Open Access Article
Open Access ArticleVisualizing antithrombin-binding 3-O-sulfated heparan sulfate motifs on cell surfaces†
Mausam
Kalita
 a,
Jie Shi
Chua
a,
Rio S.
Boothello
a,
Jie Shi
Chua
a,
Rio S.
Boothello
 bc,
April
Joice
a,
Orlando
Antelope
a,
Anindita
Roy
a,
Pon Velayutham
Anandh Babu
bc,
April
Joice
a,
Orlando
Antelope
a,
Anindita
Roy
a,
Pon Velayutham
Anandh Babu
 d,
Yukio
Saijoh
d,
Yukio
Saijoh
 e,
Umesh R.
Desai
e,
Umesh R.
Desai
 *bc and
Balagurunathan
Kuberan
*bc and
Balagurunathan
Kuberan
 *a
*a
aDepartments of Medicinal Chemistry and Bioengineering, University of Utah, Salt Lake City, UT 84112, USA. E-mail: kuby@pharm.utah.edu
bDepartment of Medicinal Chemistry, School of Pharmacy, Virginia Commonwealth University, Richmond, VA 23298, USA. E-mail: urdesai@vcu.edu
cInstitute for Structural Biology, Drug Discovery and Development, Virginia Commonwealth University, Richmond, VA 23219, USA
dDepartment of Nutrition and Integrative Physiology, University of Utah, Salt Lake City, UT 84112, USA
eDepartment of Neurobiology and Anatomy, University of Utah School of Medicine, Salt Lake City, UT 84112, USA
First published on 26th October 2020
Abstract
To map the cellular topography of the rare 3-O-sulfated structural motif of heparan sulfate (HS), we constructed quantum dot-based probes for antithrombin and FGF2, which reveal widely different distribution of the targeted HS motifs. The technology helps show that old and young aortic endothelia display widely different levels of the antithrombin-binding 3-O-sulfated HS motif.
Heparan sulfate proteoglycans (HSPGs), such as syndecans and glypicans, are present on virtually all mammalian cell surfaces and modulate a number of processes such as growth, differentiation and morphogenesis.1 The glycosaminoglycan (GAG) component of HSPGs is heparan sulfate (HS), a polysaccharide of alternating glucosamine and uronic acid residues‡ that are not uniformly sulfated resulting in a diverse array of structural and conformational microdomains within the same chain. The diversity of microdomains results in a diversity of biological functions, owing to direct engagement of proteins such as plasma proteins, extracellular matrix (ECM) proteins and cell surface receptors. Classic examples of this include the HS–antithrombin (AT)2–4 and HS–fibroblast growth factor 2 (FGF2)5–8 systems that have yielded key insights into recognition and regulation by GAGs. Human plasma AT binds tightly to a rare 3-O-sulfated pentasaccharide motif in HS,3,4 whereas FGF2 is known to bind to common tetrasaccharide motifs in HS6–8 with good affinity (Fig. 1).
 | ||
| Fig. 1 Structures of the rare 3-O-sulfated motif binding to antithrombin and the common tetrasaccharide motif binding to FGF2. | ||
The rare 3-O-sulfate (3-OS) group of HS is introduced by one of the six human 3-O-sulfotransferases (3OSTs), which possess differing substrate specificities, thereby generating different 3-O-sulfated microdomains in HS.3,9 Interestingly, 3-O-sulfation of HS chains is tissue-type dependent, e.g., endothelial cells have approximately one 3-OS group per 100 HS disaccharides, whereas follicular fluid has nearly six and Reichert's basement membrane contains five to ten 3-OS groups.10–12 Yet, are these 3-OS groups uniformly distributed or are they present in select locales in vivo remains unknown. Considering that growing evidence suggests spatial and temporal regulation of enzymes involved in HS biosynthesis,13,14 it is critical to develop readily applicable tools to help elucidate topography of HS, especially for rare structural motifs.
Although several probes for spatio-temporal visualization of DNA, mRNA and proteins have been developed,15–17 probes for monitoring spatiotemporal changes in GAGs remain limited. Few examples known in the literature relate to tracking proteins, such as HIT-1 Tat or HSV gB, surfing over HSPGs18,19 or tracking common HS motifs through the use of antibodies.20 Likewise, a radioactive probe (125I-AT) has also been explored for imaging HS too with limited applicability.21 Most importantly, none of these probes offer insight into spatio-temporal topography of rare and specific HS sequences. This is critical for elucidating fundamental biochemical principles that regulate HS biosynthesis as well as for deciphering spatio-temporal changes in sequence composition in vivo, which could help pinpoint strategies for addressing novel therapeutics.
Here, we report the construction of a quantum dot (QD)-based fluorescent probe that reveals the spatio-temporal topography of a rare HS motif. The QD-based probes are non-radioactive, photostable, highly sensitive (10–100-fold more than Alexa Fluor dyes), compatible with both optical and electron microscopy, and very useful for cell and in vivo imaging.17,22 We show that this readily-applicable QD-based technology can dramatically enhance the precision of localization of rare motifs present in HSPGs on cell surfaces.
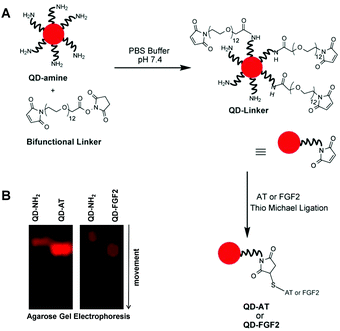
We synthesized two QD-based probes to visualize, compare and quantify the topography of rare and common HS motifs (Fig. 2). The first probe targeted the specific 3-O-sulfated pentasaccharide motif, which binds human plasma AT with high affinity, whereas an analogous QD-conjugated FGF2 probe assessed the common tetrasaccharide sequences of HS. Because neither AT nor FGF2 possess a free cysteine, we utilized a cysteine (or TCEP) reduction methodology to release –SH groups for bioconjugation with QDs (see Fig. S1, ESI† for mechanism), thereby ascertaining minimal alteration of HS recognition. In fact, fluorescence titrations with the high-affinity heparin pentasaccharide (H5) showed a KD of 38 nM for the QD-AT:H5 complex, which was similar to that of the AT:H5 complex (∼10 nM) under comparable conditions at pH 7.4, I 0.85 and 25 °C (Fig. S3, ESI†).23 We also measured a stoichiometry of nearly two AT molecules per every QD molecule (Fig. S4, ESI†), which explains the significant difference in molecular size between native and conjugated QD species (Fig. 2B).
The specificity of the QD-AT probe was first assessed in wild-type Chinese hamster ovary (CHO-K1) cells that do not express 3-OST-1 enzyme, which results in cell surface HSPGs that are devoid of 3-OS motifs. No QD-AT fluorescence (λEM = 655 nm) was observed for these cells (Fig. 3A). In contrast, treatment of the CHO-K1 cells with 3-OST-1 enzyme in the presence of the sulfate donor PAPS (3′-phosphoadenosine-5′-phosphosulfate), which is known to generate 3-OS motifs on cell surface HSPGs,24,25 resulted in ∼614-fold increase in the QD-AT fluorescence (p = 0.0008) (Fig. 3C).
Interestingly, the new motifs formed by 3-OST1/PAPS were bundled at specific loci on the CHO-K1 cells. Recent results show that 3-OST-1 recognizes substrate sequences very selectively,3 which is one of the major causes behind the rarity of this 3-OS structural motif. Further, the QD-AT-labelled HS subpopulation also overlapped with 10E4, an anti-HS epitope antibody that does not recognize any other GAGs, supporting the location of 3-OS motifs within HS chains. Finally, treatment of cells with a cocktail of heparitinases (Hep I, II and III), which degrade HS, resulted in a precipitous loss of the QD-AT signal (Fig. 3A).
Following validation in the 3-OST-1 lacking cell line (CHO-K1), we studied wild-type bovine lung microvasculature endothelial cells (BLMVECs). We observed occasional labelling of BLMVECs (Fig. 3B) with <1% 3-OS motifs present on these cells, as documented in the literature.11 A 17-fold rise in 3-OS labelling was observed with 3-OST-1 modification (p = 0.0042) (Fig. 3D). Once again, the newly introduced 3-OS motifs were localized in clusters on BLMVECs. Finally, the removal of HS chains from cell surface HSPG with heparitinase treatment ablated QD-AT fluorescence signals by ∼22-fold (p = 0.0072). This deduction was also supported by a similar reduction in signal from anti-HS antibody (10E4) (see Fig. S5, ESI†).
Next, we used the QD-FGF2 probe to visualize the common and ubiquitous tetrasaccharide motif on CHO-K1 cells and BLMVECs (Fig. S6, ESI†). Both cell types, when treated with QD-FGF2 probe, revealed a ubiquitous expression of the common tetrasaccharide motif all over the cell surface. As expected, the 3-OST-1/PAPS introduction hardly changed the QD-FGF2 signal suggesting that the 3-OS groups do not impact FGF2 recognition (Fig. S6, ESI†). In contrast, heparitinase cocktail treatment ablated probe fluorescence for both cell types. Thus, the QD-FGF2 probe demonstrates a completely distinct ability to visualize HS motifs from the QD-AT probe.
The ability to visualize and quantify rare 3-OS motifs on cell surfaces led to the question whether the QD-AT probe would help visualize in vivo temporal changes, if any, of this rare population of HS motifs. We selected two C57BL/6 mouse groups, a two-weeks and a six-months old group (N = 3 each), which would correspond to ∼2–6§ and ∼20–69 human years,§ respectively.26 We sought to visualize changes arising from aging, if any, in the distribution and abundance of 3-OS motifs on the aorta and vasa vasorum, which deliver nutrients to the walls of blood vessels.27 The aortas of each animal groups were surgically removed and divided into two treatment cohorts – QD-AT treatment and heparitinase followed by QD-AT treatment (see ESI†). Fig. 4 shows representative results for the two groups and four cohorts. The six-months old group showed a 79-fold less fluorescence of the QD-AT probe in comparison to the two-weeks old group (p < 0.0001). When treated with the heparitinase cocktail, a dramatic reduction in fluorescence intensities for both groups was observed, as expected. The loss in QD-AT fluorescence highlights the significant difference in the levels of 3-OS motifs present on HSPGs lining the aortic and vasa vasorum endothelium as a function of age.
The high fidelity of QD-AT fluorescence quantitation also pointed to that possible difference in level of heparitinase action on the ECM of a formative young aorta versus the older aorta. Whereas heparitinase essentially eliminated QD-AT fluorescence in the young animal, some 3-OS motifs appeared to persist for the older aorta. It is possible that the extracellular matrix (ECM) architecture of the young and old aortic tissue differ. In fact, ECM stiffens with age,28,29 which may impede enzymatic action of heparitinases.
Overall, we have demonstrated that using simple chemistry and QD-based imaging technology, it is possible to visualize the location as well as quantify the levels of rare 3-OS motifs of HS at cell surfaces. Our work elegantly shows that the technique is specific and robust for evaluating samples from live animals.
Our work also highlights two highly interesting biological results that will require further studies. (1) The 3-OS motifs were localized in distinct clusters on both CHO-K1 cells as well as BLMVECs. Although noted earlier,21 it would be important to assess its generality across a range of cells, especially from different organs. This observation could have major bearing on the selectivity, regulation and outcome of HS biosynthesis.1,3,13 (2) The 3-OS motifs were quantitatively down regulated in aged animals as compared to younger animals. It is well-known that the incidences of thrombosis and coronary arterial disease increase with aging.30,31 The underlying causes for these are many;31,32 yet, this study presents a new possibility. The reduction in AT-binding 3-OS motifs may reduce the anti-coagulation potential of the cell surface, thereby contributing to pro-thrombotic risk. Although much experimentation would be needed to test this hypothesis, our work demonstrates for the first time that the QD-based probes should be extremely useful in identifying dynamic changes in HS motifs arising from aging. This could have significant impact on diseases associated with 3-OS motifs such as thrombosis and neurodegeneration.4,9,33
Finally, the QD-based technology could visualize majority of rare HS motifs for which proteins are known to exhibit high selectivity, e.g., heparin cofactor II.34 Considering that a large number of proteins could bind HS selectively,35 QD-based probes should find use in elucidating the spatio-temporal dynamics of distinct HS motifs in vivo.
Conflicts of interest
There are no conflicts to declare.Notes and references
- J. R. Bishop, M. Schuksz and J. D. Esko, Nature, 2007, 446, 1030–1037 CrossRef CAS.
- A. R. Rezaie and H. Giri, J. Thromb. Haemostasis, 2020 DOI:10.1111/jth.15052.
- N. V. Sankaranarayanan, Y. Bi, B. Kuberan and U. R. Desai, Comput. Struct. Biotechnol. J., 2020, 18, 933–941 CrossRef CAS.
- U. R. Desai, Med. Res. Rev., 2004, 24, 151–181 CrossRef CAS.
- C. Lanzi and G. Cassinelli, Biochem. Pharmacol., 2020, 178, 114084, DOI:10.1016/j.bcp.2020.114084.
- V. H. Pomin, Biochimie, 2016, 127, 214–216 CrossRef CAS.
- R. Goetz and M. Mohammadi, Nat. Rev. Mol. Cell Biol., 2013, 14, 166–180 CrossRef CAS.
- J. Schlessinger, A. N. Plotnikov, O. A. Ibrahimi, A. V. Eliseenkova, B. K. Yeh, A. Yayon, R. J. Linhardt and M. Mohammadi, Mol. Cell, 2000, 6, 743–750 CrossRef CAS.
- B. E. Thacker, D. Xu, R. Lawrence and J. D. Esko, Matrix Biol., 2014, 35, 60–72 CrossRef CAS.
- A. I. de Agostini, J.-C. Dong, C. de Vantéry Arrighi, M.-A. Ramus, I. Dentand-Quadri, S. Thalmann, P. Ventura, V. Ibecheole, F. Monge, A.-M. Fischer, S. H. Mohammadi, N. W. Shworak, L. Zhang, Z. Zhang and R. J. Linhardt, J. Biol. Chem., 2008, 283, 28115–28124 CrossRef CAS.
- J. A. Marcum, D. H. Atha, L. M. Fritze, P. Nawroth, D. Stern and R. D. Rosenberg, J. Biol. Chem., 1986, 261, 7507–7517 CAS.
- G. Pejler, G. Bäckström, U. Lindahl, M. Paulsson, M. Dziadek, S. Fujiwara and R. Timpl, J. Biol. Chem., 1987, 262, 5036–5043 CAS.
- J.-P. Li and M. Kusche-Gulberg, Int. Rev. Cell Mol. Biol., 2016, 325, 215–273 CAS.
- J. Kreuger and L. Kjellén, J. Histochem. Cytochem., 2012, 60, 898–907 CrossRef.
- Z. Zou, Z. L. Luo, X. Xu, S. Yang, Z. H. Qing, J. W. Liu and R. Yang, TrAC, Trends Anal. Chem., 2020, 125, 115811 CrossRef CAS.
- B. N. G. Giepmans, S. R. Adams, M. H. Ellisman and R. Y. Tsien, Science, 2006, 312, 217–224 CrossRef CAS.
- X. Michalet, F. F. Pinaud, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Sundaresan, A. M. Wu, S. S. Gambhir and S. Weiss, Science, 2005, 307, 538–544 CrossRef CAS.
- Y. Suzuki, C. N. Roy, W. Promjunyakul, H. Hatakeyama, K. Gonda, J. Imamura, B. Vasudevanpillai, N. Ohuchi, M. Kanzaki, H. Higuchi and M. Kaku, Mol. Cell. Biol., 2013, 33, 3036–3049 CrossRef CAS.
- M. J. Oh, J. Akhtar, P. Desai and D. Shukla, Biochem. Biophys. Res. Commun., 2010, 391, 176–181 CrossRef CAS.
- N. C. Smits, S. Kurup, A. L. Rops, G. B. ten Dam, L. F. Massuger, T. Hafmans, J. E. Turnbull, D. Spillmann, J. P. Li, S. J. Kennel, J. S. Wall, N. W. Shworak, P. N. Dekhuijzen, J. van der Vlag and T. H. van Kuppevelt, J. Biol. Chem., 2010, 285, 41143–41151 CrossRef CAS.
- A. I. de Agostini, S. C. Watkins, H. S. Slayter, H. Youssoufian and R. D. Rosenberg, J. Cell Biol., 1990, 111, 1293–1304 CrossRef CAS.
- U. Resch-Genger, M. Grabolle, S. Cavaliere-Jaricot, R. Nitschke and T. Nann, Nat. Methods, 2008, 5, 763–775 CrossRef CAS.
- U. R. Desai, M. Petitou, I. Björk and S. T. Olson, J. Biol. Chem., 1998, 273, 7478–7487 CrossRef CAS.
- L. Zhang, D. L. Beeler, R. Lawrence, M. Lech, J. Liu, J. C. Davis, Z. Shriver, R. Sasisekharan and R. D. Rosenberg, J. Biol. Chem., 2001, 276, 42311–42321 CrossRef CAS.
- B. Kuberan, D. L. Beeler, M. Lech, Z. L. Wu and R. D. Rosenberg, J. Biol. Chem., 2003, 278, 52613–52621 CrossRef CAS.
- S. Dutta and P. Sengupta, Life Sci., 2016, 152, 244–248 CrossRef CAS.
- J. K. Williams and D. D. Heistad, Trends Cardiovasc. Med., 1996, 6, 53–57 CrossRef CAS.
- C. M. Kelleher, S. E. McLean and R. P. Mecham, Curr. Top. Dev. Biol., 2004, 62, 153–188 CrossRef CAS.
- J. C. Kohn, M. C. Lampi and C. A. Reinhart-King, Front. Genet., 2015, 6, 112 CrossRef.
- E. J. Favaloro, M. Franchini and G. Lippi, Semin. Thromb. Hemostasis, 2014, 40, 621–633 CrossRef CAS.
- C. Shao, J. Wang, J. Tian and Y. Tang, Adv. Exp. Med. Biol., 2020, 1177, 1–36 CrossRef.
- K. Murata, J. Mol. Cell. Cardiol., 1981, 13, 281–292 CrossRef CAS.
- J. Zhao, Y. Zhu, X. Song, Y. Xiao, G. Su, X. Liu, Z. Wang, Y. Xu, J. Liu, D. Eliezer, T. F. Ramlall, G. Lippens, J. Gibson, F. Zhang, R. J. Linhardt, L. Wang and C. Wang, Angew. Chem., Int. Ed., 2020, 59, 1818–1827 CrossRef CAS.
- N. V. Sankarayanarayanan, T. R. Strebel, R. S. Boothello, K. Sheerin, A. Raghuraman, F. Sallas, P. D. Mosier, N. D. Watermeyer, S. Oscarson and U. R. Desai, Angew. Chem., Int. Ed., 2017, 56, 2312–2317 CrossRef CAS.
- N. V. Sankarayanarayanan, B. Nagarajan and U. R. Desai, Curr. Opin. Struct. Biol., 2018, 50, 91–100 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc05893a |
| ‡ Residues include N-sulfoglucosamine (GlcNS), N-acetylglucosamine (GlcNAc), glucuronic acid (GlcA) and iduronic acid (IdoA) that may be sulfated at 2-, 3-, or 6-positions. |
| § Mice and humans have different rates of changes in different metabolic processes, which have been used to calculate an estimate of variation in correspondence between mouse and human years. |
| This journal is © The Royal Society of Chemistry 2020 |