 Open Access Article
Open Access ArticleUnravelling the cellular emission fingerprint of the benchmark G-quadruplex-interactive compound Phen-DC3†
Marco
Deiana
 ,
Jan
Jamroskovic
,
Jan
Jamroskovic
 ,
Ikenna
Obi
,
Ikenna
Obi
 and
Nasim
Sabouri
and
Nasim
Sabouri
 *
*
Department of Medical Biochemistry and Biophysics, Umeå University, Umeå 90187, Sweden. E-mail: nasim.sabouri@umu.se
First published on 16th October 2020
Abstract
Phen-DC3 is among the most commonly used G-quadruplex (G4)-stabilizers in vitro and in cells. Here, we show that the G4-interactive binding interactions enable one to tune the optical properties of Phen-DC3 allowing the detection of G4 structures in cancer cells. This work opens up new directions for the use of Phen-DC3 as a selective G4 fluorescent reporter.
G-Quadruplexes (G4s) are non-canonical four-stranded DNA and RNA structures formed by co-planar guanine-quartets stabilized through Hoogsteen-type hydrogen bonding and centrally octa-coordinated by monovalent cations.1 Recent evidences suggest the involvement of G4 structures in key biological functions, such as replication, transcription, translation, and telomere maintenance.2 Moreover, bioinformatics and sequencing studies have highlighted the widespread distribution of evolutionary conserved putative G4 sites in the genomes of humans and different model organisms.2–5 Collectively, these findings have stimulated research probing G4-associated mechanisms and consequent opportunities for therapeutic intervention. This has led to the development of small-molecule therapeutics that can selectively bind, detect and stabilize these non-B DNA globular structures.2 The majority of current G4 binders contain polyaromatic heterocyclic ring systems functionalized with flexible substituents.2 This simple but efficient molecular design approach ensures both π–π stacking interactions between the terminal G-quartets and the central ligand's core and additionally electrostatic or ionic attractions between the pendant ligand's arms and the G-quadruplex’s grooves. The phenanthroline dicarboxamide bisquinolinium compound, Phen-DC3, is one of the most potent G4-binders reported so far and is used as a benchmark compound in a wide number of studies to probe G4 formation in vitro and in cellular systems.6–16 Its overall crescent-like bent shape ensures an extensive overlap with the guanines on the terminal G-quartets and prevents intercalation between the Watson–Crick base pairs in duplex DNA, therefore limiting off-target binding interactions (Fig. 1).2
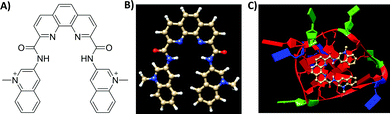 | ||
| Fig. 1 (A and B) Chemical structure of Phen-DC3 and related ball-and-stick model. (C) Solution structure of c-MYC Pu24T–Phen-DC3 complex (PDB code: 2MGN).6 Guanine, thymine and adenine bases are shown in red, green and blue, respectively. | ||
The weak fluorescence emission of Phen-DC3 has been proven to be the major bottleneck of this compound to diagnostic transformation.8 Indeed, in situ post-labeling by copper(I)-catalyzed 1,3-dipolar azide–alkyne cycloaddition (CuAAC) and strain-promoted azide–alkyne cycloaddition (SPAAC) click reactions were used to probe the cellular emission fingerprint of Phen-DC3-based compounds in cancer cells.8 These methods did not provide unambiguous evidences of Phen-DC3's cellular localization and showed inherent drawbacks including the need for ad hoc Phen-DC3 structural optimization through laborious multi-step synthetic procedures as well as the possibility of artefactual staining by intermediate species when using ligands post-labeled by CuAAC. Therefore, there is still an urgent need to decipher the intracellular localization of Phen-DC3 and answer open questions concerning its ability to track DNA and RNA G4 structures in cancer cells. Herein, we shed light on the ability of Phen-DC3 to be used as an intrinsically fluorescent probe and we unravel its in-cell fate in the particular context of G4 recognition, opening new routes to advance future studies of G4 biology.
The absorption spectrum of Phen-DC3 was monitored by UV-VIS spectroscopy and showed an intense well-structured band in the UV-spectral region with the maximum (λmax) centered at 350 nm and extinction coefficient ε ∼ 2.0 × 104 M−1 cm−1 (Fig. 2). The addition of an equimolar concentration of c-MYC Pu24T, a widely used model parallel G4 structure derived from the oncogenic c-MYC promoter6 (Fig. S1, ESI†), to Phen-DC3 resulted in the concomitant bathochromic shift of its absorption maximum (∼355 nm) and the appearance of a long-wavelength tail that extended over the blue region of the spectrum (Fig. 2).
Similar spectral changes were observed for Phen-DC3 complexed with a hybrid telomeric G4-forming sequence, Tel-22, and a parallel RNA G4 structure, 4G3U3 (Fig. S1–S3, ESI†).9 This evolution pattern suggests that Phen-DC3 may be sensitive to viscosity and/or polarity changes. To test this hypothesis, we recorded the UV-VIS absorption spectra of Phen-DC3 in pure water as well as in a highly viscous H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol (50
glycerol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) binary mixture (Fig. S4, ESI†). No relevant spectral changes were observed in the absorption band of Phen-DC3 ruling out spectral modifications associated with viscosity changes. On the other hand, Phen-DC3, in methanol (MeOH), showed similar spectral changes to those observed with DNA or RNA G4 structures (Fig. S5, ESI†). In particular, the appearance of a long-wavelength tail suggests that Phen-DC3 is sensitive to the matrix polarity and its accommodation within the hydrophobic G4 scaffold well matched a relative polarity value (
50) binary mixture (Fig. S4, ESI†). No relevant spectral changes were observed in the absorption band of Phen-DC3 ruling out spectral modifications associated with viscosity changes. On the other hand, Phen-DC3, in methanol (MeOH), showed similar spectral changes to those observed with DNA or RNA G4 structures (Fig. S5, ESI†). In particular, the appearance of a long-wavelength tail suggests that Phen-DC3 is sensitive to the matrix polarity and its accommodation within the hydrophobic G4 scaffold well matched a relative polarity value (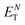 ) of 0.76.17 We also recorded the fluorescence spectrum of Phen-DC3, and upon photoexcitation at 350 nm, an emission maximum (λem) at ∼405 nm was observed (Fig. 2). The addition of an equimolar concentration of c-MYC Pu24T significantly quenched and red-shifted the emission band of Phen-DC3 (Fig. 2). Similar optical changes were observed for Phen-DC3 complexed with Tel-22 or 4G3U3 (Fig. S6, ESI†). Due to the resulting low molecular brightness of the Phen-DC3–G4 complexes, excitation at 405 nm did not provide enough sensitivity to detect the associated luminescence band of Phen-DC3's bound states by using conventional steady-state emission spectroscopy. Nevertheless, this outcome did not preclude its implementation as a cellular G4-selective fluorescent reporter in confocal laser scanning microscopy (CLSM, vide infra).
) of 0.76.17 We also recorded the fluorescence spectrum of Phen-DC3, and upon photoexcitation at 350 nm, an emission maximum (λem) at ∼405 nm was observed (Fig. 2). The addition of an equimolar concentration of c-MYC Pu24T significantly quenched and red-shifted the emission band of Phen-DC3 (Fig. 2). Similar optical changes were observed for Phen-DC3 complexed with Tel-22 or 4G3U3 (Fig. S6, ESI†). Due to the resulting low molecular brightness of the Phen-DC3–G4 complexes, excitation at 405 nm did not provide enough sensitivity to detect the associated luminescence band of Phen-DC3's bound states by using conventional steady-state emission spectroscopy. Nevertheless, this outcome did not preclude its implementation as a cellular G4-selective fluorescent reporter in confocal laser scanning microscopy (CLSM, vide infra).
In order to further examine the interaction of c-MYC Pu24T DNA and Phen-DC3, we determined Phen-DC3's stabilization properties of c-MYC Pu24T and a non-G4 DNA control. We performed the Taq-polymerase stop assay, a type of primer extension assay that determines DNA replication progression and pausing at single nucleotide resolution.18 We found that increasing the concentration of Phen-DC3 induced an increased G4 stabilization response, which resulted in the inhibition of DNA replication and a clear pausing site one nucleotide before the first G-tract (Fig. 3A). The pausing site on the c-MYC Pu24T template was already detected at 8 nM, while the synthesis of the non-G4 template was unaffected at this concentration. High concentrations of Phen-DC3 (2 and 5 μM) caused non-specific inhibition of DNA replication on both the G4 and non-G4 DNA templates showing a wide window of selectivity between G4 and non-G4 DNA structures (Fig. 3A and B). To further investigate the ability of Phen-DC3 to induce G4 stabilization on the c-MYC Pu24T template, we monitored the replication progression by primer extension assay over time (Fig. 3C and D). G4-Mediated formation of pausing sites was visible after 1 minute in both untreated and Phen-DC3-treated samples. With time progression, the pausing site in the control sample slowly disappeared, showing that allowing multiple attempts by the DNA polymerase to bypass the G4 structure induces G4 destabilization and eventually results in bypass of the G4 structures by the DNA polymerase. However, in Phen-DC3-treated samples, the pausing site was detected even after 60 mins of incubation and the synthesis of the full-length product was completely inhibited (Fig. 3C and D), indicating that Phen-DC3 is indeed a highly potent G4 stabilizer.
The G4-interactive binding properties of Phen-DC3 along with its optical changes characterized by a long-wavelength absorption tail and a red-shifted emission maximum, prompted us to study the cellular localization of Phen-DC3 in human cervical carcinoma (HeLa) cells by CLSM. Indeed, the spectral changes of Phen-DC3 in the presence of DNA and RNA G4s that led to a considerable increase in absorption at 405 nm, matches well to the excitation laser line setting used in CLSM (Fig. 2 and Fig. S2, S3, ESI†). This “double-check” detection mechanism based on binary conditions (OFF–ON absorption states at 405 nm) exclusively enables the cellular visualization of the active fluorescent bound states (Phen-DC3–G4 complexes) limiting issues related to off-target effects and low signal-to-noise ratio. A limit of detection (LOD) of 582 ± 9 nM was calculated by monitoring the changes in the optical density at 405 nm as a function of G4 concentration (Fig. S7, ESI†). This LOD value, even if slightly higher compared to the most sensitive light-up G4 probes reported so far, still falls within the nanomolar range and provides possibilities to probe G4 formation in a cellular context.16,19,20 The treatment of PFA-fixed HeLa cells with Phen-DC3 showed an intense fluorescence signal in the cytoplasmic and nuclear regions with clear peaks in the subnuclear compartments whose appearance is compatible with nucleoli (Fig. 4A and B).16,19,21,22 The observed staining pattern was independent of the fixation method, as Phen-DC3-treated cells fixed with MeOH showed a similar staining pattern but with reduced cytoplasmic signal (Fig. S8, ESI†).
Under the same experimental conditions used to image the Phen-DC3-stained cells, untreated cells did not show any fluorescence signal (Fig. 4A), showing that the fluorescence observed in Phen-DC3-treated cells is not due to auto-fluorescence of cells. Together, these data show that Phen-DC3 can be visualized in fixed cells (Fig. 4A, B and Fig. S8, ESI†).
The G4-specific BG4 antibody showed the occurrence of G4 structures at both cytoplasmic and nuclear regions indicating cellular formation of RNA and DNA G4s, respectively.23,24 Phen-DC3 targets both RNA and DNA G4 structures with nanomolar dissociation constants.9,25 Therefore, the staining pattern of Phen-DC3 may be directly related to its ability to target RNA G4s in the cytoplasm and DNA G4s in the nucleus. To elucidate the nature of the main cellular binding targets of Phen-DC3, we pretreated the cells with either RNase or DNase before incubation with Phen-DC3. RNase pretreatment did not affect the nuclear staining pattern of Phen-DC3 but reduced its cytoplasmic (p = 1.1 × 10−56) and nucleolar signal, supporting binding of Phen-DC3 to RNA molecules in these two cellular regions (Fig. 4A, B and Fig. S8, ESI†). RNase treatment caused almost complete disappearance of the Phen-DC3-associated fluorescence signal in the nucleoli. Conversely, DNase treatment significantly reduced the nuclear staining (p = 7.4 × 10−30) and to some extent the nucleolar signal (p = 5.7 × 10−12) by Phen-DC3, but its cytoplasmic signal was almost unperturbed (Fig. 4A, B and Fig. S8, ESI†). Together, these RNase/DNase-treated cell data suggest that Phen-DC3 mainly targets RNA G4s in both cytoplasmic and nucleolar regions and DNA G4s in the nucleus. In order to confirm that only the bound Phen-DC3–DNA/RNA G4s states were responsible for the fluorescence emission observed in cells, we pre-treated the cells simultaneously with both DNase and RNase (Fig. S8, ESI†). No fluorescence signal was observed for Phen-DC3 under these experimental conditions, suggesting that the detected fluorescence observed in cells is due to Phen-DC3 being in complex with DNA and RNA G4s.
In order to further probe the sensing ability of Phen-DC3 when targeting G4 structures in cells, we performed two independent fluorescence displacement assays using the well-known G4-binders BRACO-192 and TmPyP42 (Fig. 4C and D). Direct competition performed by adding BRACO-19 to Phen-DC3-pretreated cells showed a clear reduction of the cytoplasmic and nucleolar Phen-DC3 fluorescence signal (p = 2.2 × 10−21), indicating the effective competition between these two molecules for the same G4-sites, mostly at the RNA level (Fig. 4C and D). TmPyP4 is a porphyrin-based fluorescent duplex/G4 binder, and its absorption and emission spectra are more red-shifted than Phen-DC3. Cells treated with TmPyP4 demonstrated a similar staining pattern as Phen-DC3. Indeed, the addition of Phen-DC3 to cells pretreated with TmPyP4, resulted in an overall fluorescence signal reduction (p = 6.1 × 10−57) at both the extranuclear and nuclear level, demonstrating the high degree of co-localization between these two G4-binders (Fig. 4C and D). The remaining weak nuclear fluorescence signal that still occurred upon Phen-DC3-mediated TmPyP4 displacement may be ascribed to the well-known ability of the porphyrin ligand to intercalate duplex DNA, making these DNA sites highly inaccessible to Phen-DC3.2
Non-cancerous human embryonic kidney (HEK 293) cells were also used to ascertain that the Phen-DC3 emission recorded was not strictly correlated to the cell-type used (Fig. S9, ESI†). Under the same experimental conditions used to image HeLa cells, HEK 293 cells treated with Phen-DC3 showed a very similar staining pattern with most of the associated signal confined within the nucleolar sites. Direct quantitative comparison of the G4 levels present in these two cell lines was not performed because the two cell types are not derived from the same tissue, and therefore the G4 levels cannot be compared.
In conclusion, we have reported for the first time, the cellular localization of one of the most potent and widely used G4 ligands, Phen-DC3. We demonstrate that Phen-DC3 can be used to image cells without the need of any chemical modification. A set of cellular-based fluorescence experiments clearly showed the sensing ability of Phen-DC3 when targeting RNA G4s in both the cytoplasm and nucleoli, and DNA G4s in the nucleus. This report opens up new directions and possibilities for the use of Phen-DC3 to track non-canonical DNA structure formations in different cellular contexts.
This work was supported by the Knut and Alice Wallenberg Foundation (KAW2015-0189), the Swedish Research Council (VR-MH 2018-02651), and the Swedish Cancer Society (CAN19 0126). MD was supported by a fellowship from the MIMS Excellence by Choice Postdoctoral Program. We thank Dr Tamilselvi Shanmugam and Dr Irene Martinez Carrasco for their expert advice regarding microscopy. The authors thank the Biochemical Imaging Center at Umeå University and the National Microscopy Infrastructure (VR-RFI 2016-00968). We thank Dr Paulina Wanrooij for the kind gift of the HEK 293 cells.
Conflicts of interest
There are no conflicts to declare.Notes and references
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS.
- S. Neidle, J. Med. Chem., 2016, 59, 5987–6011 CrossRef CAS.
- V. S. Chambers, G. Marsico, J. M. Boutell, M. Di Antonio, G. P. Smith and S. Balasubramanian, Nat. Biotechnol., 2015, 33, 877–881 CrossRef.
- G. Marsico, V. S. Chambers, A. B. Sahakyan, P. McCauley, J. M. Boutell, M. Di Antonio and S. Balasubramanian, Nucleic Acids Res., 2019, 47, 3862–3874 CrossRef CAS.
- N. Sabouri, J. A. Capra and V. A. Zakian, BMC Biol., 2014, 12, 101 CrossRef.
- W. J. Chung, B. Heddi, F. Hamon, M.-P. Teulade-Fichou and A. T. Phan, Angew. Chem., Int. Ed., 2014, 53, 999–1002 CrossRef CAS.
- A. De Cian, E. DeLemos, J.-L. Mergny, M.-P. Teulade-Fichou and D. Monchaud, J. Am. Chem. Soc., 2007, 129, 1856–1857 CrossRef CAS.
- J. Lefebvre, C. Guetta, F. Poyer, F. Mahuteau-Betzer and M.-P. Teulade-Fichou, Angew. Chem., Int. Ed., 2017, 56, 11365–11369 CrossRef CAS.
- K. Halder, E. Largy, M. Benzler, M.-P. Teulade-Fichou and J. S. Hartig, ChemBioChem, 2011, 12, 1663–1668 CrossRef CAS.
- A. Piazza, J.-B. Boulé, J. Lopes, K. Mingo, E. Largy, M.-P. Teulade-Fichou and A. Nicolas, Nucleic Acids Res., 2010, 38, 4337–4348 CrossRef CAS.
- L. T. Gray, E. Puig Lombardi, D. Verga, A. Nicolas, M.-P. Teulade-Fichou, A. Londoño Vallejo and N. Maizels, Cell Chem. Biol., 2019, 26, 1681–1691.e5 CrossRef CAS.
- K. G. Zyner, D. S. Mulhearn, S. Adhikari, S. Martínez Cuesta, M. Di Antonio, N. Erard, G. J. Hannon, D. Tannahill and S. Balasubramanian, eLife, 2019, 8, e46793 CrossRef.
- I. Obi, M. Rentoft, V. Singh, J. Jamroskovic, K. Chand, E. Chorell, F. Westerlund and N. Sabouri, Nucleic Acids Res., 2020, gkaa820, DOI:10.1093/nar/gkaa820.
- M. Wallgren, J. B. Mohammad, K.-P. Yan, P. Pourbozorgi-Langroudi, M. Ebrahimi and N. Sabouri, Nucleic Acids Res., 2016, 44, 6213–6231 CrossRef CAS.
- P. Prorok, M. Artufel, A. Aze, P. Coulombe, I. Peiffer, L. Lacroix, A. Guédin, J.-L. Mergny, J. Damaschke, A. Schepers, C. Cayrou, M. -P. Teulade-Fichou, B. Ballester and M. Méchali, Nat. Commun., 2019, 10, 3274 CrossRef CAS.
- M. Deiana, K. Chand, J. Jamroskovic, I. Obi, E. Chorell and N. Sabouri, Angew. Chem., Int. Ed., 2020, 59, 896–902 CrossRef CAS.
- C. Reichardt, Chem. Rev., 1994, 94, 2319–2358 CrossRef CAS.
- J. Jamroskovic, M. Livendahl, J. Eriksson, E. Chorell and N. Sabouri, Chem. – Eur. J., 2016, 22, 18932–18943 CrossRef CAS.
- M. Deiana, K. Chand, J. Jamroskovic, R. N. Das, I. Obi, E. Chorell and N. Sabouri, Nanoscale, 2020, 12, 12950–12957 RSC.
- M. Zuffo, A. Guédin, E.-D. Leriche, F. Doria, V. Pirota, V. Gabelica, J.-L. Mergny and M. Freccero, Nucleic Acids Res., 2018, 46, e115 CrossRef.
- J. Jamroskovic, M. Doimo, K. Chand, I. Obi, R. Kumar, K. Brännström, M. Hedenström, R. N. Das, A. Akhunzianov, M. Deiana, K. Kasho, S. Sulis Sato, P. L. Pourbozorgi, J. E. Mason, P. Medini, D. Öhlund, S. Wanrooij, E. Chorell and N. Sabouri, J. Am. Chem. Soc., 2020, 142, 2876–2888 CrossRef CAS.
- J. Li, X. Yin, B. Li, X. Li, Y. Pan, J. Li and Y. Guo, Anal. Chem., 2019, 91, 5354–5361 CrossRef CAS.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS.
- G. Biffi, M. Di Antonio, D. Tannahill and S. Balasubramanian, Nat. Chem., 2014, 6, 75–80 CrossRef CAS.
- L. Bonnat, L. Bar, B. Gennaro, H. Bonnet, O. Jarjayes, F. Thomas, J. Dejeu, E. Defrancq and T. Lavergne, Chem. – Eur. J., 2017, 23, 5602–5613 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc05483f |
| This journal is © The Royal Society of Chemistry 2020 |



