Open Access Article
Open Access Article17O NMR spectroscopy as a tool to study hydrogen bonding of cholesterol in lipid bilayers†
Lucy J.
Rowlands
 ab,
Adam
Marks
ab,
Adam
Marks
 b,
John M.
Sanderson
b,
John M.
Sanderson
 c and
Robert V.
Law
c and
Robert V.
Law
 *ab
*ab
aInstitute of Chemical Biology, Imperial College London, Molecular Sciences Research Hub, W12 0BZ London, UK. E-mail: r.law@imperial.ac.uk
bDepartment of Chemistry, Imperial College London, Molecular Sciences Research Hub, W12 0BZ London, UK
cChemistry Department, Durham University, Durham, DH1 3LE, UK
First published on 5th November 2020
Abstract
Cholesterol is a crucial component of biological membranes and can interact with other membrane components through hydrogen bonding. NMR spectroscopy has been used previously to investigate this bonding, however this study represents the first 17O NMR spectroscopy study of isotopically enriched cholesterol. We demonstrate the 17O chemical shift is dependent on hydrogen bonding, providing a novel method for the study of cholesterol in bilayers.
Cholesterol is an important constituent of lipid bilayers both in biological and synthetic membranes. It modifies bilayer fluidity and rigidity and can lead to the formation of phase separated or micro domains.1–3 Additionally, it is a key component of liposomal formulations for drug delivery, as it reduces drug leakage.4 Cholesterol is capable of forming hydrogen bonds with other lipids and water through the hydroxyl (3β-OH) group. Understanding the nature of these bonds is crucial to understanding membrane structure and bilayer physical properties.5,6
In a phospholipid bilayer cholesterol is orientated perpendicular to the plane of the bilayer, with the hydroxyl group aligned with the carbonyl of the ester linkage in phospholipids.7,8 There have been multiple modes of hydrogen bonding proposed, some of which can be seen in Fig. 1. The two main proposed modes of cholesterol to lipid hydrogen bonding: a direct lipid-cholesterol hydrogen bond, and hydrogen bonding through a bridging water molecule.6,9,10
 | ||
| Fig. 1 Diagram of previously proposed models of cholesterol hydrogen bonding, blue lines indicate hypothetical water–bridge bonds and red indicates direct lipid–cholesterol hydrogen bonds.6,9,10 | ||
Hydrogen bonding of cholesterol in membranes has been investigated using several techniques including: infrared and Raman spectroscopy;11–14 molecular dynamics;9,10,15,16 and NMR.16–20 NMR spectroscopy studies are particularly useful due to the detailed molecular picture they can provide. These usually rely on nuclei not directly involved, such as 13C or on atoms that are exchangeable such as in the case of proton NMR.16–1817O NMR spectroscopy has been used to great effect in investigations of hydrogen bonds in both organic small molecule and biological systems.21–24 Observing 17O rather than the 1H nucleus can lead to more accurate results due to the non-exchangeable nature of the oxygen site, and wide chemical shift range.25 However, there have been no reports of cholesterol hydrogen bonding using 17O NMR spectroscopy, mainly owing to the difficulties of observing unenriched cholesterol (17O natural abundance = 0.0373%), and the practicalities of measuring quadrupolar NMR. In this work 17O enriched cholesterol was synthesised using previously published conditions,26 and utilised in solid state 17O NMR spectroscopy studies of hydrogen bonding. This exploits the single oxygen site on the cholesterol to provide information about hydrogen bonding. The chemical shift of 17O cholesterol in three different lipid bilayers was investigated. Under MAS regimes, the chemical shift can be observed to change depending on the other lipid moiety in the bilayer, which indicated the extent of hydrogen bonding within the bilayer.
The isotropic 17O chemical shift of the enriched cholesterol in toluene, acetone, and chloroform are shown in Table 1. Previously, it has been reported that the 17O NMR chemical shift of cholesterol in acetonitrile at 75 °C was 38.8 ppm with a full width half maximum of 0.7 kHz.27 However, this temperature is not physiologically relevant and is often incompatible with some phospholipids. Therefore, the chemical shifts in this instance were measured at room temperature. The chemical shift is consistent with previous reports and shows the 17O NMR chemical shift of cholesterol is between 30–40 ppm with a small solvent dependence. The spectrum of cholesterol in acetone exhibits a narrower line width than in chloroform and toluene. This may be related to cholesterol self-association. Previous reports have indicated that cholesterol can form dimers in chloroform and toluene.28,29 However, acetone is a proton acceptor, and can interrupt self-association by forming hydrogen bonds with cholesterol, leading to smaller line widths.
| Solvent | 17O NMR chemical shift (ppm) | Full width half maximum (kHz) |
|---|---|---|
| Toluene | 31.2 | 2.37 |
| Acetone | 33.3 | 1.30 |
| Chloroform | 35.3 | 2.48 |
The solid state NMR spectrum of the anhydrous cholesterol is shown in Fig. 2. The anhydrous nature of the cholesterol was determined from microscope images (S1, ESI†), as anhydrous crystalline cholesterol forms needle shaped crystals.30 In the NMR spectrum of the anhydrous cholesterol, the characteristic quadrupolar lineshape can be seen. Fitting of this peak reveals the quadrupolar coupling constant (Cq) is 9.1 MHz, the asymmetry constant (η) is 0.76 and the isotropic chemical shift is 34.6 ppm. As this is the only known example of a 17O NMR spectrum of a steroid, there are limited options for comparison spectrum available. Values for the hydroxyl peak on carbohydrate rings found by Sefzig et al. range from 8.76–9.51 MHz for Cq and 0.83–1.00 for η which are similar to the values found in this study.31
 | ||
| Fig. 2 Real (top) and simulated fitted (bottom) 17O NMR spectrum of anhydrous enriched cholesterol spun at 20 kHz. Spinning sidebands are indicated by *. | ||
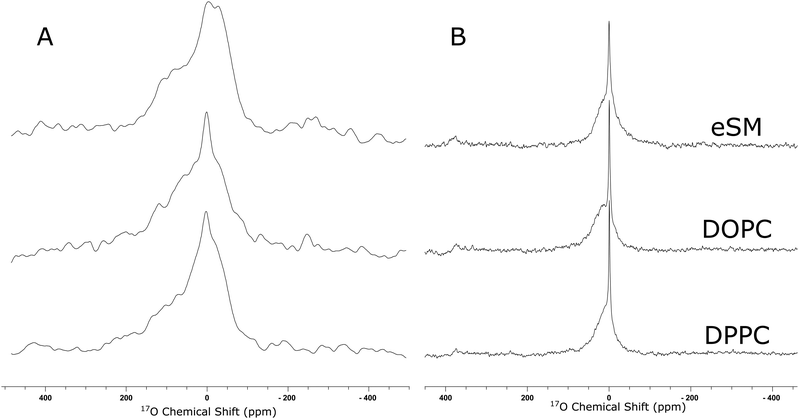
In order to use the enriched cholesterol to understand hydrogen bonding of cholesterol in bilayers, three 50 mol% cholesterol bilayers were made with DPPC, DOPC, and eSM. The static and MAS spectra can be seen in Fig. 3. The static 17O NMR spectra (Fig. 3A) of cholesterol in a bilayer possess an asymmetric line shape overlain with a sharper peak from residual 17O NMR spectrum of water, whereas, the MAS spectra (Fig. 3B) appear as two Lorentzian peaks from water and cholesterol. Cholesterol in a bilayer is a viscous gel, and all lipid bilayer systems used in this work are in the liquid ordered phase.32,33 Within this phase there is lateral motion and axial rotation in the bilayer, meaning that for this specific system, the second order quadrupolar and CSA contributions are minimal. Fitting the static 17O NMR line shapes produces small values of Cq (<1 MHz), and η values of 0 (Table 2). This indicates that the major influence on the line shape is due to a distribution of isotropic chemical shifts because of the different chemical environments of the hydroxyl group. Therefore, upon spinning the loss of the first order quadrupolar and CSA effects leaves a Lorentzian peak.
| δ(iso)/ppm | δ(11)/ppm | δ(22)/ppm | δ(33)/ppm | C q/MHz | η | |
|---|---|---|---|---|---|---|
| eSM | 15.9 | 140.6 | 140.7 | −65.5 | 0.40 | 0 |
| DOPC | 26.8 | 140.3 | 0.8 | −60.7 | 0.93 | 0 |
| DPPC | 25.4 | 152.0 | −21.7 | −54.0 | 0.00 | 0 |
The fitting of the static peaks produces values shown in Table 2. Values of isotropic chemical shift vary according to the other lipid components in the bilayer and are further upfield than the values found from the cholesterol in solvent and the MAS spectrum of the anhydrous crystalline cholesterol alone. Previously, it has been found that the CSA and quadrupolar values of some functional groups vary depending on hydrogen bond length.24,34 However, this has not been characterised for this functional group, and there does not appear to be a trend in this data set.
Fitting the two Lorentzian peaks in the MAS spectra (Fig. 3B) enables discernment of the cholesterol peak and Table 3 shows the chemical shift and full width half maximum of both the cholesterol and water peaks. To indicate a fully hydrated sample, crystalline cholesterol monohydrate was used as a control. The X-ray crystal structure of cholesterol monohydrate has been reported previously as possessing three hydrogen bonds per oxygen atom making it ideal to represent an extensively hydrogen bonded cholesterol molecule.35 The unusually narrow line width of crystalline monohydrate may derive from the crystal structure. In the unit cell of cholesterol monohydrate, the 17O of the hydroxyl has eight inequivalent crystallographic sites.35 This sometimes appears as multiple sites in the MAS NMR. We can only hypothesise that this was the case here, and has manifested as a broad distribution of sites, of which only a few are readily observed.
| Cholesterol | Water | |||
|---|---|---|---|---|
| Chemical shift/ppm | FWHM/kHZ | Chemical shift/ppm | FWHM/kHz | |
| Cholesterol monohydrate | 2.74 | 3.09 | −3.30 | 0.23 |
| eSM | 3.82 | 6.54 | 0.12 | 0.54 |
| DOPC | 18.13 | 6.54 | −0.37 | 0.50 |
| DPPC | 11.42 | 5.59 | −0.43 | 0.40 |
All chemical shift values of cholesterol in bilayers (static and MAS) are further upfield than that of cholesterol in solution and the isotropic chemical shift found from the solid anhydrous cholesterol, closer to the chemical shift of crystalline cholesterol monohydrate. However, there is a difference in chemical shift values found between the static and MAS samples. This is likely due to the poor signal-noise ratio in the static samples causing a lower quality fit and less reliable values. Errors for MAS values can be found in S7 (ESI†). The upfield shift of the NMR signal of the enriched cholesterol in bilayers is attributed to the cholesterol engaging in extensive hydrogen bonding. Whilst cholesterol can form a variety of different types of hydrogen bond when in a phospholipid bilayer, they are predominantly through the proton on the hydroxyl group.10 The hydrogen bond causes an increase in electron density around the oxygen, and an upfield shift closer to zero ppm. This is consistent with previous reports indicating that hydrogen bonding via the proton causes an upfield shift in the 17O NMR signal of the attached oxygen.25 If cholesterol engages in hydrogen bonding it shifts upfield, providing a simple method of measuring the hydrogen bonding in bilayers.
Whilst the cholesterol in all three lipid systems has a more upfield chemical shift than that of the cholesterol in the solution state, there also appears to be differences between the cholesterol in the egg sphingomyelin and the phosphocholine membranes. The NMR chemical shift of cholesterol in the sphingomyelin bilayer is further upfield than that of the phosphocholines. This is likely due to stronger cholesterol–lipid interactions. In some cases, cholesterol has been shown to preferentially interact with sphingomyelin over phosphocholines.6,36,37 This is due to the extra hydrogen bonding site, originating from the amide on sphingomyelin.
The NMR chemical shift of the water peak also moves depending on bilayer composition as can be seen from Table 3. Pure water has an 17O NMR chemical shift of 0.0 ppm, however this signal depends on the environment that the water is in, and the presence of dissolved salts can cause this to change.38 The chemical shift of water in the monohydrate sample shows a value of −3.30 ppm, which is primarily from the bulk water surrounding the hydrated crystal. The chemical shift of water in the samples containing a bilayer has a more mixed character because these samples are hydrated at the minimum required to achieve excess hydration (65 wt%). The chemical shift of water in the sphingomyelin sample is 0.12 ppm which is higher than the phosphocholine samples at −0.37 ppm and −0.43 ppm for DOPC and DPPC respectively. This could indicate the extent that water is involved in the hydrogen bonding.
Previous work has already demonstrated the power of 17O NMR spectroscopy to elucidate hydrogen bonding in biological systems.21–24 This work has shown that it can also be used to clearly demonstrate this interaction in lipid bilayers. After enrichment of cholesterol, the 1D 17O NMR spectra provide a measure of hydrogen bonding from chemical shift alone, where an upfield shift indicates more hydrogen bonding. This allowed for simple comparison of three lipid systems, which supported previous ascertains that cholesterol binds more strongly to sphingolipids. A limitation of this, however, is that this method alone does not indicate of the mode of hydrogen bonding, and future studies incorporating 2D-NMR spectroscopy or isotopically enriched phospholipids would provide further detail and may be able to elucidate the nature of the hydrogen bonds.
Thanks are given to Dr Garry Pairaudeau, Dr Jonathan Wingfield and Professor Ramon Vilar for supervisory assistance. Additional thanks are given to Peter Haycock for carrying out the solution state spectroscopy. Collaborative assistance from the 850 MHz Facility Manager (Dr Dinu Iuga, University of Warwick) is acknowledged. The UK 850 MHz solid-state NMR Facility used in this research was funded by EPSRC and BBSRC (contract reference PR140003), as well as the University of Warwick including via part funding through Birmingham Science City Advanced Materials Projects 1 and 2 supported by Advantage West Midlands (AWM) and the European Regional Development Fund (ERDF). This work was supported by an Engineering and Physical Sciences Research Council (EPSRC) Centre for Doctoral Training Studentship from the Institute of Chemical Biology (Imperial College London) with additional funding from AstraZeneca.
Conflicts of interest
There are no conflicts to declare.References
- R. Dimova, Adv. Colloid Interface Sci., 2014, 208, 225–234 CrossRef CAS.
- I. Levental, F. J. Byfield, P. Chowdhury, F. Gai, T. Baumgart and P. A. Janmey, Biochem. J., 2009, 424, 163–167 CrossRef CAS.
- K. Simmons and E. Ikonen, Nature, 1997, 387, 569–572 CrossRef.
- M. L. Briuglia, C. Rotella, A. McFarlane and D. A. Lamprou, Drug Delivery Transl. Res., 2015, 5, 231–242 CrossRef CAS.
- J. P. Slotte, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 304–310 CrossRef CAS.
- H. Ohvo-Rekilä, B. Ramstedt, P. Leppimäki and J. Peter Slotte, Prog. Lipid Res., 2002, 41, 66–97 CrossRef.
- N. P. Franks and W. R. Lieb, J. Mol. Biol., 1979, 133, 469–500 CrossRef CAS.
- D. Marquardt, N. Kučerka, S. R. Wassall, T. A. Harroun and J. Katsaras, Chem. Phys. Lipids, 2016, 199, 17–25 CrossRef CAS.
- S. A. Pandit, D. Bostick and M. L. Berkowitz, Biophys. J., 2004, 86, 1345–1356 CrossRef CAS.
- M. Pasenkiewicz-Gierula, T. Róg, K. Kitamura and A. Kusumi, Biophys. J., 2000, 78, 1376–1389 CrossRef CAS.
- S. F. Bush, H. Levin and I. W. Levin, Chem. Phys. Lipids, 1980, 27, 101–111 CrossRef CAS.
- P. T. T. Wong, S. E. Capes and H. H. Mantsch, Biochim. Biophys. Acta, 1987, 980, 37–41 CrossRef.
- Z. Arsov and L. Quaroni, Chem. Phys. Lipids, 2007, 150, 35–48 CrossRef CAS.
- J. Villalain and J. C. Gomez-Fernandez, Biochem. Soc. Trans., 1992, 20, 122S CrossRef CAS.
- J. Hénin and C. Chipot, Chem. Phys. Lett., 2006, 425, 329–335 CrossRef.
- F. Jolibois, O. Soubias, V. Réat and A. Milon, Chem. – Eur. J., 2004, 10, 5996–6004 CrossRef CAS.
- A. K. Lala, Int. J. Quantum Chem., 1981, 20, 93–97 CrossRef CAS.
- O. Soubias, F. Jolibois, V. Réat and A. Milon, Chem. – Eur. J., 2004, 10, 6005–6014 CrossRef CAS.
- D. L. Gater, V. Réat, G. Czaplicki, O. Saurel, A. Milon, F. Jolibois and V. Cherezov, Langmuir, 2013, 29, 8031–8038 CrossRef CAS.
- M. B. Sankaram and T. E. Thompson, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 8686–8690 CrossRef CAS.
- A. Brinkmann and A. P. M. Kentgens, J. Am. Chem. Soc., 2006, 128, 14758–14759 CrossRef CAS.
- F. G. Vogt, H. Yin, R. G. Forcino and L. Wu, Mol. Pharmaceutics, 2013, 10, 3433–3446 CrossRef CAS.
- G. Wu, Modern Magnetic Resonance., Springer, Cham, 2017 Search PubMed.
- V. Lemaître, M. E. Smith and A. Watts, Solid State Nucl. Magn. Reson., 2004, 26, 215–235 CrossRef.
- J. Reuben, J. Am. Chem. Soc., 1969, 91, 5725–5729 CrossRef CAS.
- C. De La Calle Arregui, J. A. Purdie, C. A. Haslam, R. V. Law and J. M. Sanderson, Chem. Phys. Lipids, 2016, 195, 58–62 CrossRef CAS.
- L. L. Smith, J. Herz and E. L. Ezell, Steroids, 1993, 58, 260–267 CrossRef CAS.
- B. W. Foster, J. Robeson, N. Tagata, J. M. Beckerdite, R. L. Huggins and E. T. Adams, J. Phys. Chem. B, 1981, 85, 3715–3720 CrossRef CAS.
- M. Senegačnik and C. Klofutar, Spectrochim. Acta, Part A, 1998, 54, 709–717 CrossRef.
- C. R. Loomis, G. G. Shipley and D. M. Small, J. Lipid Res., 1979, 20, 525–535 CAS.
- T. H. Sefzik, J. B. Houseknecht, T. M. Clark, S. Prasad, T. L. Lowary, Z. Gan and P. J. Grandinetti, Chem. Phys. Lett., 2007, 434, 312–315 CrossRef CAS.
- J. A. Clarke, A. J. Heron, J. M. Seddon and R. V. Law, Biophys. J., 2006, 90, 2383–2393 CrossRef CAS.
- D. Marsh, Biochim. Biophys. Acta, Biomembr., 2010, 1798, 688–699 CrossRef CAS.
- I. P. Gerothanassis, Prog. Nucl. Magn. Reson. Spectrosc., 2010, 57, 1–110 CrossRef CAS.
- B. M. Craven, Nature, 1976, 260, 727–729 CrossRef CAS.
- J. P. Slotte, Chem. Phys. Lipids, 1999, 102, 13–27 CrossRef CAS.
- T. P. W. McMullen and R. N. McElhaney, Curr. Opin. Colloid Interface Sci., 1996, 1, 83–90 CrossRef CAS.
- V. Maemets and I. Koppel, J. Chem. Soc., Faraday Trans., 1998, 94, 3261–3269 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc05466f |
| This journal is © The Royal Society of Chemistry 2020 |