- T. Du, W. Yuan, Z. Zhao and S. Liu, Chem. Commun., 2019, 55, 3658 RSC.
- W. Yuan, J. Ma, Z. Zhao and S. Liu, Macromol. Rapid Commun., 2020, 41, 2000022 CrossRef CAS.
- Z. Li, S. Sun, Z. Yang, S. Zhang, H. Zhang, M. Hu, J. Cao, J. Wang, F. Liu, F. Song, J. Fan and X. Peng, Biomaterials, 2013, 34, 6473 CrossRef CAS.
- P. A. Sharp, RNA interference – 2001, Genes Dev., 2001, 15, 485 CrossRef CAS.
- B. Hu, L. Zhong, Y. Weng, L. Peng, Y. Huang, Y. Zhao and X.-J. Liang, Signal Transduction Targeted Ther., 2020, 5, 101 CrossRef CAS.
- F. Li, M. Wang, S. Guan, Z. Huang, S. Liu, X. Li, X. Jiang, Q. Luo, J. Xu and J. Liu, Polym. Chem., 2019, 10, 5659 RSC.
- Y.-M. Zhang, Y. Yang, Y.-H. Zhang and Y. Liu, Sci. Rep., 2016, 6, 28848 CrossRef CAS.
- (a) K. D. Warner, C. E. Hajdin and K. M. Weeks, Nat. Rev. Drug Discovery, 2018, 17, 547 CrossRef CAS; (b) N. F. Rizvi and G. F. Smith, Bioorg. Med. Chem. Lett., 2017, 27, 5083 CrossRef CAS; (c) J. R. Thomas and P. J. Hergenrother, Chem. Rev., 2008, 108, 1171 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |