Metal–organic framework based nanozymes: promising materials for biochemical analysis
Xiangheng
Niu
 ab,
Xin
Li
ab,
Zhaoyuan
Lyu
a,
Jianming
Pan
b,
Shichao
Ding
a,
Xiaofan
Ruan
a,
Wenlei
Zhu
a,
Dan
Du
ab,
Xin
Li
ab,
Zhaoyuan
Lyu
a,
Jianming
Pan
b,
Shichao
Ding
a,
Xiaofan
Ruan
a,
Wenlei
Zhu
a,
Dan
Du
 *a and
Yuehe
Lin
*a and
Yuehe
Lin
 *a
*a
aSchool of Mechanical and Materials Engineering, Washington State University, Pullman, WA 99164, USA. E-mail: annie.du@wsu.edu; yuehe.lin@wsu.edu
bInstitute of Green Chemistry and Chemical Technology, School of Chemistry and Chemical Engineering, Jiangsu University, Zhenjiang 212013, P. R. China
First published on 18th August 2020
Abstract
In recent years, there has been rapid growth of enzyme-mimicking catalytic nanomaterials (nanozymes). Compared with biological enzymes, nanozymes exhibit several superiorities, including robust activity, easy production, and low cost, which endow them with promising applications in biochemical analysis. As an emerging member of nanozymes, metal–organic framework (MOF) nanozymes are attracting growing attention because of their composition and structural characteristics. Rationally designing MOFs with enzyme-like catalytic ability is opening up a new avenue for biochemical detection. In this Feature Article, we summarize the latest developments of MOF nanozymes and their applications in biochemical sensing. First, the types of nanozymes derived from MOFs are categorized, and effective strategies to improve the weak activity inherent in MOF nanozymes are introduced. Then, the multi-functionalization of MOFs with enzyme mimic activity and other attractive properties is emphasized. After that, the typical applications of MOF nanozymes in the detection of various analytes are rigorously reviewed. Finally, the current challenges and some development directions in this field are discussed. It is believed that the versatile nature of MOFs will bring a bright future for MOF nanozymes in biochemical analysis.
1. Introduction
Enzymes are biocatalysts having the power of accelerating one or a class of reactions without consuming themselves. Currently, most enzymes commercially available are separated and purified from organisms. Thanks to the merits of excellent substrate selectivity and activity, these biological enzymes have been extensively applied in various fields, including biomedicine, biochemical detection, agricultural science, food processing, and environmental remediation. Nonetheless, they usually suffer from poor stability in harsh environments, difficulty in recycling, and complicated and high-cost production. Instead, in the past decades scientists have been exploring appropriate artificial enzymes as potential alternatives to natural enzymes, such as porphyrins, cyclodextrins, and supramolecules.1Since Yan's group pioneeringly found in 2007 that some peroxidase-like catalytic features were hidden in common magnetic nanoparticles,2 the past several years have witnessed the rapid rise and prosperity of enzyme-like catalytic nanomaterials (defined as ‘nanozymes’).3,4 Up to today, hundreds of nanosized materials mimicking several types of enzyme activities have been reported and widely applied to medicine, sensing, catalysis, and environmental engineering.5–7 In comparison with natural enzymes, nanozymes show a few attractive merits, including low cost, ease of preparation, good stability, and tailored design. Especially in the analytical field, the flexible adjustment of nanozyme activity offers rich principles and methods for analyte detection, and nanozyme catalysis provides amplified signals for high-sensitivity quantitative determination, both of which make nanozymes emerging and promising materials for biochemical sensing.
Metal–organic frameworks (MOFs) are a class of materials composed of organic ligands and metal nodes. The strong coordination interaction between metal ions and the corresponding organic ligands enables the formation of unique framework structures with versatile properties: (1) the porous structure of MOFs offers rich surfaces and channels for fast mass transfer; (2) the specific pore size in MOFs benefits the adsorption, loading, and separation of targets; (3) the organic ligands in MOFs provide attractive optical, electrical, and thermal characteristics as well as abundant functional groups for chemical modification; and (4) the presence of metal nodes contributes possible active sites for catalysis. As a consequence, MOFs are regarded as star materials and extensively used in a variety of fields, including gas separation, adsorption removal, chemical catalysis, drug delivery, and biochemical analysis. Especially in the sensing field, MOFs with attractive porosity, luminescence or/and electrical conductivity have found intensive use in detecting various analytes ranging from gases, ions and small molecules to aromatic compounds and bioactive species based on optical, electrochemical, mechanical or other sensing modes.8–10 In the past few years, a few MOF materials have been found exhibiting catalytic activities similar to biological enzymes in function. For instance, some Zr-containing MOFs are able to act as organophosphorus hydrolase mimics to degrade organophosphate warfare agents.11–13 In comparison with nanozymes based on carbon materials, noble metals or transition metal compounds, aside from the common merits of relatively low cost, easy preparation and tailored design, MOF nanozymes show some special characteristics thanks to their versatile structures and functions. For example, MOF materials can be employed as both peroxidase mimics and natural enzyme carriers,14 enabling cascade reactions with high catalytic efficiency; some MOFs exhibit enzyme-mimicking catalytic capacity as well as optical responses,15 thus providing integrated sensing platforms. The attractive enzyme-mimetic feature of these MOFs, together with their versatile nature, has rapidly endowed them with growing applications, especially in the biochemical detection field.
In this Feature Article, we aim at presenting a critical review of MOF nanozymes and their emerging applications in biochemical sensing. First, typical types of enzymes mimicked by MOF materials are discussed, followed by the introduction of strategies currently explored to enhance the intrinsic poor activity of MOF nanozymes. After that, the integration of enzyme-mimicking activity with other attractive functions to design and develop multifunctional MOF materials is specially highlighted. Also, applications of MOF nanozymes in detecting various analytes are summarized according to the different analytical principles. Finally, the challenges and perspectives in this field are given for future investigation.
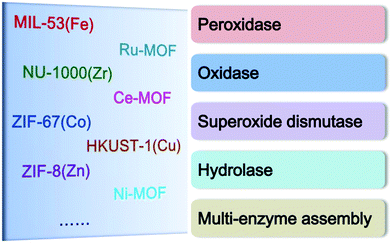
2. Types of MOF nanozymes
In a typical MOF, metal nodes and organic ligands exist simultaneously. Therefore, the enzyme-like catalytic capacity of MOFs can originate from the following two aspects: on the one hand, MOFs containing Fe, Cu, Co, Ni or Ce nodes can provide enzyme-mimicking catalytic activity due to the existence of these metal redox couples, and, on the other hand, some special organic ligands in MOFs act as electron mediators to accept electrons from a substrate and then donate the electrons to another substrate,16 thus catalyzing reactions similarly to natural enzymes. In the past few years, a number of MOFs have been explored as promising nanozymes with different types of enzyme activities, including peroxidase, oxidase, superoxide dismutase, and hydrolase (Scheme 1). Also, some MOFs are found to exhibit two or more kinds of enzyme activities under the same conditions or in different environments.2.1. Peroxidases
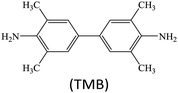
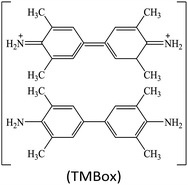
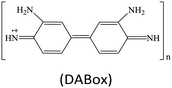
Peroxidases are a type of natural enzymes having the ability of catalyzing the H2O2 substrate to H2O with the participation of another reducing substrate, where the reducing substrate is oxidized to its corresponding oxide. As a typical peroxidase, horseradish peroxidase (HRP) is widely applied to catalyze H2O2 for various applications. In HRP, the active hemin as a redox cofactor is bound by a large helical protein. Similar to HRP, several MOFs containing Fe redox nodes have been verified to show peroxidase-mimetic catalytic ability.15,17–31 Aside from Fe-based MOFs, some other transition metal-containing MOFs, including Cu-based,32–35 Ru-based36 and Ni-based,37 have also been explored as peroxidase mimics.For biochemical analytical applications, a suitable substrate often cooperates with nanozyme catalysis to establish sensing systems. At present, several substrates are generally applied to provide colorimetric, fluorescent, Raman, chemiluminescent or/and electrochemical responses for biochemical detection (Table 1). For instance, colorless 3,3′,5,5′-tetramethylbenzidine (TMB) can be oxidized to its corresponding oxide TMBox during the peroxidase mimic catalyzed H2O2 reaction, where the product TMBox shows a blue color and finally turns yellow after acid treatment, providing the base for colorimetric measurement.38 Besides, the TMBox species with a biphenyl structure easily quenches other fluorescent labels, making it possible to use in fluorescence sensing.39 Also, compared to the original TMB, the TMBox product can provide characteristic Raman signals for surface enhanced Raman scattering (SERS) detection.40 These changes afford versatile detection modes for biochemical analysis.
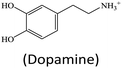
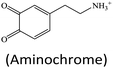
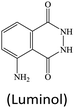
Although lots of peroxidase mimics, including peroxidase-like MOFs, have been developed for various applications, a majority of them exhibit catalytic activity only in a narrow pH range. Common peroxidase mimics provide the maximum activity in a moderately acidic medium (pH ∼ 4), while they have very weak or even no catalytic effect under neutral conditions. This defect limits their potential use in biochemical analysis, because many biochemical reactions should occur in physiological environments. Taking glucose detection as a typical example, it first requires a neutral medium for the oxidation of glucose to produce H2O2 under the catalysis of glucose oxidase (GOx), and then the amount of H2O2 produced is quantified by peroxidase mimic catalysis in a moderately acidic environment. The mismatch of the working conditions of the two enzymes/mimics not only complicates the detection operation but may also impact the analytical performance. Thus, exploring nanozymes with favorable activity in a wide pH range is always desired.41,42 Very recently, He and co-workers proposed Ru(III)-based metal–organic gels (Ru-MOGs) with favorable peroxidase-like activity in both acidic and alkaline solutions.36 They found that in the presence of H2O2 the Ru-MOGs not only exhibited good ability of catalyzing dopamine to aminochrome in a pH 5.5 buffer (Fig. 1A and B) but also showed the peroxidase-mimetic catalytic capacity to trigger the luminol chemiluminescence response in a pH 9.0 medium (Fig. 1C and D). This wide pH applicability would bring great convenience to biochemical detection.
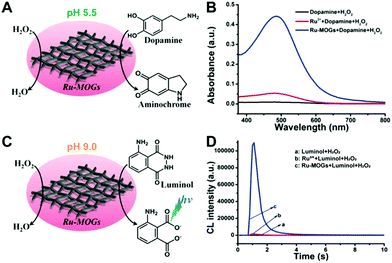 | ||
| Fig. 1 (A) and (B) Verify the peroxidase-like activity of Ru-MOGs towards the oxidation of dopamine to aminochrome in an acidic solution, and (C) and (D) demonstrate that Ru-MOGs also have peroxidase-mimicking ability to catalyze the chemiluminescence of luminol in an alkaline medium (reused with permission from ref. 36). | ||
2.2. Oxidases
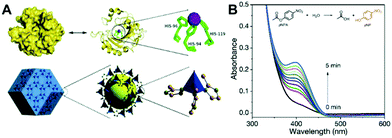
In addition to peroxidases, another main type of MOF nanozymes is oxidases. Typically, some Ce-based,43–46 Co-based47 and Cu-based48,49 MOFs are found to exhibit oxidase-like catalytic activity. They can directly catalyze the oxidization of substrates without the participation of H2O2. Instead, dissolved oxygen is often involved in these oxidase mimic catalyzed reactions. To some extent, oxidase mimics are preferable compared to the peroxidase ones for biochemical detection thanks to their simpler reaction conditions.50It is known that the catalytic activity of a nanozyme highly depends on its shape, size, crystal face, and surface chemistry. Apart from these physicochemical factors, the enzyme-like capacity of nanozymes can also be regulated by external stimuli like light. In 2019, Wei's group reported a photosensitized MOF (PSMOF) whose oxidase-mimetic activity could be stimulated by light irradiation.51 The PSMOF was obtained by introducing a Ru(bpy)32+ derivative as photoactive ligands within a Zn-based MOF (Fig. 2A). With no light irradiation, the mixing of the PSMOF with the TMB substrate induced neither a solution color change nor characteristic absorption (Fig. 2B). When the mixture was irradiated with light, the PSMOF rapidly catalyzed the oxidation of TMB and gave a blue color. With the external light irradiation control, one could start and stop the nanozyme catalytic reaction easily (Fig. 2C), which would find promising applications in developing controllable sensors and biosensors. For more information about light-responsive nanozymes, one can refer to the recent review from Wei's group.52
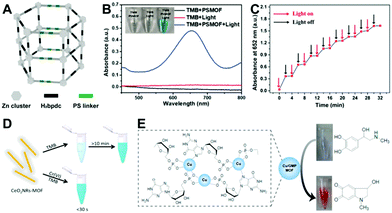 | ||
| Fig. 2 (A) Shows the structure of the PSMOF, and (B) and (C) verify the light-stimulated oxidase-mimetic activity of the PSMOF (reused with permission from ref. 51). (D) presents the Cr(VI)-boosted oxidase-mimetic ability of the CeO2NR-MOF (reused with permission from ref. 43). (E) depicts the laccase-like capacity of the Cu/GMP MOF (reused with permission from ref. 48). | ||
Aside from light stimulation, some MOF-based oxidase mimics can be triggered by certain chemicals. Very recently, Cr(VI) was reported to accelerate the oxidase-mimicking activity of a ceria nanorod-templated metal–organic framework (CeO2NR-MOF) significantly in a pH 4 buffer.43 Wang et al. found that the TMB oxidation reaction was rapidly catalyzed in a short time (less than 30 s) by the CeO2NR-MOF with the presence of Cr(VI), while the chromogenic reaction occurred very slowly (more than 10 min) with only the CeO2NR-MOF (Fig. 2D). Similar phenomena are also observed in Au@Hg and polyethylenimine-stabilized silver nanoclusters (PEI-AgNCs),53,54 which can further be applied to photometrically detect toxic Cr(VI). However, the real functions of these materials are controversial. Wang's study43 reported that the CeO2NR-MOF acted as an oxidase mimic, while other work53,54 proposed Au@Hg and PEI-AgNCs as oxidoreductase mimics. Besides, the reason why the enzyme-mimicking activities of these materials can be promoted by Cr(VI) selectively over other ions is still unknown, and the underlying mechanism of the Cr(VI)-boosted nanozyme catalysis is also unclear.
As a special oxidase, laccase is capable of catalyzing the oxidation of phenols to benzoquinone compounds. With this property, laccase is often used to detect and degrade phenolic pollutants. Inspired by the composition and structure of natural laccase, Liang and co-workers developed a laccase mimic composed of guanosine monophosphate (GMP) coordinated Cu.48 The formed Cu/GMP MOF with a Cu-to-GMP ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 exhibited superior activity over natural laccase to catalyze the conversion of phenol-containing substrates (Fig. 2E). They further employed the Cu/GMP MOF laccase mimic to establish a colorimetric assay of epinephrine. By using a cysteine–histidine dipeptide to replace the GMP ligands, Wang et al. fabricated another Cu-based laccase mimic for the degradation of phenolic pollutants.49
4 exhibited superior activity over natural laccase to catalyze the conversion of phenol-containing substrates (Fig. 2E). They further employed the Cu/GMP MOF laccase mimic to establish a colorimetric assay of epinephrine. By using a cysteine–histidine dipeptide to replace the GMP ligands, Wang et al. fabricated another Cu-based laccase mimic for the degradation of phenolic pollutants.49
2.3. Superoxide dismutase
Superoxide dismutase (SOD) is a type of enzyme capable of catalyzing the dismutation of unstable superoxide radicals to O2 and H2O2. Inspired by the active composition of natural CuZn-SOD, Qu's group designed a Cu-TCPP MOF with SOD-like activity.55 To further improve its catalytic activity, they sonicated the Cu-TCPP nanosheets into ultra-small nanodots. Compared to the nanosheets, these dots, featuring a size similar to that of natural enzymes, not only offered more accessible catalytic sites for substrates with smaller diffusion barriers but also enabled themselves to be cleared out via renal clearance, which greatly reduced the nanozyme's toxicity. As validated by in vitro and in vivo experiments, the Cu-TCPP MOF nanodots acting as an artificial SOD exhibited high efficiency in alleviating endotoxemia while retaining very low cytotoxicity themselves.2.4. Hydrolases
Hydrolases are an assembly of enzymes that are able to catalyze a variety of hydrolysis reactions. Up to today, according to the different kinds of target substrates, there are three main types of hydrolases that some MOF materials can mimic, namely organophosphorus hydrolase, protease, and esterase. A number of studies have demonstrated that Zr-containing MOFs can effectively break the phosphonate ester bond.11–13,56–58 It is generally thought that the hydrolase-mimetic activity of these Zr-based MOFs should be attributed to the strong and specific interaction between Zr–O clusters and the phosphate group in substrates. Typically, Farha's group reported the efficient hydrolysis of chemical warfare agents containing phosphonate ester bonds catalyzed by a Zr6-based MOF named NU-1000.11 NU-1000 was composed of eight connected Zr6(μ3-O)4(μ3-OH)4(H2O)4(OH)4 nodes and tetratopic 1,3,6,8(p-benzoate)pyrene linkers. The proposed NU-1000 had ultra-wide pores and channels, allowing bulky phosphate ester molecules to permeate the whole framework for the reaction. With dimethyl 4-nitrophenyl phosphate (DMNP) as a substrate, it was hydrolyzed into p-nitrophenoxide anions and phosphate at pH 10 under the catalysis of the NU-1000 nanozyme, giving a turnover frequency (TOF) as high as 0.06 s−1.Aside from organophosphorus hydrolase, some MOFs are found to show the capacity of catalyzing the hydrolysis of proteins. Li's group developed two Cu-based MOFs and found that they had intrinsic protease-like catalytic activity for protein hydrolysis.59,60 It was reported that the HKUST-1 MOF exhibited much stronger affinity towards bovine serum albumin (BSA) with a Michaelis constant (Km) 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold smaller than that of trypsin.59 Besides, the insoluble MOF nanozyme was easy to recycle via simple centrifugation and reused with no obvious activity loss.
000-fold smaller than that of trypsin.59 Besides, the insoluble MOF nanozyme was easy to recycle via simple centrifugation and reused with no obvious activity loss.
Recently, inspired by the composition and structure of human carbonic anhydrase II (hCAII), Chen et al. designed and developed a series of zeolitic imidazolate frameworks (ZIFs) that were able to efficiently catalyze the hydrolysis of esters.61 In hCAII, the active centre consisted of a tetrahedrally coordinated Zn2+ ion with three symmetric histidine imidazoles and a H2O molecule (Fig. 3A). Like hCAII, the ZIF-8 material consisted of a central Zn2+ ion and four 2-methylimidazolate (2-mIM) ligands. The tetrahedrally coordinated Zn2+ ion was interconnected via 2-mIM to form a microporous framework. With the similarities in composition and geometric structure, the MOF material exhibited comparable esterase-like ability to catalyze the hydrolysis of p-nitrophenyl acetate (pNPA) (Fig. 3B).
 | ||
| Fig. 3 (A) Shows the bio-inspired design of ZIF-8 as a hydrolase mimic, and (B) verifies that the designed ZIF-8 can hydrolyze the pNPA substrate to produce yellow pNP (reused with permission from ref. 61). | ||
2.5. Multi-enzyme assembly
Apart from the above mentioned MOFs exhibiting single-enzyme activity, some MOF materials are able to show multi-enzyme activities under different conditions or even in the same environment.62–64 This character, with two or more enzyme-like activities in one MOF, makes the material a multi-enzyme assembly, which provides one with an integrated tool for cascade catalysis and biomedical therapy. Typically, our group designed a CuBDC MOF with both cysteine oxidase- and peroxidase-mimetic activities.63 In the MOF, the Cu–O clusters not only exhibited the ability to catalyze the oxidation of cysteine to produce cystine and H2O2 with the participation of dissolved oxygen but also showed the peroxidase-mimicking power to catalyze the produced H2O2 to generate hydroxide radicals in the same buffer, which further oxidized the BDC ligands in the material to give a fluorescence signal (Fig. 4). Such a cascade reaction catalyzed by the CuBDC MOF, together with its stimulus-responsive fluorescence, enabled us to detect the cysteine species with simple operation and high performance.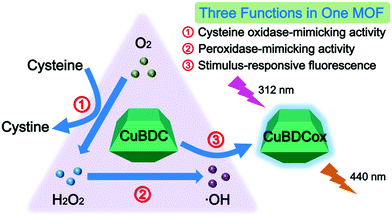 | ||
| Fig. 4 Diagram of the cascade cysteine oxidase- and peroxidase-mimetic activities and stimulus-responsive fluorescence of CuBDC for cysteine sensing (reused with permission from ref. 63). | ||
3. Strategies for improving the MOF nanozyme activity
Although a growing number of MOF materials are reported to act as some natural enzymes, the catalytic activity of the currently developed MOFs is still much less than the latter. The intrinsic poor enzyme-like activity of a MOF should be attributed to several factors: (1) The size of the prepared MOFs is often hundreds of nanometers or even on the micrometer scale. Such a large size greatly decreases the active surfaces exposed for substrate adsorption and reaction. (2) In a MOF, the majority of active sites are buried inside the material, and only a very small proportion of surface sites are available for substrate catalysis. (3) Although MOFs feature rich pores, only pores large enough are accessible for substrate molecules. Therefore, the mass transfer resistance is supposed to be large, greatly affecting the free diffusion of substrates, intermediates, and products. Therefore, low activity is one of the most significant deficiencies in MOF nanozymes. To address this issue, researchers have explored several strategies to enhance the enzyme-like catalytic activity of MOFs, such as 2D MOF nanosheets, bimetal MOFs, valence state regulation, and single-atom nanozymes.3.1. 2D MOF nanosheets
Traditional MOFs have interconnected 3D structures with large sizes, which not only decrease the exposed surface sites but also increase the diffusion barriers. Compared to 3D MOFs, 2D nanosheets or nanoflakes are supposed to provide larger surfaces, more exposed active sites, and lower diffusion barriers. Based on this, researchers have developed several 2D MOF nanosheets and obtained enhanced enzyme-mimetic activity.20,35,37,65–67 For instance, Cheng et al. employed binuclear paddle-wheel metal clusters and metalated tetrakis(4-carboxyphenyl)porphyrin (TCPP) ligands as precursors to prepare MOFs with PVP confining the growth of the MOF crystal into 2D nanosheets (Fig. 5A).20 The synthesized 2D MOF exhibited a well-defined ultrathin sheet-like structure (Fig. 5B). This structure would expose large surfaces with more accessible active sites for catalysis. As verified, compared to bulky Zn-TCPP(Fe), the proposed 2D Zn-TCPP(Fe) exhibited faster peroxidase-like catalytic kinetics towards the oxidation of TMB with the existence of H2O2 (Fig. 5C). The 2D strategy is a relatively simple but efficient route to obtain high-activity MOF nanozymes. It is expected to develop more 2D MOF nanozymes with favorable activity for applications in the near future. Besides, converting 2D MOFs into lower dimensional structures (e.g., nanodots) may be feasible for further improving their enzyme-mimicking activity.55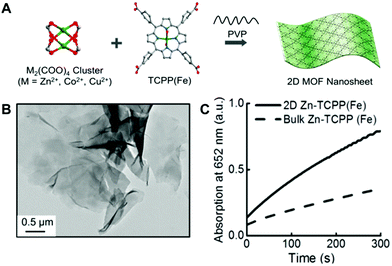 | ||
| Fig. 5 (A) Shows the synthetic process of 2D MOF nanosheets, (B) is the TEM image of 2D Zn-TCPP(Fe), and (C) compares the peroxidase-mimicking catalytic activity of 2D Zn-TCPP(Fe) and bulky Zn-TCPP(Fe) towards the TMB color reaction (reused with permission from ref. 20). | ||
3.2. Bimetal MOFs
In comparison with single-metal MOFs, bimetal coordination with organic ligands is supposed to have better catalytic effects: on the one hand, the different potentials of the two metals lead to a difference in potential, which plays a vital role in promoting electron transport;68 on the other hand, the introduction of bimetal nodes into one MOF can generate defects, which may also increase the active sites and further improve the catalytic performance. With this consideration, several bimetal MOFs have been explored as nanozymes.64,68,69 Some of them are reported to exhibit better enzyme-mimicking catalytic activities than the corresponding single-metal MOFs. Despite this, the enhancement should be treated with caution: (1) in some of these studies, no strict comparisons between bimetal MOFs and corresponding single-metal MOFs are made to demonstrate the enhanced activity of the former; and (2) although an enhancement in catalytic activity is observed in several studies, the mechanism for this enhancement is still unclear. Commonly, one tries to use the synergistic effect to explain the activity enhancement in bimetal MOFs. However, how the two metal nodes synergistically interact to improve the nanozyme activity is still vague. Once the underlying mechanism is uncovered, it is believed that more bimetal or multi-metal MOFs with desired activity will be rationally designed and developed.3.3. Valence state regulation
In most currently developed MOF nanozymes, the activity origin comes from the metal nodes in the MOFs. More specifically, the metal redox couple in a MOF plays the enzyme-like catalytic role. The chemical valence states of the metal redox couple and their ratio determine the catalytic activity of the MOF nanozyme. Thus, engineering the valence state regulation of MOF nanozymes is supposed to be a feasible strategy to regulate their catalytic activity.44,47,62 With this strategy, Ye's group developed a mixed valence Ce-MOF (MVCM) as an oxidase mimic.44 They obtained the MVCM through a simple and fast partial oxidation treatment of a common Ce-MOF in a mixture of NaOH and H2O2 (Fig. 6A). The treatment increased the ratio of Ce4+ to Ce3+. The MVCM was demonstrated to exhibit remarkable oxidase-like catalytic ability to trigger the TMB color reaction, while the original Ce-MOF showed very weak activity (Fig. 6B). Inversely, our group found that when the MVCM was incubated with ascorbic acid (AA), a reducing substance, the high Ce4+/Ce3+ ratio in the MVCM could be down-regulated (Fig. 6C).46 The resulting MVCM with a low Ce4+/Ce3+ ratio showed much less oxidase-mimetic activity. With this finding, an alkaline phosphatase (ALP) activity colorimetric assay was fabricated based on the analyte-induced valence state regulation of the oxidase-like MVCM.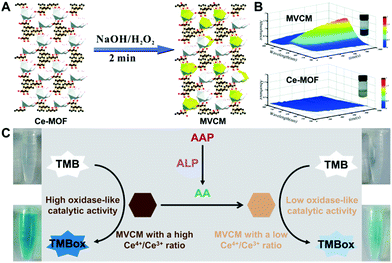 | ||
| Fig. 6 (A) Shows the transformation process of the Ce-MOF to the MVCM, and (B) compares the oxidase-mimicking activity of the Ce-MOF and MVCM (reused with permission from ref. 44). (C) presents the ALP activity colorimetric assay based on the analyte-triggered valence state regulation of the oxidase-mimicking MVCM (reused with permission from ref. 46). | ||
3.4. MOF-based single-atom nanozymes
In addition to the above strategies, single atomization of catalytically active sites is another efficient route to obtain high-activity nanozymes.70 It is generally thought that reducing the size of nanostructured catalysts is able to improve their catalytic activity, because the proportion of surface atoms exposed is significantly increased with the decreased catalyst size. Besides, the size change may alter the catalyst's surface atomic and electronic structures and surface defects.71 Thus, it is expected that further adjusting the catalyst's structure at the atomic level and decreasing its size down to a single-atom scale will maximize the atomic utilization and boost the catalytic activity. As catalysts with single-atom dispersion have been demonstrated to have high activity in electrocatalysis,72–79 according to the ‘single-atom catalyst (SAC)’ concept80 and inspired by SACs, natural enzymes and nanozymes (Fig. 7), in the last three years scientists have paid much attention to designing and developing high-activity single-atom nanozymes (SANs),81–90 most of which are derived from MOF materials. In general, SANs can be obtained by pyrolyzing a MOF material at high temperature, followed by achieving single-atom entities via appropriate acid pickling.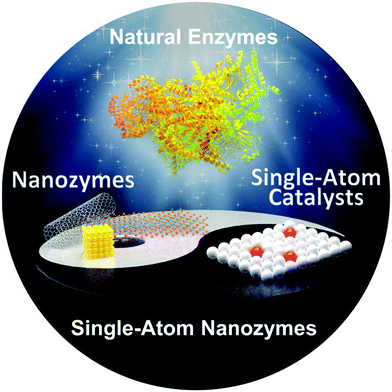 | ||
| Fig. 7 Design and development of high-activity single-atom nanozymes inspired by natural enzymes, nanozymes and single-atom catalysts (reused with permission from ref. 91). | ||
Typically, our group obtained a SAN with atomically dispersed Fe–Nx units supported by a MOF-derived porous carbon.85 The SAN was prepared with a relatively simple and low-cost procedure (Fig. 8A): first, ZIF-8 doped with a small amount of Fe was synthesized in a one-pot way; then, the obtained Fe-doped ZIF-8 was calcinated in an inert atmosphere (N2); and, after that, the material was pyrolyzed in NH3 to get the SAN. Thanks to both the maximized active Fe dispersion and the large surface area of the porous carbon support, the SAN exhibited a specific activity as high as 57.76 U mg−1 measured according to the standardized protocol (Fig. 8B and C),92 almost at the same level as natural HRP. Other studies also demonstrate that the single atomization of active sites derived from MOFs can prominently increase the enzyme-like catalytic activity.84 For more information about SANs, one can refer to our recent review.91
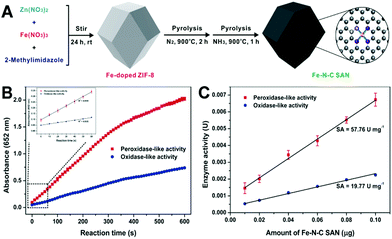 | ||
| Fig. 8 (A) Shows the preparation process of the Fe–N–C SAN, (B) presents the absorbance–time curves of the TMB chromogenic reaction catalyzed by the Fe–N–C SAN, and (C) exhibits the relationship between the enzyme-like activity of the Fe–N–C SAN and its amount (reused with permission from ref. 85). | ||
It should be pointed out that, although converting MOFs into other materials via calcination and further achieving single-atom entities can efficiently improve the enzyme-mimicking catalytic activity, the featured framework structure of MOFs could be irretrievably destroyed and some of their intrinsic properties may be lost. Realizing the single atomization of active sites in MOF nanozymes while retaining their attractive framework structures and versatile properties is still challenging.
4. Multi-functionalization of MOF nanozymes
Undoubtedly, recent years have seen the rapid rise of MOF materials in various fields ranging from gas separation and catalysis to drug delivery and biomedicine. One of the main driving forces is their flexible design and versatile functions. For a MOF, it not only acts as a nanozyme but also plays some other roles. Typically, MOFs with rich channels, pores and functional groups can be employed as carriers for natural enzyme loading and encapsulation. Also, the flexible design of organic ligands and metal nodes makes MOF materials signal sources for biochemical detection.4.1. MOF nanozymes act as natural enzyme carriers
In many biological systems, cascade catalytic reactions occur in a confined space. This requires multiple enzymes confined in sub-cellular compartments as an enzyme assembly. Some MOFs can act as not only enzyme mimics but also as supports for natural enzyme loading due to their unique porous structures.14,66,93–96 In general, there are two strategies to obtain a MOF nanozyme with natural enzyme loading: one is to encapsulate biological enzymes within MOFs during their synthesis, and the other is to post-modify enzymes onto MOF materials via chemical bonding. Wei's group proposed an in situ method to fabricate an integrated nanozyme by simultaneously embedding two cascade catalysts (a molecular catalyst hemin and a biological enzyme GOx) inside the ZIF-8 nanostructure93 (Fig. 9A). Such a design endowed the integrated MOF with merits in improving the overall catalytic efficiency, where the first enzymatic reaction occurred in proximity to the second enzyme mimic. The products generated from the first reaction could be utilized immediately as substrates for the second reaction, which overcame the diffusion-limited kinetics and product instability problems.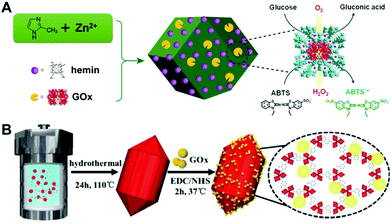 | ||
| Fig. 9 (A) Presents the one-step synthetic approach to GOx/hemin@ZIF-8 for the one-pot detection of glucose (reused with permission from ref. 93). (B) illustrates the post-modification of GOx onto the Fe-MOF via chemical bonding towards the cascade catalytic determination of glucose (reused with permission from ref. 14). | ||
Different from Wei's method, our group loaded GOx onto the surface of a MOF via a post-modification strategy14 (Fig. 9B). In our work, an Fe-MOF (Fe-MIL-88B-NH2) with intrinsic peroxidase-mimicking activity and good biocompatibility was employed as a GOx carrier. With rich amino groups in the Fe-MOF, GOx could be immobilized onto the Fe-MOF surface with the help of cross-linking agents to initiate the cascade reaction for glucose determination. Thanks to the ‘nanoscale proximity’ effect, H2O2 produced from the GOx catalyzed oxidation of glucose with dissolved O2 would be in situ oxidized by the peroxidase-like Fe-MOF, such that the effect of diffusion resistance was diminished and the spontaneous decomposition of H2O2 was minimized. As a consequence, in comparison with the free system (Fe-MOF/GOx), the integrated system (Fe-MOF-GOx) for glucose analysis showed not only strong acid–base tolerance and temperature adaptation but also good reusability. Very recently, Ye's group employed the boronate affinity of a boronic acid-functionalized MOF peroxidase mimic towards GOx to anchor the latter.96 In their strategy, Fe-MIL-88B-NH2 was first post-modified with 4-formylphenylboronic acid to introduce interaction sites, and then the strong boronate affinity between the 4-formylphenylboronic acid modifier and the cis-diol group on the GOx surface enabled the encapsulation of GOx inside the MOF pores and channels.
No matter which strategy (the in situ immobilization method or the post-modification one) is used to load enzymes, two important challenges need to be overcome: (1) how to keep the activity of the immobilized natural enzyme as high as possible; and (2) how to make the loaded natural enzyme close enough to the nanozyme active sites for efficient cascade reactions and lower diffusion resistance. For the two purposes, more efficient and convenient methods of loading natural enzymes within MOF nanozymes are still desired.
4.2. MOF nanozymes act as analytical signal sources
In a typical nanozyme detection process, signaling substrates are often added to produce a response, which not only complicates the detection operation but also weakens the catalytic reaction. Instead, these signaling substrates can also be skillfully integrated into MOF nanozymes.15,29,35,97,98 In these MOFs, they not only play an enzyme-like catalytic role but also act as analytical signal sources for detection. As a typical example, Lin et al. designed a bifunctional MOF (MIL-53(Fe)) with both peroxidase-mimicking activity and oxidation-responsive fluorescence15 (Fig. 10A). In MIL-53(Fe), the Fe nodes had the peroxidase-mimetic catalytic function, and the terephthalic acid (TA) ligands acted as a potential fluorescent source. When the MOF was incorporated with GOx, glucose was first oxidized under the catalysis of GOx to produce H2O2, and the generated H2O2 was then catalyzed by the peroxidase-like MOF to generate rich hydroxyl radicals, which further oxidized the TA ligands in the MOF to form a blue-fluorescent product, thus giving the signal for glucose detection. The use of such a bifunctional MOF nanozyme avoids the addition of foreign substrates and labels, simplifying the detection operation to some extent.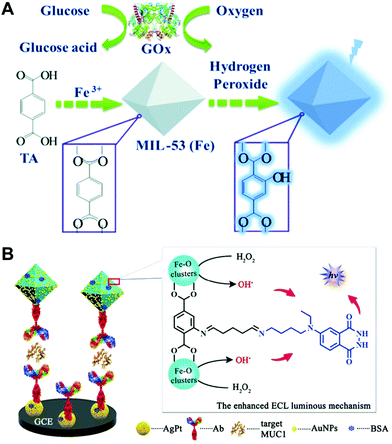 | ||
| Fig. 10 (A) Presents the label-free glucose sensing strategy using MIL-53(Fe) with peroxidase-mimicking activity and oxidation-stimulated self-fluorescence (reused with permission from ref. 15). (B) Shows the peroxidase-mimetic ABEI/MIL-101(Fe) with a stimulus-triggered electrochemical chemiluminescence response for the fabrication of a MUC1 immunosensor (reused with permission from ref. 97). | ||
In addition to fluorescence signals, some MOF nanozymes themselves can also provide other signals. For instance, Wang et al. developed an N-(4-aminobutyl)-N-(ethylisoluminol) modified metal–organic framework (ABEI/MIL-101(Fe)) with peroxidase-mimicking catalytic capacity and used it as an efficient electrochemiluminescent (ECL) indicator for immunoassays97 (Fig. 10B). The Fe–O clusters catalyzed the decomposition of H2O2 to form hydroxyl radicals, and the isoluminol molecule linked onto the ligands of the MOF could be stimulated to produce the ECL signal.
5. Biochemical sensing applications
With the attractive features of MOF nanozymes, they have found wide applications especially in the biochemical sensing field. Generally, these analytical applications can be classified into the following four categories (Scheme 2): (1) enzymatic substrates (H2O2, phenols, etc.) and substrate-related molecules (e.g., glucose) can be detected using MOFs as nanozymes; (2) the nanozyme activity of MOFs is easily impacted by foreign molecules such as inorganic ions and DNA, such that the sensing of these substances can be realized via the modulation of the nanozyme activity; (3) some reducing species, including biothiols, AA and dopamine (DA), can have effects on nanozyme catalyzed systems, and thus these species can be sensed employing MOF nanozymes; and (4) as potential alternatives to natural enzymes, MOF nanozymes can also produce amplified signals in immunoassays and aptamer sensors. With the above principles, a number of analytes can be efficiently detected with the participation of MOFs as nanozymes.5.1. Detection of substrates and substrate-related molecules
In a common peroxidase catalyzed system, H2O2 is often used as an enzymatic substrate, where the H2O2-involved oxidation of a substrate is catalyzed by a peroxidase mimic. By measuring the production of oxidized substrate, H2O2 can be determined. With the system, several MOFs have been used as peroxidase mimics to sense the H2O2 analyte.15,37,64,99 As H2O2 is an important intermediate in many cascade biochemical reactions, MOF nanozymes can also be integrated with natural enzymes for the sensing of H2O2-related molecules. A typical example is the detection of glucose.14,15,23,26,66,93–95,99,100 Qin and co-workers utilized a hemin-containing MOF as an efficient peroxidase mimic to establish a colorimetric assay of glucose by incorporating the mimic with GOx.100 As shown in Fig. 11A, glucose was first specifically catalyzed by GOx to generate the H2O2 intermediate, and then the intermediate was catalyzed by the MOF peroxidase mimic, inducing the TMB chromogenic reaction for quantitative determination. Besides glucose, MOF nanozymes can also be combined with cholesterol oxidase, lactate oxidase or urate oxidase to detect cholesterol,101 lactate102 or uric acid,27 respectively.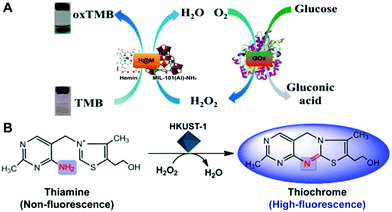 | ||
| Fig. 11 (A) Displays the colorimetric sensing of glucose by integrating GOx with hemin@MOF as a mimetic peroxidase (reused with permission from ref. 100). (B) Illustrates the oxidation of thiamine to thiochrome under the peroxidase-mimicking catalysis of HKUST-1 for the fluorescence sensing of thiamine (reused with permission from ref. 32). | ||
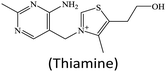
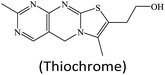
In addition to H2O2 as an enzymatic substrate, other organic species can also be used as nanozyme substrates for their detection.32,48,49,98 Tan's group reported a high-performance fluorescent assay of thiamine using a MOF nanozyme (HKUST-1) catalyzed system.32 In their design, the analyte thiamine was directly employed as a peroxidase substrate. Under the peroxidase-like catalysis of HKUST-1, the thiamine molecule was oxidized to produce a thiochrome (Fig. 11B), giving high fluorescence around 435 nm with an excitation wavelength of 370 nm. With this strategy, wide-range and high-sensitivity fluorescence detection of thiamine was realized.
5.2. Sensing based on nanozyme activity modulation
In nature, nanozyme catalysis is a heterogeneous surface reaction process, and factors affecting the surface properties of nanozymes can impact the activity of nanozymes. Currently, many species, including ions, small molecules, DNA, and proteins, have been found to exhibit various effects on the nanozyme activity. This provides one with new principles and strategies to establish effective methods for the sensing of these species based on nanozyme activity modulation.Currently, detection of phosphates,35,65 Cr6+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 and ALP activity29 has been realized based on MOF nanozyme activity modulation. Typically, Wei's group fabricated 2D MOF nanozyme sensor arrays by adjusting their peroxidase-like activity with several phosphates, namely phosphate, pyrophosphate, ATP, ADP and AMP65 (Fig. 12A). The fabricated sensor arrays were successfully employed to differentiate the five phosphates. The practical application of the sensor arrays was demonstrated with double blind experiments, where unknown samples containing phosphates were identified with good accuracy. Furthermore, the sensor arrays were utilized to probe the hydrolytic processes of PPi and ATP.
43 and ALP activity29 has been realized based on MOF nanozyme activity modulation. Typically, Wei's group fabricated 2D MOF nanozyme sensor arrays by adjusting their peroxidase-like activity with several phosphates, namely phosphate, pyrophosphate, ATP, ADP and AMP65 (Fig. 12A). The fabricated sensor arrays were successfully employed to differentiate the five phosphates. The practical application of the sensor arrays was demonstrated with double blind experiments, where unknown samples containing phosphates were identified with good accuracy. Furthermore, the sensor arrays were utilized to probe the hydrolytic processes of PPi and ATP.
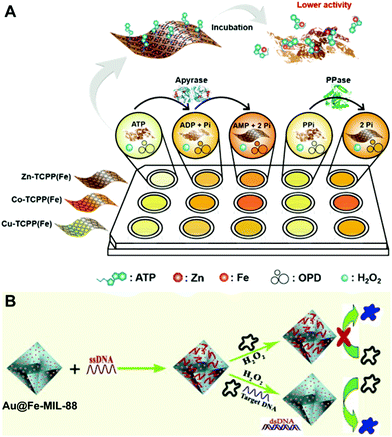 | ||
| Fig. 12 (A) Illustrates the 2D MOF nanozyme sensor arrays for analyzing phosphates and their hydrolytic processes based on peroxidase-like activity modulation (reused with permission from ref. 65). (B) presents the principle of Au@Fe-MIL-88 as a peroxidase mimic for the colorimetric detection of DNA (reused with permission from ref. 103). | ||
Aside from ions, DNA chains can also impact the catalytic properties of nanozymes,104 providing the foundation for DNA and DNA-related analyses. Li's group constructed a DNA sensor based on the activity modulation of a Au-decorated Fe-MOF (Au@Fe-MIL-88)103 (Fig. 12B). In their sensor, Fe-MIL-88 exhibited peroxidase-mimetic activity, and the AuNPs had different adsorption properties towards double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA). In detail, ssDNA was easily adsorbed onto the Au@Fe-MIL-88 surface, leading to the decrease of the hybrid peroxidase-like activity. However, it was recovered by adding target DNA. Moreover, the recovery degree highly depended on the concentration of target DNA added. In contrast, base mismatched DNA could not trigger the recovery of the peroxidase-mimicking activity. Based on this, they developed a colorimetric method for DNA hybridization detection. Besides, appropriate sensing methods can also be designed and performed for the detection of heparin20 and ALP activity46 based on the MOF nanozyme activity modulation principle.
5.3. Detection of species impacting nanozyme catalyzed systems
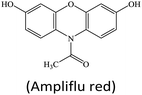
Apart from the sensing principle depending on the direct effect of foreign species on the nanozyme activity, many foreign species can make some impacts on nanozyme catalyzed reaction systems. Typically, some reducing species, including biothiols,25,44,45,47,51,69,105 AA17,22,24,62 and DA,34 can inhibit or hinder the nanozyme catalyzed TMB chromogenic reaction. On the one hand, these species with certain reducibility are able to be competitively oxidized with the TMB substrate. On the other hand, they can reduce the TMBox product to TMB again. As a result, the presence of these reducing species suppresses the TMB color reaction. With this principle, nanozyme catalyzed systems can be extensively used to detect these reducing species. For instance, Wang et al. synthesized a MOF composed of Cu2+ and 1,10-phenanthroline-2,9-dicarboxylic acid (H2PDA).34 The [Cu(PDA)(DMF)] MOF exhibited high peroxidase-mimetic activity and could catalytically oxidize the colorless substrate TMB to a blue product with the participation of H2O2. Nonetheless, the peroxidase mimic catalyzed reaction was dramatically suppressed in the presence of DA. According to this finding, the colorimetric detection of DA was realized (Fig. 13A). Another typical sample ZIF-67 as an oxidase mimic can catalyze Amplex red (AR) oxidation (Fig. 13B).47 During the process, two consecutive redox reactions of AR are observed, which can be applied for the fluorescence detection of biothiols like L-cysteine (L-Cys).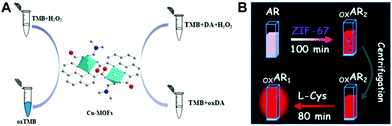 | ||
| Fig. 13 (A) Presents the strategy for DA detection based on the target impacting the peroxidase-like Cu-MOF catalyzed TMB color reaction (reused with permission from ref. 34). (B) shows the sensing principle for L-Cys based on oxidase-mimicking ZIF-67 catalyzed AR oxidation (reused with permission from ref. 47). | ||
What should be stated is that, theoretically, any species with stronger reducibility than the substrate used (e.g., TMB) can affect the nanozyme catalyzed system. Therefore, this sensing principle highly relies on the reducibility of the analyte and that of the substrate used. As a result, the strategy lacks enough specificity for quantitative detection. To overcome this problem, very recently the fabrication of nanozyme sensor arrays combining principal component analysis (PCA) has been proposed for differentiating and quantitating these reducing species.106–109
5.4. Replacing natural enzymes to generate amplified signals
Initially, nanozymes are designed and developed as alternatives to natural enzymes. Naturally, they can be used to replace natural enzyme to generate amplified signals, while retaining good stability and low cost. Typically, nanozymes can be applied as natural enzyme alternatives in enzyme-linked immunosorbent assays (ELISA).21,30,97,110 Currently, there are a growing number of studies on nanozyme-linked immunoassays. As mentioned above, although nanozymes perform better in terms of stability in harsh environments and production cost, most of them exhibit lower catalytic activity, which has negative effects on the immunosensing of trace targets. To improve the sensitivity of nanozyme-linked immunoassays, recently we proposed a ‘nanozyme nest’ that was integrated with immunosensing for the detection of a woodsmoke exposure biomarker.21 As compared in Fig. 14A, the first-generation labels employed for ELISA are natural enzymes like HRP. Their deficiencies, including low stability and environment-dependent performance, greatly restrict their applications in real conditions. Nanozymes as second-generation labels afford options to develop assays with good stability and low cost, while their catalytic capacity is inferior in comparison with that of natural enzymes, resulting in challenges for ultrasensitive detection. Besides, the poor solubility of nanomaterial-based artificial enzymes is another considerable issue. In our ‘nanozyme nest’, a number of peroxidase-like active Fe-MOF particles were loaded on a GO support. This construction not only provided dual-amplified catalytic ability but also overcame the poor water solubility of the active Fe-MOF. Thus, it could be regarded as a third-generation label offering amplified catalytic ability and improved water solubility. After bioconjugation with antibodies, the ‘nanozyme nest’ could provide ultrasensitive detection performance for the biomarker. For more information about nanozyme-based immunoassays and immunosensors, one can refer to our recent review.110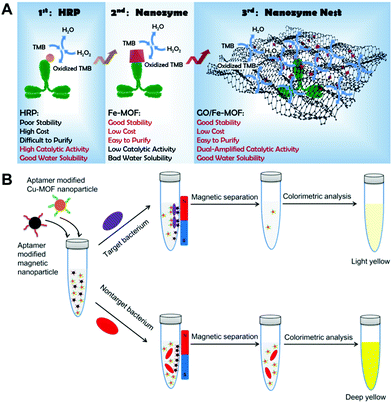 | ||
| Fig. 14 (A) Compares the GO/Fe-MOF nanozyme nest with the conventional HRP and Fe-MOF as a nanozyme for immunoassays (reused with permission from ref. 21). (B) illustrates the detection procedure of bacteria using aptamer recognition, magnetic separation and nanozyme catalysis (reused with permission from ref. 33). | ||
Besides immunoassays, nanozymes are also combined with aptamers to realize the selective and high-sensitivity sensing of analytes. In a nanozyme-based aptamer sensor, the aptamer component plays the specific recognition role towards targets, and the nanozyme one gives amplified optical or electrochemical readouts.33,111 As a typical example, Wang and co-authors proposed a colorimetric protocol for Staphylococcus aureus sensing by using aptamer-modified Cu-MOF nanoparticles as a peroxidase mimic (Fig. 14B).33 In the protocol, both aptamer-decorated Cu-MOF nanoparticles and aptamer-decorated magnetic nanoparticles (MNPs) simultaneously recognized target bacteria in a test tube. After magnetic separation, the Cu-MOF nanoparticles bound to bacteria were removed from the supernatant, and the number of Cu-MOF nanoparticles in the supernatant decreased. With the existence of non-target bacteria, the Cu-MOF nanoparticles could not be removed from the supernatant due to the lack of specific recognition. With this protocol, target bacteria could be selectively detected by measuring the residual Cu-MOF nanoparticles in the supernatant with the Cu-MOF catalyzed color reaction.
6. Conclusions and perspectives
Unquestionably, the past few years have witnessed the rapid rise of nanozymes in terms of both the exploration of various materials and the extensive applications of these nanozymes. Among these nanozymes, MOF nanozymes are arousing increasing interest because of their unique features. They integrate the versatile properties of MOF materials with attractive enzyme-like ability together and have found growing use ranging from sensing and biomedicine to environmental engineering and catalytic synthesis. Especially in the biochemical analytical community, MOF nanozymes are applied to establish several principles and strategies for the determination of a variety of analytes. Although in recent years many breakthroughs have been made in design, synthesis, and performance study, MOF nanozymes and their applications in biochemical detection are still in their infancy. Some issues should be taken into consideration for their future development in this attractive field.(1) Design and development of MOF nanozymes with desired activity. As mentioned above, the currently developed MOF nanozymes still exhibit very low catalytic activity due to their relatively large size and limited accessible active sites. Although some strategies have been proposed and demonstrated to effectively enhance the intrinsic poor activity of MOF nanozymes, the enhancement is still far from satisfactory. As a result, their catalytic capacity is still much lower than that of natural enzymes. In this respect, more efficient means of dramatically improving the poor activity of MOF nanozymes are required. Recently, single-atom nanozymes derived from MOF materials have been reported to exhibit impressive activity.81,85,91 Single atomization may be a feasible route to efficiently enhance the nanozyme activity. However, how to realize the single atomization of active sites in MOF nanozymes while retaining their attractive structures and properties is a great challenge.
(2) Stability of MOFs in aqueous solution. Another challenge for MOFs used as nanozymes is their stability in aqueous solution. Currently, the majority of MOFs are synthesized in organic solvents. When they are employed as nanozymes to catalyze reactions, their stability in aqueous solution should be taken into consideration. Nanozymes are expected to be recyclable and reusable. However, in most current analytical studies neither the reusable performance of MOF nanozymes nor their structural stability after the reaction is investigated. Especially, the framework structures of these MOFs are prone to be damaged during nanozyme catalysis. Therefore, developing MOF nanozymes highly stable in aqueous solution will benefit both their reuse and applications in biochemical sensing.
(3) Exploration of multifunctional MOF nanozymes. With diverse metal nodes and organic ligands, MOF materials can exhibit versatile properties, including an enzyme-like feature. Integrating the enzyme-mimicking catalytic ability and other attractive functions into one MOF material is a direction for future development, because such a multifunctional MOF will find more interesting applications in various fields. Up to today, MOFs with enzyme-like catalytic activity as well as optical properties (fluorescence, electrochemiluminescence, etc.) have been designed.15,29 Despite these advances, there are still some challenges that need to be overcome, including how to ensure that the multiple roles of a MOF nanozyme work efficiently without mutual interferences, and how to enable all the functions to synergistically serve a specific purpose. Aside from luminescent MOF nanozymes, it is also highly expected to design novel MOF nanozymes with interesting thermal, electrochemical, and magnetic properties.112
(4) Exploring new sensing principles and strategies using MOF nanozymes. Although a number of nanozyme-based detection systems have been constructed for the sensing of certain analytes, the type and number of targets that can be analyzed by nanozymes are still very limited.113 Currently, nanozymes are mainly employed to detect ions and small molecules using optical means. Their applications in protein detection, nucleic acid analysis, aptamer sensors, and immunoassays are still in the early stage. Given the catalytic nature of nanozymes to offer amplified signals, they are expected to find extensive use in ELISA, paper-based sensors, and electrochemical sensors for environmental monitoring, food safety control, biomarker detection and disease diagnosis, including the possible application in rapidly testing the COVID-19 spreading around the whole world. Further exploring novel principles and strategies and combining nanozymes with other advanced biochemical detection technologies for wider detection applications will be a hot topic in the near future.
(5) Selective detection. Lack of substrate specificity is one of the greatest barriers impeding the applications of nanozymes in the biochemical analytical field.114 Currently, achieving the selective detection of analytes using nanozyme catalysis relies on the specific interactions between nanozymes and analytes or the employment of biological recognition elements (antibodies, aptamers, etc.). Discovering more specific interactions of targets with nanozymes is a research trend. Besides, improving the catalytic specificity of nanozymes via appropriate means (e.g., artificial molecular imprinting) is also a general route to realize selective detection.115 It is known that the high catalytic selectivity of natural enzymes mainly depends on their specific spatial structures. Therefore, more attention should be paid to designing nanozymes mimicking biological enzymes not only in catalytic function but also in spatial structure. Only in this way can the catalytic selectivity problem of nanozymes be completely solved.
Taken together, the enzyme-like catalytic feature of MOFs together with their versatile nature is attracting more and more attention in the scientific community. It is expected that an increasing number of new MOFs mimicking natural enzymes not only in function but also in spatial structure will be designed and developed. Also, the poor activity problem of MOF nanozymes will be addressed with advanced nanotechnology and molecular engineering. More amazing designs of multifunctional MOF nanozymes will appear in the near future. These advances will greatly expand their application promise in various fields, including the biochemical detection field.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work is supported by a WSU startup fund. X. Niu and X. Li appreciate the support from the China Scholarship Council (Grant No. 201808695015 and 201908320292).References
- J. Bjerre, C. Rousseau, L. Marinescu and M. Bols, Appl. Microbiol. Biotechnol., 2008, 81, 1–11 CrossRef.
- L. Z. Gao, J. Zhuang, L. Nie, J. B. Zhang, Y. Zhang, N. Gu, T. H. Wang, J. Feng, D. L. Yang, S. Perrett and X. Y. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef.
- H. Wei and E. K. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- J. J. X. Wu, X. Y. Wang, Q. Wang, Z. P. Lou, S. R. Li, Y. Y. Zhu, L. Qin and H. Wei, Chem. Soc. Rev., 2019, 48, 1004–1076 RSC.
- Y. Y. Huang, J. S. Ren and X. G. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS.
- M. M. Liang and X. Y. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 CrossRef CAS.
- Y. B. Zhou, B. W. Liu, R. H. Yang and J. W. Liu, Bioconjugate Chem., 2017, 28, 2903–2909 CrossRef.
- P. Kumar, A. Deep and K. H. Kim, TrAC, Trends Anal. Chem., 2015, 73, 39–53 CrossRef.
- D. J. Wales, J. Grand, V. P. Ting, R. D. Burke, K. J. Edler, C. R. Bowen, S. Mintova and A. D. Burrows, Chem. Soc. Rev., 2015, 44, 4290–4321 RSC.
- J. P. Lei, R. C. Qian, P. H. Ling, L. Cui and H. X. Ju, TrAC, Trends Anal. Chem., 2014, 58, 71–78 CrossRef.
- J. E. Mondloch, M. J. Katz, W. C. I. Ill, P. Ghosh, P. Liao, W. Bury, G. W. Wagner, M. G. Hall, J. B. DeCoste, G. W. Peterson, R. Q. Snurr, C. J. Cramer, J. T. Hupp and O. K. Farha, Nat. Mater., 2015, 14, 512–516 CrossRef.
- E. Lýpez-Maya, C. Montoro, L. M. Rodriguez-Albelo, S. D. A. Cervantes, A. A. Lozano-Perez, J. L. Cenis, E. Barea and J. A. R. Navarro, Angew. Chem., Int. Ed., 2015, 54, 6790–6794 CrossRef.
- S. Y. Moon, G. W. Wagner, J. E. Mondloch, G. W. Peterson, J. B. DeCoste, J. T. Hupp and O. K. Farha, Inorg. Chem., 2015, 54, 10829–10833 CrossRef CAS.
- W. Q. Xu, L. Jiao, H. Y. Yan, Y. Wu, L. J. Chen, W. L. Gu, D. Du, Y. H. Lin and C. Z. Zhu, ACS Appl. Mater. Interfaces, 2019, 11, 22096–22101 CrossRef CAS.
- T. R. Lin, Y. M. Qin, Y. L. Huang, R. T. Yang, L. Hou, F. G. Ye and S. L. Zhao, Chem. Commun., 2018, 54, 1762–1765 RSC.
- L. J. Wang, Z. Hu, S. W. Wu, J. M. Pan, X. C. Xu and X. H. Niu, Anal. Chim. Acta, 2020, 1121, 26–34 CrossRef CAS.
- C. J. Gao, H. M. Zhu, J. Chen and H. D. Qiu, Chin. Chem. Lett., 2017, 28, 1006–1012 CrossRef CAS.
- P. H. Ling, J. P. Lei, L. Zhang and H. X. Ju, Anal. Chem., 2015, 87, 3957–3963 CrossRef CAS.
- L. Cui, J. Wu, J. Li and H. X. Ju, Anal. Chem., 2015, 87, 10635–10641 CrossRef CAS.
- H. J. Cheng, Y. F. Liu, Y. H. Hu, Y. B. Ding, S. C. Lin, W. Cao, Q. Wang, J. J. X. Wu, F. Muhammad, X. Z. Zhao, D. Zhao, Z. Li, H. Xing and H. Wei, Anal. Chem., 2017, 89, 11552–11559 CrossRef.
- X. F. Ruan, D. Liu, X. H. Niu, Y. J. Wang, C. D. Simpson, N. Cheng, D. Du and Y. H. Lin, Anal. Chem., 2019, 91, 13847–13854 CrossRef.
- L. H. Ai, L. L. Li, C. H. Zhang, J. Fu and J. Jiang, Chem. – Eur. J., 2013, 19, 15105–15108 CrossRef.
- Y. L. Liu, X. J. Zhao, X. Y. Yang and Y. F. Li, Analyst, 2013, 138, 4526–4531 RSC.
- J. W. Zhang, H. T. Zhang, Z. Y. Du, X. Q. Wang, S. H. Yu and H. L. Jiang, Chem. Commun., 2014, 50, 1092–1094 RSC.
- Z. W. Jiang, Y. L. Liu, X. L. Hu and Y. F. Li, Anal. Methods, 2014, 6, 5647–5651 RSC.
- W. F. Dong, X. D. Liu, W. B. Shi and Y. M. Huang, RSC Adv., 2015, 5, 17451–17457 RSC.
- J. Y. Lu, Y. H. Xiong, C. J. Liao and F. G. Ye, Anal. Methods, 2015, 7, 9894–9899 RSC.
- D. M. Chen, B. Li, L. Jiang, D. L. Duan, Y. Z. Li, J. Q. Wang, J. He and Y. B. Zeng, RSC Adv., 2015, 5, 97910–97917 RSC.
- K. Ye, L. J. Wang, H. W. Song, X. Li and X. H. Niu, J. Mater. Chem. B, 2019, 7, 4794–4800 RSC.
- N. Cheng, C. Z. Zhu, Y. L. Wang, D. Du, M. J. Zhu, Y. B. Luo, W. T. Xu and Y. H. Lin, J. Anal. Test., 2019, 3, 99–106 CrossRef.
- D. W. Feng, Z. Y. Gu, J. R. Li, H. L. Jiang, Z. W. Wei and H. C. Zhou, Angew. Chem., Int. Ed., 2012, 51, 10307–10310 CrossRef CAS.
- H. L. Tan, Q. Li, Z. C. Zhou, C. J. Ma, Y. H. Song, F. G. Xu and L. Wang, Anal. Chim. Acta, 2015, 856, 90–95 CrossRef CAS.
- S. Q. Wang, W. F. Deng, L. Yang, Y. M. Tan, Q. J. Xie and S. Z. Yao, ACS Appl. Mater. Interfaces, 2017, 9, 24440–24445 CrossRef CAS.
- J. Wang, Y. Y. Hu, Q. Zhou, L. Z. Hu, W. S. Fu and Y. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 44466–44473 CrossRef CAS.
- S. S. Hu, L. Zhu, C. W. Lam, L. H. Guo, Z. Y. Lin, B. Qiu, K. Y. Wong, G. N. Chen and Z. H. Liu, Microchim. Acta, 2019, 186, 190 CrossRef.
- L. He, Y. Li, Q. Wu, D. M. Wang, C. M. Li, C. Z. Huang and Y. F. Li, ACS Appl. Mater. Interfaces, 2019, 11, 29158–29166 CrossRef CAS.
- J. Y. Chen, Y. Shu, H. L. Li, Q. Xu and X. Y. Hu, Talanta, 2018, 189, 254–261 CrossRef CAS.
- X. H. Niu, Y. F. He, J. M. Pan, X. Li, F. X. Qiu, Y. S. Yan, L. B. Shi, H. L. Zhao and M. B. Lan, Anal. Chim. Acta, 2016, 947, 42–49 CrossRef CAS.
- H. Q. Liu, J. N. Rong, G. Q. Shen, Y. Song, W. Gu and X. Liu, Dalton Trans., 2019, 48, 4168–4175 RSC.
- S. Sloan-Dennison, S. Laing, N. C. Shand, D. Grahama and K. Faulds, Analyst, 2017, 142, 2484–2490 RSC.
- X. H. Niu, X. C. Xu, X. Li, J. M. Pan, F. X. Qiu, H. L. Zhao and M. B. Lan, Chem. Commun., 2018, 54, 13443–13446 RSC.
- Y. F. He, X. Li, X. C. Xu, J. M. Pan and X. H. Niu, J. Mater. Chem. B, 2018, 6, 5750–5755 RSC.
- Y. Wang, R. P. Liang and J. D. Qiu, Anal. Chem., 2020, 92, 2339–2346 CrossRef CAS.
- Y. H. Xiong, S. H. Chen, F. G. Ye, L. L. Su, C. Zhang, S. F. Shen and S. L. Zhao, Chem. Commun., 2015, 51, 4635–4638 RSC.
- R. Dalapati, B. Sakthivel, M. K. Ghosalya, A. Dhakshinamoorthy and S. Biswas, CrystEngComm, 2017, 19, 5915–5925 RSC.
- H. W. Song, K. Ye, Y. X. Peng, L. J. Wang and X. H. Niu, J. Mater. Chem. B, 2019, 7, 5834–5841 RSC.
- T. Jin, Y. L. Li, W. J. Jing, Y. C. Li, L. Z. Fan and X. H. Li, Chem. Commun., 2020, 56, 659–662 RSC.
- H. Liang, F. F. Lin, Z. J. Zhang, B. W. Liu, S. H. Jiang, Q. P. Yuan and J. W. Liu, ACS Appl. Mater. Interfaces, 2017, 9, 1352–1360 CrossRef CAS.
- J. H. Wang, R. L. Huang, W. Qi, R. X. Su, B. P. Binks and Z. M. He, Appl. Catal., B, 2019, 254, 452–462 CrossRef CAS.
- D. Li, B. W. Liu, P. J. J. Huang, Z. J. Zhang and J. W. Liu, Chem. Commun., 2018, 54, 12519–12522 RSC.
- Y. F. Liu, M. Zhou, W. Cao, X. Y. Wang, Q. Wang, S. R. Li and H. Wei, Anal. Chem., 2019, 91, 8170–8175 CrossRef CAS.
- Y. F. Liu, X. Y. Wang and H. Wei, Analyst, 2020, 145, 4388–4397 RSC.
- X. H. Zhang, W. Liu, X. M. Li, Z. Zhang, D. L. Shan, H. Xia, S. T. Zhang and X. Q. Lu, Anal. Chem., 2018, 90, 14309–14315 CrossRef CAS.
- Q. S. Xue, X. Li, Y. X. Peng, P. Liu, H. B. Peng and X. H. Niu, Microchim. Acta, 2020, 187, 263 CrossRef CAS.
- L. Zhang, Y. Zhang, Z. Z. Wang, F. F. Cao, Y. J. Sang, K. Dong, F. Pu, J. S. Ren and X. G. Qu, Mater. Horiz., 2019, 6, 1682–1687 RSC.
- P. Li, R. C. Klet, S. Y. Moon, T. C. Wang, P. Deria, A. W. Peters, B. M. Klahr, H. J. Park, S. S. Al-Juaid, J. T. Hupp and O. K. Farha, Chem. Commun., 2015, 51, 10925–10928 RSC.
- H. Y. Chen, P. L. Liao, M. L. Mendonca and R. Q. Snurr, J. Phys. Chem. C, 2018, 122, 12362–12368 CrossRef CAS.
- H. J. Park, J. K. Jang, S. Y. Kim, J. W. Ha, D. Moon, I. N. Kang, Y. S. Bae, S. Kim and D. H. Hwang, Inorg. Chem., 2017, 56, 12098–12101 CrossRef CAS.
- B. Li, D. M. Chen, J. Q. Wang, Z. Y. Yan, L. Jiang, D. L. Duan, J. He, Z. R. Luo, J. P. Zhang and F. G. Yuan, Sci. Rep., 2015, 4, 6759 CrossRef.
- L. L. Li, B. Li, D. M. Chen, J. C. Zhao, D. Q. Yang, D. H. Ma, L. Jiang, Y. P. Yang, Y. Z. Li and J. Q. Wang, J. Biosci. Med., 2019, 7, 222–230 CAS.
- J. X. Chen, L. Huang, Q. Q. Wang, W. W. Wu, H. Zhang, Y. X. Fang and S. J. Dong, Nanoscale, 2019, 11, 5960 RSC.
- L. P. Luo, L. J. Huang, X. N. Liu, W. T. Zhang, X. L. Yao, L. N. Dou, X. Zhang, Y. Nian, J. Sun and J. L. Wang, Inorg. Chem., 2019, 58, 11382–11388 CrossRef CAS.
- X. Li, H. Zhou, F. Qi, X. H. Niu, X. C. Xu, F. X. Qiu, Y. F. He, J. M. Pan and L. Ni, J. Mater. Chem. B, 2018, 6, 6207–6211 RSC.
- H. G. Yang, R. T. Yang, P. Zhang, Y. M. Qin, T. Chen and F. G. Ye, Microchim. Acta, 2017, 184, 4629–4635 CrossRef CAS.
- L. Qin, X. Y. Wang, Y. F. Liu and H. Wei, Anal. Chem., 2018, 90, 9983–9989 CrossRef CAS.
- N. F. Zhu, L. T. Gu, J. Wang, X. S. Li, G. X. Liang, J. H. Zhou and Z. Zhang, J. Phys. Chem. C, 2019, 123, 9388–9393 CrossRef CAS.
- Y. X. Wang, M. T. Zhao, J. F. Ping, B. Chen, X. H. Cao, Y. Huang, C. L. Tan, Q. L. Ma, S. X. Wu, Y. F. Yu, Q. P. Lu, J. Z. Chen, W. Zhao, Y. B. Ying and H. Zhang, Adv. Mater., 2016, 28, 4149–4155 CrossRef CAS.
- X. H. Qi, H. M. Tian, X. M. Dang, Y. F. Fan, Y. B. Zhang and H. M. Zhao, Anal. Methods, 2019, 11, 1111–1124 RSC.
- Y. Zhang, C. L. Dai, W. Liu, Y. Y. Wang, F. Ding, P. Zou, X. X. Wang, Q. B. Zhao and H. B. Rao, Microchim. Acta, 2019, 186, 340 CrossRef.
- S. C. Lin and H. Wei, Sci. China: Life Sci., 2019, 62, 710–712 CrossRef.
- C. Z. Zhu, S. F. Fu, Q. R. Shi, D. Du and Y. H. Lin, Angew. Chem., Int. Ed., 2017, 56, 13944–13960 CrossRef CAS.
- C. Z. Zhu, S. F. Fu, J. H. Song, Q. R. Shi, D. Su, M. H. Engelhard, X. L. Li, D. D. Xiao, D. S. Li, L. Estevez, D. Du and Y. H. Lin, Small, 2017, 13, 1603407 CrossRef.
- S. F. Fu, C. Z. Zhu, D. Su, J. H. Song, S. Y. Yao, S. Feng, M. H. Engelhard, D. Du and Y. H. Lin, Small, 2018, 14, 1703118 CrossRef.
- C. Z. Zhu, Q. R. Shi, B. Z. Xu, S. F. Fu, G. Wan, C. Yang, S. Y. Yao, J. H. Song, H. Zhou, D. Du, S. P. Beckman, D. Su and Y. H. Lin, Adv. Energy Mater., 2018, 8, 1801956 CrossRef.
- J. C. Li, M. Cheng, T. Li, L. Ma, X. F. Ruan, D. Liu, H. M. Cheng, C. Liu, D. Du, Z. D. Wei, Y. H. Lin and M. H. Shao, J. Mater. Chem. A, 2019, 7, 14478–14482 RSC.
- D. Liu, J. C. Li, Q. R. Shi, S. Feng, Z. Y. Lyu, S. C. Ding, L. D. Hao, Q. Zhang, C. H. Wang, M. J. Xu, T. Li, E. Sarnello, D. Du and Y. H. Lin, ACS Appl. Mater. Interfaces, 2019, 11, 39820–39826 CrossRef CAS.
- D. Liu, J. C. Li, S. C. Ding, Z. Y. Lyu, S. Feng, H. Y. Tian, C. X. Huyan, M. J. Xu, T. Li, D. Du, P. Liu, M. H. Shao and Y. H. Lin, Small Methods, 2020, 4, 1900827 CrossRef.
- W. L. Zhu, J. J. Fu, J. Liu, Y. Chen, X. Li, K. K. Huang, Y. M. Cai, Y. M. He, Y. Zhou, D. Su, J. J. Zhu and Y. H. Lin, Appl. Catal., B, 2020, 264, 118502 CrossRef.
- C. Z. Zhu, Q. R. Shi, S. Feng, D. Du and Y. H. Lin, ACS Energy Lett., 2018, 3, 1713–1721 CrossRef CAS.
- B. T. Qiao, A. Q. Wang, X. F. Yang, L. F. Allard, Z. Jiang, Y. T. Cui, J. Y. Liu, J. Li and T. Zhang, Nat. Chem., 2011, 3, 634–641 CrossRef CAS.
- L. Huang, J. X. Chen, L. F. Gan, J. Wang and S. J. Dong, Sci. Adv., 2019, 5, 5490 CrossRef.
- W. J. Ma, J. J. Mao, X. T. Yang, C. Pan, W. X. Chen, M. Wang, P. Yu, L. Q. Mao and Y. D. Li, Chem. Commun., 2019, 55, 159–162 RSC.
- B. L. Xu, H. Wang, W. W. Wang, L. Z. Gao, S. S. Li, X. T. Pan, H. Y. Wang, H. L. Yang, X. Q. Meng, Q. W. Wu, L. R. Zheng, S. M. Chen, X. H. Shi, K. L. Fan, X. Y. Yan and H. Y. Liu, Angew. Chem., Int. Ed., 2019, 58, 4911–4916 CrossRef CAS.
- C. Zhao, C. Xiong, X. K. Liu, M. Qiao, Z. J. Li, T. W. Yuan, J. Wang, Y. T. Qu, X. Q. Wang, F. Y. Zhou, Q. Xu, S. Q. Wang, M. Chen, W. Y. Wang, Y. F. Li, T. Yao, Y. E. Wu and Y. D. Li, Chem. Commun., 2019, 55, 2285–2288 RSC.
- X. H. Niu, Q. R. Shi, W. L. Zhu, D. Liu, H. Y. Tian, S. F. Fu, N. Cheng, S. Q. Li, J. N. Smith, D. Du and Y. H. Lin, Biosens. Bioelectron., 2019, 142, 111495 CrossRef CAS.
- N. Cheng, J. C. Li, D. Liu, Y. H. Lin and D. Du, Small, 2019, 15, 1901485 CrossRef CAS.
- Y. Wu, L. Jiao, X. Luo, W. Q. Xu, X. Q. Wei, H. J. Wang, H. Y. Yan, W. L. Gu, B. Z. Xu, D. Du, Y. H. Lin and C. Z. Zhu, Angew. Chem., Int. Ed., 2019, 15, 1903108 CAS.
- L. Jiao, W. Q. Xu, H. Y. Yan, Y. Wu, C. R. Liu, D. Du, Y. H. Lin and C. Z. Zhu, Anal. Chem., 2019, 18, 11994–11999 CrossRef.
- W. L. Gu, H. J. Wang, L. Jiao, Y. Wu, Y. X. Chen, L. Y. Hu, J. M. Gong, D. Du and C. Z. Zhu, Angew. Chem., Int. Ed., 2020, 59, 3534–3538 CrossRef CAS.
- Y. Wu, J. B. Wu, L. Jiao, W. Q. Xu, H. J. Wang, X. Q. Wei, W. L. Gu, G. X. Ren, N. Zhang, Q. H. Zhang, L. Huang, L. Gu and C. Z. Zhu, Anal. Chem., 2020, 92, 3373–3379 CrossRef CAS.
- L. Jiao, H. Y. Yan, Y. Wu, W. L. Gu, C. Z. Zhu, D. Du and Y. H. Lin, Angew. Chem., Int. Ed., 2019, 132, 2585–2596 CrossRef.
- B. Jiang, D. M. Duan, L. Z. Gao, M. J. Zhou, K. L. Fan, Y. Tang, J. Q. Xi, Y. H. Bi, Z. Tong, G. F. Gao, N. Xie, A. F. Tang, G. H. Nie, M. M. Liang and X. Y. Yan, Nat. Protoc., 2018, 13, 1506–1520 CrossRef CAS.
- H. J. Cheng, L. Zhang, J. He, W. J. Guo, Z. Y. Zhou, X. J. Zhang, S. M. Nie and H. Wei, Anal. Chem., 2016, 88, 5489–5497 CrossRef CAS.
- C. Hou, Y. Wang, Q. H. Ding, L. Jiang, M. Li, W. W. Zhu, D. Pan, H. Zhu and M. Z. Liu, Nanoscale, 2015, 7, 18770–18779 RSC.
- Z. H. Zhao, J. H. Pang, W. W. Liu, T. R. Lin, F. G. Ye and S. L. Zhao, Microchim. Acta, 2019, 186, 295 CrossRef.
- Z. H. Zhao, Y. J. Huang, W. R. Liu, F. G. Ye and S. L. Zhao, ACS Sustainable Chem. Eng., 2020, 8, 4481–4488 CrossRef CAS.
- Z. L. Wang, X. Y. Jiang, R. Yuan and Y. Q. Chai, Biosens. Bioelectron., 2018, 121, 250–256 CrossRef CAS.
- L. Wang and Y. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 8351–8358 CrossRef CAS.
- X. Q. Tang, Y. D. Zhang, Z. W. Jiang, D. M. Wang, C. Z. Huang and Y. F. Li, Talanta, 2018, 179, 43–50 CrossRef.
- F. X. Qin, S. Y. Jia, F. F. Wang, S. H. Wu, J. Song and Y. Liu, Catal. Sci. Technol., 2013, 3, 2761–2768 RSC.
- Y. Z. Wu, Y. J. Ma, G. H. Xu, F. D. Wei, Y. S. Ma, Q. Song, X. Wang, T. Tang, Y. Y. Song, M. L. Shi, X. M. Xu and Q. Hu, Sens. Actuators, B, 2017, 249, 195–202 CrossRef CAS.
- Y. H. Hu, H. J. Cheng, X. Z. Zhao, J. J. X. Wu, F. Muhammad, S. C. Lin, J. He, L. Q. Zhou, C. P. Zhang, Y. Deng, P. Wang, Z. Y. Zhou, S. M. Nie and H. Wei, ACS Nano, 2017, 11, 5558–5566 CrossRef CAS.
- Y. L. Liu, W. L. Fu, C. M. Li, C. Z. Huang and Y. F. Li, Anal. Chim. Acta, 2015, 861, 55–61 CrossRef CAS.
- B. W. Liu and J. W. Liu, Nanoscale, 2015, 7, 13831–13835 RSC.
- Y. Zhang, W. T. Zhang, K. Chen, Q. F. Yang, N. Hu, Y. R. Suo and J. L. Wang, Sens. Actuators, B, 2018, 262, 95–101 CrossRef CAS.
- J. S. Lin, Q. Wang, X. Y. Wang, Y. Y. Zhu, X. Zhou and H. Wei, Analyst, 2020, 145, 3916–3921 RSC.
- Y. Y. Zhu, J. J. X. Wu, L. J. Han, X. Y. Wang, W. Li, H. C. Guo and H. Wei, Anal. Chem., 2020, 92, 7444–7452 CrossRef CAS.
- X. C. Xu, S. W. Wu, D. Z. Guo and X. H. Niu, Anal. Chim. Acta, 2020, 1107, 203–212 CrossRef CAS.
- S. W. Wu, D. Z. Guo, X. C. Xu, J. M. Pan and X. H. Niu, Sens. Actuators, B, 2020, 303, 127225 CrossRef CAS.
- X. H. Niu, N. Cheng, X. F. Ruan, D. Du and Y. H. Lin, J. Electrochem. Soc., 2020, 167, 037508 CrossRef CAS.
- Y. Wang, Y. J. Zhu, A. Binyam, M. Liu, Y. N. Wu and F. T. Li, Biosens. Bioelectron., 2016, 86, 432–438 CrossRef CAS.
- J. J. X. Wu, S. R. Li and H. Wei, Nanoscale Horiz., 2018, 3, 367–382 RSC.
- X. Li, L. J. Wang, D. Du, L. Ni, J. M. Pan and X. H. Niu, TrAC, Trends Anal. Chem., 2019, 120, 115653 CrossRef CAS.
- J. J. Gooding, ACS Sens., 2019, 4, 2213–2214 CrossRef CAS.
- Z. J. Zhang, X. H. Zhang, B. W. Liu and J. W. Liu, J. Am. Chem. Soc., 2017, 139, 5412–5419 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |