Open Access Article
Open Access ArticleControlling receptor function from the extracellular vestibule of G-protein coupled receptors†
Attila
Egyed
a,
Katalin
Domány-Kovács
b,
Bence
Koványi
b,
Ferenc
Horti
b,
Dalma
Kurkó
 b,
Dóra Judit
Kiss
b,
Dóra Judit
Kiss
 a,
Gáspár
Pándy-Szekeres
a,
Gáspár
Pándy-Szekeres
 a,
István
Greiner
a,
István
Greiner
 b and
György M.
Keserű
b and
György M.
Keserű
 *a
*a
aMedicinal Chemistry Research Group, Research Center for Natural Sciences, Magyar Tudósok krt. 2, Budapest, H-1117, Hungary. E-mail: keseru.gyorgy@ttk.hu
bGedeon Richter plc, Gyömrői út 19-21, Budapest, H-1103, Hungary
First published on 13th October 2020
Abstract
Receptor function is traditionally controlled from the orthosteric binding site of G-protein coupled receptors. Here, we show that the functional activity and signalling of human dopamine D2 and D3 receptor ligands can be fine-tuned from the extracellular secondary binding pocket (SBP) located far from the signalling interface suggesting optimization of the SBP binding part of bitopic ligands might be a useful strategy to develop GPCR ligands with designed functional and signalling profile.
G-protein coupled receptors (GPCRs) are considered important therapeutic targets due to their pathophysiological significance and pharmacological relevance.1 The large number of GPCR ligands reported have different efficacy profiles for specific signalling pathways and their various functional activity profiles are associated with diverse therapeutic effects. Aminergic GPCRs are considered as clinically validated drug targets in multiple psychiatric indications. Arylpiperazines and arylpiperidines mimicking aminergic neurotransmitters were confirmed as orthosteric ligands by competitive binding experiments. Most of the atypical antipsychotics contain these privileged structures located at the same orthosteric binding pocket (OBP) as seen in the recently published structure of the D2 antagonist risperidone – receptor complex.2 During the last years, partial agonists attracted particular attention since they are usually better tolerated than full agonists or antagonists.3 Third generation atypical antipsychotics such as aripiprazole4 and cariprazine5 are both partial agonists at dopamine hD2 and hD3 receptors (Fig. 1) and they share the orthosteric dichlorophenyl-piperazine substructure. The risperidone-D2 receptor structure confirmed that the drug also occupies an extracellular binding site. This secondary binding pocket (SBP) is much less conserved and suggested to contribute significantly to the affinity and selectivity of bitopic ligands occupying both the OBP and the SBP.6 Consequently, the markedly different receptor profile of cariprazine7 can be traced back to its unique aliphatic SBP moiety that differs from that applied in any other atypical antipsychotics.
Functional activity of GPCR ligands has traditionally been connected to structural moieties bound in the OBP. Using specifically designed bitopic ligands we suggest that functional profiles can be controlled from the secondary site. This opportunity has been shown on the dopamine hD2 and hD3 receptors that are targets for next generation treatment options for schizophrenia, mania, bipolar disorders and depression. Cariprazine5 (2), the D3 receptor preferring D2/D3 partial agonist showed efficacy on both positive and negative symptoms of schizophrenia8 and it has been approved for the full spectrum of bipolar disorders.9
As a part of the mechanistic studies10 on cariprazine action we systematically compared the effect of allosteric modifications on the binding and functional activity of 9 designed, synthesized and tested ligands (Fig. 1) containing three OBP and two SBP motifs together with the OBP binding cores.
First, we tested all ligands in binding assays on the hD2 and hD3 receptors; the pKi values and the selectivity are reported in Table S1 (ESI†). Extending the ligands towards the extracellular vestibule enhances the affinity of all ligands both in the hD2 and hD3 receptors. Even though none of these ligands are considered highly selective, in accordance with previous studies11,12 introducing allosteric interactions helps to fine-tune subtype-selectivity as well as affinity, however, the improvement of the selectivity ratio depends on both OBP and SBP binding moieties.
Biased agonists might offer a potential therapeutic route to specifically interfere with malfunctioning signalling pathways while avoid adverse effects caused by the non-selective activation of downstream signalling. Therefore, developing ligands with specific signalling profiles are in the forefront of drug discovery.13 To demonstrate the pathway specific control of the functional outcome from the extracellular vestibule we carried out functional assays on both the G-protein (Go) mediated and the β-arrestin mediated pathways of the hD2 and hD3 receptor.
On the Go mediated signalling pathway (Table 1) 15 is a partial agonist on both of the receptors. The N,N-dimethyl head-group (SBP1) resulted in a partial agonist (cariprazine, 2) on both receptors with efficacy (Emax – maximal achievable response) values of 77.4% and 27%, while the pEC50 increased to 8.85 and 8.58, respectively. The addition of the O-tBu head group (SBP2) leads to a full agonist (3)5. The improvement in potency was more significant on the hD2 receptor reaching the pEC50 of 8.64 as compared to the pEC50 measured on the hD3 (8.09). With changing the OBP binding moiety to 2-methoxyphenyl piperazine (OBP2, 4), no significant change in the functional activity was observed, the 4 OPB core and the bitopic ligands (5 and 6) were all partial agonist. Comparing this observation to that found with OBP1 (1) and OBP3 (7) moieties it suggests that OBP and SBP effects might be cooperative. At hD2 receptors we found that both their potency and efficacy were much improved. Interestingly, the potency improvement at hD3 receptors was not coupled to significant changes in Emax values. Similarly to OBP1(1), the N,N-dimethyl head group (8) did not change the functional outcome of 75, both ligands showed antagonist effect on the G-protein mediated pathway at hD2 and hD3 receptors, however the potency increased from pIC50 = 4.72 to 6.10 and pIC50 = 5.01 to 7.56, respectively with the increment of the ligand size. The SBP2 group, however, tuned the function of OBP3 (7) resulting in a weak partial agonist (9)5 at both hD2 and hD3 receptors. At hD2 receptors we found 9 less potent but more efficacious as compared to hD3 receptors, however, differences in Emax values were statistically significant (Student's t-test, p = 0.0106) on both receptors.
In contrast with the G-protein mediated signalling we observed smaller effects on the β-arrestin mediated pathways (Table 2). All three ligands containing OBP1 displayed partial agonism on β-arrestin at hD2 and hD3 receptors, although the addition of the two SBP moieties significantly increased the pEC50 values. Similar to the G-protein mediated pathway, SBP2 introduced the biggest increase in the Emax values up to 26% for hD2 and 61% for hD3, while SBP1 only displayed 13.9% and 32% in the case of 2. Interestingly, this suggests that cariprazine, which displays significant bias towards the G-protein mediated pathway in the hD2 receptor,14 shows a more balanced behaviour in the hD3.
All the OBP2 containing ligands (4–6) turned from being partial agonist to antagonist on β-arrestin compared to that found on the G-protein mediated pathway. This observation shows the well-known characteristic effect of OBP binders on the functional profile. All OBP3 containing ligands (7–9) remained antagonists, however with increased pIC50 values in the case of SBP1 and SBP2 moieties. In general, efficacy data measured on both receptors in both modalities followed similar trends as receptor affinities (see ESI†).
Functional modulation from the orthosteric site was already demonstrated for aminergic GPCRs.16 Also for the hD2 receptor, several biased ligands were described and developed in the last few years,14,17–19 however the hD3 receptor was less targeted in this field13,20 and the functional effect of SBP binding moieties remained unclear for both of the targets. Our data clearly show that the functional activity on the hD2 and hD3 receptors can be pathway specifically modulated from the extracellular vestibule by introducing different SBP motifs to the ligands.
In the case of the β-arrestin signalling pathway the functional profile is controlled from the orthosteric site, which is in agreement with the traditional viewpoint. In the G protein mediated pathway, however, we were able to tune the signalling from the secondary binding pocket, therefore we focused our structural investigations to the G-protein mediated pathway. To understand the structural basis of the observed functional effects we performed a series of molecular dynamics simulations on both receptors and analysed the structural behaviour of the 6 bitopic ligands in the light of mutational data. Investigating the corresponding Go coupled signalling complexes we built homology models for the hD2 and hD3 receptors. All partial or full agonists were modelled in the activated signalling complex, while the antagonist was modelled in the inactive hD2 and hD3 experimental structures.
In the G-protein activated hD2 models, the overall orientation of agonists and the antagonist are similar. All ligands are anchored to the receptor by a strong, direct H-bond with the conserved Asp1143.32, while hydrophobic interactions also add to the stabilization in the orthosteric pocket. In the extracellular vestibule, direct and water mediated hydrogen bonds form to different extent with Glu952.65 and Trp100ECL1 (Fig. S2a, ESI†). Even though we see some preference in hydrogen bond formation towards Trp100ECL1 instead of Glu952.65 in the case of the antagonist 8, we could not detect any specific interactions characteristic only to either agonists or the antagonist.
Available mutational data for key residues (Asp1143.32Ala, Phe3896.51Ala, Phe3896.51Ala, Glu952.65Ala) for different ligands like dopamine, risperidone, MLS1547, SB269652 support that these interactions are essential for binding and functional activity.2,21–23 Likewise, mutation of Trp100ECL1Ala increases the association and dissociation of antagonist ligand risperidone.24
The hD3 simulations lead to a very similar result (Fig. S2b, ESI†). One important difference is that in the hD3 structure Trp96ECL1 is positioned away from the secondary binding site therefore not available for hydrogen bond formation. Instead we observe interaction with Cys181ECL2 in the case of the antagonist, however, in less than 40% of the frames, while the main interaction partner in the secondary pocket remains Glu902.65 for both the agonists and the antagonist.
Lack of stable, agonist specific interactions in the extracellular vestibule propose the modulation through a different mechanism. Since experimental data suggested cooperative effects, we hypothesized that SBP binders might influence the functional activity through the positioning of the orthosteric binding motifs. As shown in our experimental results, changing the SBP only causes change in function when the OBP makes it possible. Therefore, we focused on finding differences between agonists and the antagonist in the OBP. As based only on the cumulated interaction diagram we were unable to see significantly different patterns, we further investigated the trajectories with cluster analysis focusing on the most interesting ligands 9 and 8 revealing that the OBP atoms of these ligands occupy different 3D positions (Fig. 2). This is in line with recent D2 structures showing that the OBP of haloperidol and risperidone, both antagonists, overlap; however, both extend to a lower subpocket compared to the agonist bromocriptine2,25,26 and the plane of the ring system rotates around 35 degrees (Fig. S3, ESI†); we observed a similar rotation in our MD studies (Fig. 2).
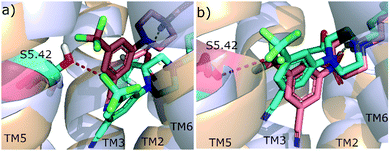
The MDs revealed that the halogen atoms of the agonists in hD3 are positioned closer to Ser1925.42 compared to the antagonist 8, however this difference is not significant and was less visible in hD2, especially for 9 (Fig. S4, ESI†). This is likely in connection with 9 showing a very weak partial agonist behaviour. This Ser5.42 forms H-bonds with several ligands in aminergic GPCR X-ray structures11,27,28 and is also expected to be involved in dopamine binding. In the case of 2, 3 and 9 a halogen bond formation with Ser5.42 might add to the stabilization of the position (Fig. 2) likely in connection with the alteration of the signalling profile. Although this is not the only interaction driving the functional response, but it indicates that 8 and 9 prefers different conformations in the OBP.
Functional modulation through a differently positioned orthosteric site instead of specific SBP interactions is also in line with 5-HT2B X-ray structure of LSD29 and ergotamine,30 which share the orthosteric binding motif but display different signalling profile (Fig. S7, ESI†). In line with the concept, both salmeterol and salbutamol display similar signalling profile in the β2 receptor, while their shared saligenin etanolamine pharmacophore completely overlaps in the orthosteric site based on their X-ray structures (β2 for salmeterol11 and β1 for salbutamol31). Recent studies on the hD2 and AT1R receptor also reveal that the positioning of the orthosteric motif influences the signalling profile, β-arrestin biased ligands tend to bind closer to the ECL2 loop compared to balanced/G-protein biased ligands in the examined cases.16,32
In this work, we have systematically investigated how introducing interactions in the extracellular SBP site can tune the functional profile of the ligands in hD2 and hD3 receptors. First, investigating the Go and β-arrestin pathways we show that the functional activity of hD2 and hD3 receptors can be pathway specifically modulated from the extracellular vestibule by introducing different secondary site binding motifs to the ligands. Second, we were able to tailor the functional profile of the ligands presenting antagonists, partial and full agonists as well. Third, we provide a plausible structural explanation for the observed effects based on molecular dynamics simulations. Together with mutational data, these simulations revealed that the SBP interactions do not directly modulate the G-protein signalling in these ligands; rather they influence the positioning of the orthosteric binding motif. These findings together might help designing high affinity and selective GPCR ligands with desired functional profiles.
The authors are grateful for the support of the National Brain Research Program (2017-1.2.1-NKP-2017-00002), for KIFÜ for providing access to the Hungarian HPC Service and to András Visegrády for fruitful discussions.
Conflicts of interest
K. D. K., B. K., F. H., D. K. and I. G. are the employees of Gedeon Richter plc, the producer of cariprazine. I. G. and G. M. K. contributed to the discovery of cariprazine.Notes and references
- A. S. Hauser, M. M. Attwood, M. Rask-Andersen, H. B. Schiöth and D. E. Gloriam, Nat. Rev. Drug Discovery, 2017, 16, 829–842 CrossRef CAS.
- S. Wang, T. Che, A. Levit, B. K. Shoichet, D. Wacker and B. L. Roth, Nature, 2018, 555, 269–273 CrossRef CAS.
- B. T. Zhu, J. Theor. Biol., 1996, 181, 273–291 CrossRef CAS.
- K. D. Burris, T. F. Molski, C. Xu, E. Ryan, K. Tottori, T. Kikuchi, F. D. Yocca and P. B. Molinoff, J. Pharmacol. Exp. Ther., 2002, 302, 381–389 CrossRef CAS.
- É. Ágai-Csongor, G. Domány, K. Nógrádi, J. Galambos, I. Vágó, G. M. Keserű, I. Greiner, I. Laszlovszky, A. Gere, É. Schmidt, B. Kiss, M. Vastag, K. Tihanyi, K. Sághy, J. Laszy, I. Gyertyán, M. Zájer-Balázs, L. Gémesi, M. Kapás and Z. Szombathelyi, Bioorg. Med. Chem. Lett., 2012, 22, 3437–3440 CrossRef.
- M. Michino, T. Beuming, P. Donthamsetti, A. H. Newman, J. A. Javitch and L. Shi, Pharmacol. Rev., 2015, 67, 198–213 CrossRef CAS.
- B. Kiss, A. Horváth, Z. Némethy, É. Schmidt, I. Laszlovszky, G. Bugovics, K. Fazekas, K. Hornok, S. Orosz, I. Gyertyán, É. Ágai-Csongor, G. Domány, K. Tihanyi, N. Adham and Z. Szombathelyi, J. Pharmacol. Exp. Ther., 2010, 333, 328–340 CrossRef CAS.
- G. Németh, I. Laszlovszky, P. Czobor, E. Szalai, B. Szatmári, J. Harsányi, Á. Barabássy, M. Debelle, S. Durgam, I. Bitter, S. Marder and W. W. Fleischhacker, Lancet, 2017, 389, 1103–1113 CrossRef.
- S. M. Stahl, S. Laredo and D. A. Morrissette, Ther. Adv. Psychopharmacol., 2020, 10, 1–11 Search PubMed.
- A. Frank, D. J. Kiss, G. M. Keserű and H. Stark, Sci. Rep., 2018, 8, 12509 CrossRef.
- M. Masureel, Y. Zou, L.-P. Picard, E. van der Westhuizen, J. P. Mahoney, J. P. G. L. M. Rodrigues, T. J. Mildorf, R. O. Dror, D. E. Shaw, M. Bouvier, E. Pardon, J. Steyaert, R. K. Sunahara, W. I. Weis, C. Zhang and B. K. Kobilka, Nat. Chem. Biol., 2018, 14, 1059–1066 CrossRef CAS.
- A. H. Newman, T. Beuming, A. K. Banala, P. Donthamsetti, K. Pongetti, A. LaBounty, B. Levy, J. Cao, M. Michino, R. R. Luedtke, J. A. Javitch and L. Shi, J. Med. Chem., 2012, 55, 6689–6699 CrossRef CAS.
- L. Tan, W. Yan, J. D. McCorvy and J. Cheng, J. Med. Chem., 2018, 61, 9841–9878 CrossRef CAS.
- J. Shonberg, C. K. Herenbrink, L. López, A. Christopoulos, P. J. Scammells, B. Capuano and J. R. Lane, J. Med. Chem., 2013, 56, 9199–9221 CrossRef CAS.
- Y. Gao, S. Peterson, B. Masri, M. T. Hougland, N. Adham, I. Gyertyán, B. Kiss, M. G. Caron and R. S. El-Mallakh, Pharmacol. Res. Perspect., 2014, 3, e00073 Search PubMed.
- J. D. McCorvy, K. V. Butler, B. Kelly, K. Rechsteiner, J. Karpiak, R. M. Betz, B. L. Kormos, B. K. Shoichet, R. O. Dror, J. Jin and B. L. Roth, Nat. Chem. Biol., 2017, 14, 126–134 CrossRef.
- J. A. Allen, J. M. Yost, V. Setola, X. Chen, M. F. Sassano, M. Chen, S. Peterson, P. N. Yadav, X. Huang, B. Feng, N. H. Jensen, X. Che, X. Bai, S. V. Frye, W. C. Wetsel, M. G. Caron, J. A. Javitch, B. L. Roth and J. Jin, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 18488–18493 CrossRef CAS.
- X. Chen, M. F. Sassano, L. Zheng, V. Setola, M. Chen, X. Bai, S. V. Frye, W. C. Wetsel, B. L. Roth and J. Jin, J. Med. Chem., 2012, 55, 7141–7153 CrossRef CAS.
- B. Männel, M. Jaiteh, A. Zeifman, A. Randakova, D. Möller, H. Hübner, P. Gmeiner and J. Carlsson, ACS Chem. Biol., 2017, 12, 2652–2661 CrossRef.
- D. Möller, R. C. Kling, M. Skultety, K. Leuner, H. Hübner and P. Gmeiner, J. Med. Chem., 2014, 57, 4861–4875 CrossRef.
- W. Cho, L. P. Taylor, A. Mansour and H. Akil, J. Neurochem., 1995, 65, 2105–2115 CrossRef CAS.
- N. Tschammer, M. Dörfler, H. Hübner and P. Gmeiner, ACS Chem. Neurosci., 2010, 1, 25–35 CrossRef CAS.
- J. R. Lane, P. Donthamsetti, J. Shonberg, C. J. Draper-Joyce, S. Dentry, M. Michino, L. Shi, L. López, P. J. Scammells, B. Capuano, P. M. Sexton, J. A. Javitch and A. Christopoulos, Nat. Chem. Biol., 2014, 10, 745–752 CrossRef CAS.
- J. R. Lane, A. M. Abramyan, P. Adhikari, A. C. Keen, K. H. Lee, J. Sanchez, R. K. Verma, H. D. Lim, H. Yano, J. A. Javitch and L. Shi, eLife, 2020, 9, e52189 CrossRef CAS.
- L. Fan, L. Tan, Z. Chen, J. Qi, F. Nie, Z. Luo, J. Cheng and S. Wang, Nat. Commun., 2020, 11, 1–11 Search PubMed.
- J. Yin, K. Y. M. Chen, M. J. Clark, M. Hijazi, P. Kumari, X. chen Bai, R. K. Sunahara, P. Barth and D. M. Rosenbaum, Nature, 2020, 584, 125 CrossRef CAS.
- E. Y. T. Chien, W. Liu, Q. Zhao, V. Katritch, G. W. Han, M. A. Hanson, L. Shi, A. H. Newman, J. A. Javitch, V. Cherezov and R. C. Stevens, Science, 2010, 330, 1091–1095 CrossRef CAS.
- A. M. Ring, A. Manglik, A. C. Kruse, M. D. Enos, W. I. Weis, K. C. Garcia and B. K. Kobilka, Nature, 2013, 502, 575–579 CrossRef CAS.
- D. Wacker, S. Wang, J. D. McCorvy, R. M. Betz, A. J. Venkatakrishnan, A. Levit, K. Lansu, Z. L. Schools, T. Che, D. E. Nichols, B. K. Shoichet, R. O. Dror and B. L. Roth, Cell, 2017, 168, 377–389.e12 CrossRef CAS.
- D. Wacker, C. Wang, V. Katritch, G. W. Han, X.-P. Huang, E. Vardy, J. D. McCorvy, Y. Jiang, M. Chu, F. Y. Siu, W. Liu, H. E. Xu, V. Cherezov, B. L. Roth and R. C. Stevens, Science, 2013, 340, 615–619 CrossRef CAS.
- T. Warne, R. Moukhametzianov, J. G. Baker, R. Nehmé, P. C. Edwards, A. G. W. Leslie, G. F. X. Schertler and C. G. Tate, Nature, 2011, 469, 241–244 CrossRef CAS.
- L. M. Wingler, M. A. Skiba, C. McMahon, D. P. Staus, A. L. W. Kleinhenz, C. M. Suomivuori, N. R. Latorraca, R. O. Dror, R. J. Lefkowitz and A. C. Kruse, Science, 2020, 367, 888–892 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc05532h |
| This journal is © The Royal Society of Chemistry 2020 |