Open Access Article
Open Access ArticleResolution enhancement in NMR spectra by deconvolution with compressed sensing reconstruction†
Krzysztof
Kazimierczuk
 a,
Paweł
Kasprzak
a,
Paweł
Kasprzak
 ab,
Panagiota S.
Georgoulia
ab,
Panagiota S.
Georgoulia
 c,
Irena
Matečko-Burmann
c,
Irena
Matečko-Burmann
 de,
Björn M.
Burmann
de,
Björn M.
Burmann
 ce,
Linnéa
Isaksson
c,
Emil
Gustavsson
ce,
Linnéa
Isaksson
c,
Emil
Gustavsson
 f,
Sebastian
Westenhoff
f,
Sebastian
Westenhoff
 c and
Vladislav Yu.
Orekhov
c and
Vladislav Yu.
Orekhov
 *cf
*cf
aCentre of New Technologies, University of Warsaw, ul. Banacha 2C, 02-097 Warsaw, Poland
bFaculty of Physics, University of Warsaw, Pasteura 5, 02-093 Warsaw, Poland
cDepartment of Chemistry and Molecular Biology, University of Gothenburg, Box 465, Gothenburg 405 30, Sweden
dDepartment of Psychiatry and Neurochemistry, University of Gothenburg, Gothenburg 405 30, Sweden
eWallenberg Centre for Molecular and Translational Medicine, University of Gothenburg, Gothenburg 405 30, Sweden
fSwedish NMR Centre, University of Gothenburg, Box 465, 405 30, Gothenburg, Sweden. E-mail: vladislav.orekhov@nmr.gu.se
First published on 28th October 2020
Abstract
NMR spectroscopy is one of the basic tools for molecular structure elucidation. Unfortunately, the resolution of the spectra is often limited by inter-nuclear couplings. The existing workarounds often alleviate the problem by trading it for another deficiency, such as spectral artefacts or difficult sample preparation and, thus, are rarely used. We suggest an approach using the coupling deconvolution in the framework of compressed sensing (CS) spectra processing that leads to a major increase in resolution, sensitivity, and overall quality of NUS reconstruction. A new mathematical description of the decoupling by deconvolution explains the effects of thermal noise and reveals a relation with the underlying assumption of the CS. The gain in resolution and sensitivity for challenging molecular systems is demonstrated for the key HNCA experiment used for protein backbone assignment applied to two large proteins: intrinsically disordered 441-residue Tau and a 509-residue globular bacteriophytochrome fragment. The approach will be valuable in a multitude of chemistry applications, where NMR experiments are compromised by the homonuclear scalar coupling.
Nuclear magnetic resonance (NMR) is among the main analytical techniques allowing atomic-level studies of proteins. The prerequisite step for most protein NMR work is a resonance-specific spectral assignment, i.e. association of resonance frequencies with atoms in the protein amino acid chain.1 The HNCA2–4 is by far the most sensitive and thus often the only feasible triple resonance experiment that provides sequential connectivities between neighbouring protein residues. In principle, a sequence-specific resonance assignment could be obtained using the HNCA experiment alone. Unfortunately, low signal resolution relative to the dispersion of the protein 13Cα resonances results in massive ambiguity of the assignment even for relatively small proteins. For large systems with many amino acid residues as well as for intrinsically disordered proteins (IDP) characterized by the particularly low resonance dispersion, one has to rely on additional experiments at the expense of sensitivity loss, a significant increase of measurement time, and more complicated and tedious analyses.
Slow transverse relaxation of the 13Cα spins, which can be further decreased by deuteration, corresponds to the natural line-width of 5–8 Hz even for relatively large protein systems. Unfortunately, the practical resolution in the HNCA spectra is usually almost ten times worse. Two main factors limit the resolution in the HNCA: (i) a large number of time-increments in the 13Cα dimension needed in the 3D experiment to achieve the high resolution. This leads to too long measurement time that can be unaffordable because of short sample stability and/or limitation on the measurement time at an NMR instrument; (ii) homonuclear one-bond coupling between 13Cα and 13Cβ spins that produces a doublet with separation of approximately 35 Hz for every 13Cα signal and thus effectively broadens the spectral line. The former issue is well addressed by using fast pulsing5,6 and non-uniform sampling (NUS) techniques.7–12 A large number of methods for handling of the 1J(Cα–Cβ) coupling had been introduced over the last decades, including biochemical unlabelling of the β carbon atom to 12C,13–18 constant time evolution,19 band-selective homonuclear decoupling,20,21 and IPAP decoupling.22,23 However, broad practical use of these techniques is hindered due to the inherent compromises in sensitivity, extra demands on sample isotope labelling, inability to deal with serine and threonine residues, and/or significant spectral distortions and artefacts.
A viable alternative to these experimental approaches is the virtual decoupling that is the post-acquisition deconvolution of the J-coupling, i.e. in- and anti-phase peak multiplets, at the signal processing stage.24–29 The aim of this communication is to investigate the possibility of effective deconvolution in compressed sensing (CS) algorithms that are among the most powerful for the NUS spectra – to propose a method of selective deconvolution of individual spectral regions; and to demonstrate the relation of the deconvolution to the cornerstone CS concept of sparseness with the resulting benefits for the effectiveness of CS.
The fundamental relation between the NMR signal f(t) detected in the time domain and the spectrum s is
| f = Fs | (1) |
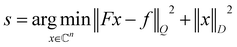 | (2) |
Assuming the same value of active 1J(Cα–Cβ) coupling for all signals, the measured in experiment 13Cα signal f and the signal without the J-coupling ![[f with combining tilde]](https://www.rsc.org/images/entities/i_char_0066_0303.gif) are related as:
are related as:
f = C![[f with combining tilde]](https://www.rsc.org/images/entities/i_char_0066_0303.gif) and and ![[f with combining tilde]](https://www.rsc.org/images/entities/i_char_0066_0303.gif) = C−1f = C−1f | (3) |
![[f with combining tilde]](https://www.rsc.org/images/entities/i_char_0066_0303.gif) has the inverse covariance matrix
has the inverse covariance matrix ![[Q with combining tilde]](https://www.rsc.org/images/entities/i_char_0051_0303.gif) = σ−2(CC). Then, the decoupled spectrum is
= σ−2(CC). Then, the decoupled spectrum is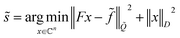 | (4) |
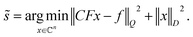 | (5) |
The last equation shows that the post-acquisition deconvolution can be achieved in IRLS and any other algorithm based on equation akin to eqn (2), e.g. maximum entropy24–27,29 and Multi-Dimensional Decomposition (MDD),9 by using measurement matrix CF instead of F. Finally, we note that the deconvoluted spectrum contains half of the peaks relative to the undecoupled spectrum. Thus, it is sparser, and in accordance with the theory of compressed sensing,33,34 it requires nearly half of the measured data points for successful reconstruction. This means that the virtual decoupling not only enhances spectral resolution but also provides conditions for higher quality CS reconstruction (see Theory in ESI†).
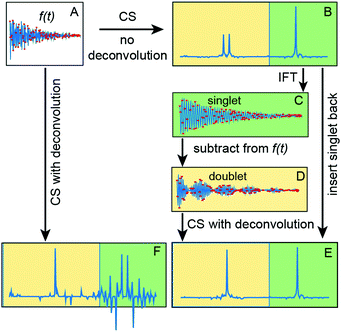
Use of the deconvolution for the HNCA experiment is based on the assumption that 1J(Cα–Cβ) coupling constants are nearly the same for all residues in the protein. The variation of the coupling values ±2.5 Hz35 is lower than the line width determined by the transverse relaxation of 13Cα spins and, thus, does not pose a problem for the reconstruction (see ESI†). However, signals (singlets) from Gly residues that do not have Cβ atoms, have no sparse representation in the columns of measurement matrix CF. Fig. 1 illustrates that this not only corrupts the Gly peaks in the deconvoluted spectrum but also affects other signals and reduces the overall quality of the reconstruction. To tackle this, we suggest a procedure of deconvolution-IRLS (D-IRLS) with the Gly-region selection as outlined in Fig. 1 (more details are found in the ESI†). We start with reconstructing the full undecoupled spectrum using matrix F. Because the 13Cα atoms usually have distinctly different chemical shifts with values lower than 45 ppm, we can subtract the well-reproduced signals in the Gly region from the original time-domain signal f, which is then used to reconstruct the spectrum with all signals except for Gly using eqn (5) with the measurement matrix CF. Finally, signals of Gly and other residues are combined into the full decoupled spectrum in the frequency domain.
Selection of the NUS acquisition schedule has a profound effect on the reconstruction quality. As f is multiplied by C−1 in eqn (3), the noise is amplified the most for the points in ![[f with combining tilde]](https://www.rsc.org/images/entities/i_char_0066_0303.gif) at times, where cos(πJt) function has small values (i.e. near t = k/(2J), k = 1, 3, 5). In the weighted least squares method used to derive eqn (4), these points are used with low weights and thus contain relatively low information value. In the NUS schedule, it is logical to avoid these points and instead invest spectrometer time into more informative measurements. We used the signal amplitude matched NUS schedule with the sampling density corresponding to |cos(πJt)| and rejecting points with probability less than 0.232,36 (Fig. S1, ESI†). Additionally, the schedule was in all cases relaxation-matched.
at times, where cos(πJt) function has small values (i.e. near t = k/(2J), k = 1, 3, 5). In the weighted least squares method used to derive eqn (4), these points are used with low weights and thus contain relatively low information value. In the NUS schedule, it is logical to avoid these points and instead invest spectrometer time into more informative measurements. We used the signal amplitude matched NUS schedule with the sampling density corresponding to |cos(πJt)| and rejecting points with probability less than 0.232,36 (Fig. S1, ESI†). Additionally, the schedule was in all cases relaxation-matched.
We demonstrate the new D-IRLS procedure using examples of two representative systems: intrinsically disordered human 441-residue Tau protein (the longest hTau40 isoform)37 and the monomeric variant of the 509-residue globular photosensory module PAS-GAF-PHY of Deinococcus radiodurans phytochrome (DrBphPPSM).38 For each protein, Fig. 2 shows the traditional low-resolution 3D HNCA spectrum superimposed with the resolution-enhanced spectrum obtained using D-IRLS with Gly-region selection. For DrBphPPSM the two experiments were reconstructed using nearly the same number of NUS points corresponding to the same measurement time; for Tau, the low resolution experiment was around two times shorter. In the shown examples, the dramatically improved resolution of the D-IRLS spectrum allows us to observe sequential connectivities that are ambiguous in the traditional spectrum.
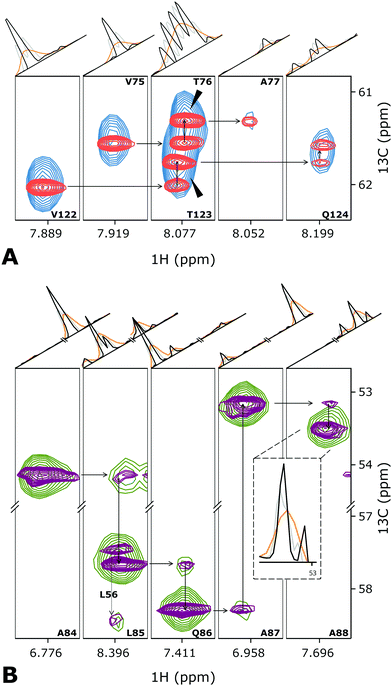 | ||
| Fig. 2 Several planes from the 3D HNCA spectra of (A) Tau and (B) DrBphPPSM showing the assignment walk for selected residues. Overlaid blue (green) and red (purple) contour levels depict traditional low-resolution and high-resolution spectra of Tau (DrBphPPSM) protein. For the peak annotations, we use the previously published assignment.37,38 The one-dimensional cross-sections above the spectra planes are taken (orange) the low-resolution, (black) high-resolution deconvoluted, and (grey) high-resolution non-deconvoluted spectra. | ||
Fig. 2 and Fig. S2, S3 (ESI†) demonstrate that, in addition to the enhanced resolution, the D-IRLS spectra show higher or similar sensitivity in comparison to both the traditional low resolution and non-deconvoluted spectra. The peak connecting A87 and A88 in DrBphPPSM spectra (Fig. 2B) provides a specific example of this. It is clearly seen in the 1D cross-sections in the D-IRLS spectrum. In the traditional experiment, the weak peak is completely masked by the slope of a stronger peak. In the non-deconvoluted spectrum, only one of the doublet components is present, which gives a completely wrong idea of the peak position.
Fig. S2 (ESI†) shows the 13C/15N projections from the spectra of both studied proteins, which confirms the superior quality of the spectra reconstructed with Gly-region selective D-IRLS. While the improved resolution in the spectra is anticipated from the deconvolution, the remarkable sensitivity of the D-IRLS spectrum can be explained by the increased sparsity favourable for the NUS reconstruction.
In order to extensively test the proposed D-IRLS method, we conducted simulations using synthetic peaks added to the 3D HNCA signal of Tau. Adding the simulated components with known positions and intensities to the time domain signal makes it possible to define the precision of the corresponding peak parameters derived from the reconstructed spectrum.39 A detailed description of the simulations can be found in the ESI.† The results shown in Fig. S3 (ESI†) confirm that the peak intensities and positions are much more accurate when the D-IRLS deconvolution is augmented with the Gly-region selective procedure (Fig. 1E).
Notably, at low sampling levels (250 and 400 NUS points) the number of detected peaks from the lowest intensity fraction of the injected peaks is significantly larger in the deconvoluted spectrum in comparison to the IRLS without the deconvolution. This is fully in line with the theoretical consideration that the deconvoluted spectrum is much more sparse and thus can be successfully reconstructed with fewer measured points. In the wide range of NUS levels, cosine J-modulated sampling scheme provides spectra with comparable or somewhat better accuracy of the peak positions and intensities than the schedules matched to the exponential relaxation decay only. However, the main practical problem with the latter scheme is the necessity to increase and carefully adjust the Tikhonov regularization parameter λ in the IRLS algorithm, whereas the cosine-modulated sampling is much less demanding in this respect and thus more robust. It is also worth noting that precision of the peak positions derived from the non-deconvoluted spectrum is somewhat better than in its decoupled counterpart provided that both components of the doublet are detected and resolved from other peaks.
In conclusion, we proposed an efficient CS method to improve the resolution, sensitivity and quality of NUS reconstruction using virtual decoupling at the processing stage. We presented a complete mathematical description of the spectrum deconvolution in terms of the generalized Tikhonov regularization formalism. We also showed that removing singlets from the spectrum before decoupling significantly improves results. The method was demonstrated on the 3D HNCA spectra of two large systems prototypical for the intrinsically disordered and globular proteins. The new CS virtual decoupling technique will enable the sequential signal assignment for many challenging proteins and will be useful for other types of NMR spectra in a variety of applications.
KK and PK thank the Foundation for Polish Science for support via the FIRST TEAM program co-financed by the European Union under the European Regional Development Fund no. (POIR.04.04.00-00-4343/17-00). BMB and SW acknowledge funding from the Knut och Alice Wallenberg Foundation. VO thanks for the support by the Swedish Research Council (Research Grant 2019-3661).
Conflicts of interest
There are no conflicts to declare.References
- M. Ikura, L. E. Kay and A. Bax, Biochemistry, 1990, 29, 4659–4667 CrossRef CAS.
- L. E. Kay, M. Ikura, R. Tschudin and A. Bax, J. Magn. Reson., 1990, 89, 496–514 CAS.
- M. Salzmann, K. Pervushin, G. Wider, H. Senn and K. Wüthrich, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 13585–13590 CrossRef CAS.
- Z. Solyom, M. Schwarten, L. Geist, R. Konrat, D. Willbold and B. Brutscher, J. Biomol. NMR, 2013, 55, 311–321 CrossRef CAS.
- K. Pervushin, B. Vögeli and A. Eletsky, J. Am. Chem. Soc., 2002, 124, 12898–12902 CrossRef CAS.
- E. Lescop, P. Schanda and B. Brutscher, J. Magn. Reson., 2007, 187, 163–169 CrossRef CAS.
- J. C. J. Barna, E. D. Laue, M. R. Mayger, J. Skilling and S. J. P. Worrall, J. Magn. Reson., 1987, 73, 69–77 CAS.
- J. C. Hoch, A. S. Stern and M. Mobli, Maximum Entropy Reconstruction, Update based on original article by Jeffrey C. Hoch, Encyclopedia of Magnetic Resonance, 1996, John Wiley & Sons Ltd., 2007 DOI:10.1002/9780470034590.emrstm0299.pub2.
- V. Orekhov and V. A. Jaravine, Prog. Nucl. Magn. Reson. Spectrosc., 2011, 59, 271–292 CrossRef CAS.
- K. Kazimierczuk and V. Y. Orekhov, Angew. Chem., Int. Ed., 2011, 50, 5556–5559 CrossRef CAS.
- M. Bostock and D. Nietlispach, Concepts Magn. Reson., Part A, 2017, 46, e21438 CrossRef.
- S. Robson, H. Arthanari, S. G. Hyberts and G. Wagner, Methods in Enzymology, Elsevier, 2019, vol. 614, pp. 263–291 Search PubMed.
- D. M. LeMaster and D. M. Kushlan, J. Am. Chem. Soc., 1996, 118, 9255–9264 CrossRef.
- P. E. Coughlin, F. E. Anderson, E. J. Oliver, J. M. Brown, S. W. Homans, S. Pollak and J. W. Lustbader, J. Am. Chem. Soc., 1999, 121, 11871–11874 CrossRef CAS.
- M. Kainosho, T. Torizawa, Y. Iwashita, T. Terauchi, A. M. Ono and P. Güntert, Nature, 2006, 440, 52–57 CrossRef CAS.
- P. Lundström, K. Teilum, T. Carstensen, I. Bezsonova, S. Wiesner, D. F. Hansen, T. L. Religa, M. Akke and L. E. Kay, J. Biomol. NMR, 2007, 38, 199–212 CrossRef.
- K. Takeuchi, Z.-Y. J. Sun and G. Wagner, J. Am. Chem. Soc., 2008, 130, 17210–17211 CrossRef CAS.
- S. A. Robson, K. Takeuchi, A. Boeszoermenyi, P. W. Coote, A. Dubey, S. Hyberts, G. Wagner and H. Arthanari, Nat. Commun., 2018, 9, 356 CrossRef.
- R. Powers, A. M. Gronenborn, G. M. Clore and A. Bax, J. Magn. Reson., 1991, 94, 209–213 CAS.
- H. Matsuo, E. Kupče, H. Li and G. Wagner, J. Magnet. Reson. Ser. B, 1996, 113, 91–96 CrossRef CAS.
- P. W. Coote, S. A. Robson, A. Dubey, A. Boeszoermenyi, M. Zhao, G. Wagner and H. Arthanari, Nat. Commun., 2018, 9, 3014 CrossRef.
- P. Andersson, J. Weigelt and G. Otting, J. Biomol. NMR, 1998, 12, 435–441 CrossRef CAS.
- M. Ottiger, F. Delaglio and A. Bax, J. Magn. Reson., 1998, 131, 373–378 CrossRef CAS.
- A. A. Bothner-By and J. Dadok, J. Magn. Reson., 1987, 72, 540–543 CAS.
- M. A. Delsuc and G. C. Levy, J. Magn. Reson., 1988, 76, 306–315 CAS.
- Z. Serber, C. Richter, D. Moskau, J.-M. Böhlen, T. Gerfin, D. Marek, M. Häberli, L. Baselgia, F. Laukien, A. S. Stern, J. C. Hoch and V. Dötsch, J. Am. Chem. Soc., 2000, 122, 3554–3555 CrossRef CAS.
- N. Shimba, A. S. Stern, C. S. Craik, J. C. Hoch and V. Dötsch, J. Am. Chem. Soc., 2003, 125, 2382–2383 CrossRef CAS.
- N. Shimba, H. Kovacs, A. S. Stern, A. M. Nomura, I. Shimada, J. C. Hoch, C. S. Craik and V. Dötsch, J. Biomol. NMR, 2004, 30, 175–179 CrossRef CAS.
- R. Kerfah, O. Hamelin, J. Boisbouvier and D. Marion, J. Biomol. NMR, 2015, 63, 389–402 CrossRef CAS.
- E. J. Candes, M. B. Wakin and S. P. Boyd, J. Fourier Anal. Appl., 2008, 14, 877–905 CrossRef.
- K. Kazimierczuk and V. Y. Orekhov, J. Magn. Reson., 2012, 223, 1–10 CrossRef CAS.
- Other functional forms are possible for different multiplet types, e.g. sine-modulation for an anti-phase doublet.
- E. Candes, J. Romberg and T. Tao, IEEE Trans. Inf. Theory, 2004, 52, 489–509 Search PubMed.
- S. Foucart and H. Rauhut, Bull. Am. Math., 2017, 54, 151–165 Search PubMed.
- J. M. Schmidt, M. J. Howard, M. Maestr-Martínez, C. S. Pérez and F. Löhr, Magn. Reson. Chem., 2009, 47, 16–30 CrossRef CAS.
- V. Jaravine, I. Ibraghimov and V. Y. Orekhov, Nat. Methods, 2006, 3, 605–607 CrossRef CAS.
- R. L. Narayanan, U. H. N. D urr, S. Bibow, J. Biernat, E. Mandelkow and M. Zweckstetter, J. Am. Chem. Soc., 2010, 132, 11906–11907 CrossRef CAS.
- E. Gustavsson, L. Isaksson, C. Persson, M. Mayzel, U. Brath, L. Vrhovac, J. A. Ihalainen, B. G. Karlsson, V. Orekhov and S. Westenhoff, Biophys. J., 2020, 118, 415–421 CrossRef CAS.
- M. A. Zambrello, A. D. Schuyler, M. W. Maciejewski, F. Delaglio, I. Bezsonova and J. C. Hoch, Methods, 2018, 138–139, 62–68 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Extended mathematical description of D-IRLS, sample preparation and experimental details, and results of simulations with injected peaks. See DOI: 10.1039/d0cc06188c |
| This journal is © The Royal Society of Chemistry 2020 |