 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Automated solid-phase concatenation of Aib residues to form long, water-soluble, helical peptides†
Francis
Zieleniewski
 a,
Derek N.
Woolfson
a,
Derek N.
Woolfson
 *abc and
Jonathan
Clayden
*abc and
Jonathan
Clayden
 *a
*a
aSchool of Chemistry, University of Bristol, Cantock's Close, Bristol, BS8 1TS, UK. E-mail: D.N.Woolfson@bristol.ac.uk; j.clayden@bristol.ac.uk
bSchool of Biochemistry, University of Bristol, Medical Sciences Building, University Walk, Bristol, BS8 1TS, UK
cBristol BioDesign Institute, University of Bristol, Life Sciences Building, Tyndall Avenue, Bristol BS8 1TQ, UK
First published on 2nd September 2020
Abstract
Iterative coupling of 2-aminoisobutyric acid (Aib) has been achieved rapidly and efficiently using automated solid-phase peptide synthesis, employing diisopropylcarbodiimide (DIC) in the presence of ethyl cyanohydroxyiminoacetate (Oxyma). This method has allowed the first total synthesis of the fungal antibiotic Cephaibol D, and enabled the synthesis of water-soluble oligomers of Aib containing up to an unprecedented sequence of 17 consecutive Aib residues. Conformational analysis of the Aib oligomers in aqueous solution shows a length dependence in their CD spectra, with oligomers of more than 14 Aib residues apparently adopting structured helical conformations.
De novo protein design using both rational and computational approaches allows a wide range a novel peptide structures to be explored.1,2 α-Helices have been widely used as design components of these novel proteins.3,4 However, the 310 helix, an analogue of the α-helix with only three residues per turn and also found in natural protein structures, has gained significantly less attention.5 A general strategy for incorporating long 310-helical modules into artificial proteins would advance de novo protein design, allowing use of a new secondary structure in synthetic proteins.
The extended 310-helical conformation is commonly adopted by peptides with a high density of α,α-disubstituted amino acids,6 the simplest of which is aminoisobutyric acid (Aib, abbreviated here as U).7 However, these 310 helices tend to collapse towards α-helical conformations in more polar solvents,8 and while the conformational properties of Aib oligomers have been studied in organic solvents9,10 and membranes,11 it is not clear whether long Aib-rich structures can be maintained in water. Indeed, there are few reported examples of water-soluble 310 helices. The gp41659–671 peptide adopts a 310 helix in water, and, remarkably, it is made up entirely of proteinogenic amino acids.12 Toniolo and co-workers report a water-soluble Aib-rich 310 helix with (L)-2-amino-3[1-(1,4,7-triazacyclononane)]propanoic acid residues placed three residues apart to allow aqueous solvation,13 but this peptide is only 7 residues in length.
Here, we show that it is possible to form long oligomers of Aib (up to 17 consecutive residues) that are soluble in water. To achieve this, we developed conditions that allow iterative successive coupling of hindered α,α-disubstituted amino acids on a solid-phase support. For the first time, this allows the synthesis of long Aib-rich sequences by automated solid-phase peptide synthesis (SPPS).
The conformational properties of Aib oligomers—typically of 4–12 monomers—have been studied in non-aqueous solvents by making use of an iterative solution-phase acylation-reduction sequence.9 However, the standard technique for the synthesis of natural and unnatural peptide sequences is SPPS,14 employing the base-labile Fmoc group to protect the α-amino group of the growing peptide chain, and orthogonal acid-labile peptide-resin linkages and side-chain protecting groups.15 Automation of SPPS16 has accelerated coupling–deprotection cycles to the extent that each single-residue chain extension may take as little as four minutes in microwave-equipped instruments.17 Although these standard conditions work well with proteinogenic and other α-amino acids, they give poor yields with sterically hindered α,α-disubstituted amino acids such as Aib.
Nonetheless, successive Aib residues may be coupled to a peptide chain using SPPS much more rapidly than in solution.7 Hara and co-workers report the use of 3-nitro-1,2,4-triazol-1-yltris(pyrrolidin-1-yl)phosphonium hexafluorophosphate (PyNTP) to couple up to 10 successive Aib residues in high yield with a coupling time of only 20 minutes at room temperature.18 The drawback of this approach is that 20 equivalents of PyNTP and of base are required. Also, the expense and instability of phosphonium and uronium salts makes them less attractive for routine use in automated synthesis.
A more-appealing approach to Aib couplings employs ethyl cyanohydroxyiminoacetate (Oxyma). Initially reported in the 1970s as a way to reduce epimerisation,19 this weakly acidic oxime has been applied in conjunction with DIC to enable difficult couplings on a solid support by Albericio and coworkers.20 Oxyma allows two consecutive Aib residues to be coupled using DIC more efficiently than with common HOAt and HOBt additives. Oxyma is also stable in DMF and does not cap the resin prematurely during synthesis; two factors that favour its utility in automated SPPS. A variety of oxonium and phosphonium coupling reagents based on Oxyma have been reported, such as COMU and PyOxim.21
The combination of DIC with Oxyma has found use in microwave-assisted automated SPPS,17 and Oxyma's superiority as an activating additive has been demonstrated for coupling two successive Aib residues during the synthesis of the peptaibols alamethicin and bergofungin.22 Using two equivalents of DIC relative to the amino acid (or 10 equivalents relative to the resin loading) significantly increases peptide purity,23 by increasing the rate of formation of O-acylurea, accelerating the activation of the amino acid. The use of DIC + Oxyma is generally considered the method of choice for difficult peptide couplings using microwave assisted automated SPPS.24
Using DIC + Oxyma as the coupling method, we explored the types of water-soluble Aib-rich sequences that could be made by automated SPPS. To validate the method for making Aib-rich structures, we targeted Cephaibol D (1, Fig. 1),25 a 15-residue peptaibol containing eight Aib (U) and two hydroxyproline (O) residues. A partial synthesis of this molecule has been reported26 but not a total synthesis. The Aib residues occur as a synthetically challenging group of four consecutive residues, a pair of consecutive Aib residues followed by hydroxyproline (UUO), and a UOUP motif towards the C terminus.
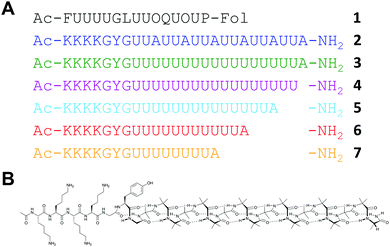 | ||
| Fig. 1 (a) Sequences of Cephaibol D 1 and other peptides synthesised (2–7). (b) Displayed structure of 3. Key: U, Aib; O, Hyp; Fol, phenylalaninol. | ||
Starting from preloaded Fmoc-phenylalaninol o-chlorotrityl resin (0.44 mmol g−1) and using Fmoc protection of the N terminus, 1 was synthesised on a 0.1 mmol scale using 5 equivalents of amino acid and Oxyma and 10 equivalents of DIC in each coupling cycle. The optimum conditions for coupling were irradiation at 90 °C for 3 min with standard amino acids, or at 100 °C for 10 min with Pro, Hyp, Aib and any amino acid required to couple to an N-terminal Aib. Each Fmoc deprotection was achieved using a 20% solution of morpholine in DMF with formation of the morpholine-fulvene adduct monitored by UV absorption at 301 nm to estimate coupling efficacy (see ESI†). On completion of the synthesis, the N terminus of the peptide was acetylated on the resin with a solution of acetic anhydride and DIPEA in DMF for 30 min. The peptide was cleaved from the resin and deprotected in dilute TFA to prevent cleavage of Aib-Pro bond.22
This approach gave Cephaibol D 1 in just over 5.5 h with high crude purity (Table 1), as determined by HPLC (Fig. 2). Purification by semipreparative HPLC gave 1 in 26% yield over 15 coupling and deprotection steps.
| Product | Synthesis time (h![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) min) min) |
Crude purity (%) | Isolated yield (%) |
|---|---|---|---|
| 1 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 37 |
40 | 26 |
| 2 | 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 22 |
67 | 41 |
| 3 | 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 19 |
59 | 21 |
| 4 | 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 03 03 |
61 | 19 |
| 5 | 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 01 01 |
61 | 33 |
| 6 | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 19 |
75 | 52 |
| 7 | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
74 | 55 |
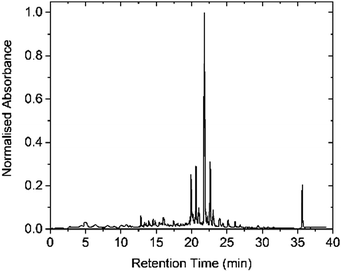 | ||
| Fig. 2 HPLC profile of crude Cephaibol D 1 (detection at 220 nm). The peak at 22 min was identified as Cephaibol D by MALDI-TOF mass spectrometry. | ||
This successful first total synthesis of Cephaibol D demonstrates that multiple Aib-on-Aib couplings are possible in high yield using SPPS without recourse to large excesses of phosphonium salts. On this basis, we designed longer sequences containing consecutive Aib residues and maintaining water solubility to allow purification by reverse-phase HPLC. The extended Aib-rich sequences 2 and 3 are similar to the Aib-rich 310 helices studied in organic solvents.9,26 However, to facilitate solubility in water, they incorporate a 7-residue cationic tag, KKKKGYG. The tag was positioned at the N terminus of the peptide to avoid interference with the folding of the Aib-rich segment and incorporated a glycine-flanked27 chromophore to allow the aqueous concentration to be quantified.
As Aib is achiral, its oligomers populate equally pairs of enantiomeric confomers.28 Therefore, observation of conformation by circular dichroism (CD) spectroscopy requires at least one chiral residue to induce a screw-sense preference. The 6-fold UUA repeat of 2 incorporates a high density of chiral residues to induce a screw sense preference. Nonetheless, the screw sense of an Aib oligomer can be propagated from a single chiral residue.29 Therefore, we made 3, which is identical in length to 2 but with a single Ala residue adjacent to 17 consecutive Aib residues. Control peptide 4, lacking the Ala residue, was made to verify the conformational influence of the cationic tag. 2, 3 and 4 were synthesised using the same conditions as 1, replacing Fmoc-phenylalaninol o-chlorotrityl resin with rink amide MHBA resin, and using more powerfully acidic conditions for resin cleavage and side-chain deprotection (see ESI†). All three peptides were synthesised in just over 8 hours with acceptable crude purities and isolated yields (Table 1).
The water-soluble peptides 2 and 3 allowed the first ever observation of the CD spectrum of Aib oligopeptides in aqueous solution (100 μM, PBS buffer). CD spectra of 2 and 3 (Fig. 3a) resembled spectra of Aib oligomers in organic solvents,9 with a negative signal at 205 nm (indicating a right-handed screw sense preference) and a positive maximum at 225 nm.30 The more-intense CD signal of 2 can be attributed to the greater density of Ala residues, giving 2 a greater average screw-sense preference than 3. Nonetheless, the comparable magnitude of their CD signals suggests that a substantial proportion of 3 adopts a right-handed screw sense, induced from the C terminus (Fig. 1b).9d Peptide 4 had no signal in the range 210–260 nm, confirming that the cationic tag does not induce a screw-sense preference.
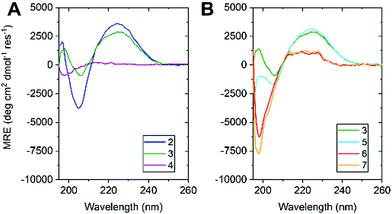 | ||
| Fig. 3 CD spectra of (A) 2, 3 & 4 and (B) 3, 5, 6 & 7 in H2O (100 μM, 1 mm pathlength, 5 °C, PBS, pH 7.4). Colour coded as in Fig. 1. | ||
Previous data recorded in organic solvents suggest that the stability of the helical conformation adopted by oligomers rich in α,α-disubstituted amino acids is length dependent.31,32 Therefore, we synthesised 5, 6 & 7, shorter homologues of 3 with 14, 11 and 8 consecutive Aib residues, respectively. 6 and 7 showed no evidence of folding, despite their C-terminal L-Ala residue (Fig. 3b), suggesting that oligomers of fewer than 11 Aib residues do not form stable helices in water.
Peptide 5, with 14 Aib residues, behaved quite differently to the others. Immediately after dissolving in PBS buffer, its CD spectrum matched closely that of 3, suggesting that a 14-mer of Aib may adopt a structured conformation in water. However, over a period of minutes at room temperature, the CD curve changed to one characteristic of a random coil (Fig. S1, ESI†). Further exploration of the details of the structure adopted by amphipathic peptides 2, 3 and 5, and of the conformational transition of 5 in water, is under way.
In summary, long chains of Aib residues can be synthesised rapidly and in high yield by automated SPPS using DIC with an Oxyma coupling cocktail. This method has allowed the first total synthesis of Cephaibol D. The coupling of 17 consecutive Aib residues has been achieved in only 8 hours, which is much more rapid than alternative methods. Capping the Aib oligomers with a cationic tag makes them water-soluble, and these water-soluble Aib oligopeptides show CD spectra similar to those seen in organic solvents. We anticipate that this work will be of value in the de novo design, synthesis and characterisation of both water-soluble and membrane-active helical peptides built from non-proteinogenic residues.
F. Z. thanks Dr Caroline Morris for helpful discussions and assistance with the peptide synthesiser. This work was supported by the EPSRC funded Bristol Chemical Synthesis Centre for Doctoral Training EP/G036764/1 (studentship to F. Z.) and programme grant EP/P027067 (awarded to JPC). DNW held a Royal Society Wolfson Research Merit Award (WM140008). We also thank the University of Bristol School of Chemistry Mass Spectrometry Facility for use of the EPSRC-funded Bruker Ultraflex MALDI-TOF/TOF instrument (EP/K03927X/1), and the BBSRC/EPSRC-funded BrisSynBio (BB/L01386X1) for access to its peptide synthesisers.
Conflicts of interest
There are no conflicts to declare.Notes and references
- L. Regan, D. Caballero, M. R. Hinrichsen, A. Virrueta, D. M. Williams and C. S. O’Hern, Biopolymers, 2015, 104, 332 CrossRef.
- P. S. Huang, S. E. Boyken and D. Baker, Nature, 2016, 537, 320 CrossRef CAS.
- D. N. Woolfson, in Fibrous Proteins: Structures and Mechanisms, ed. D. A. D. Perry and J. M Squire, Springer, Cham, 1st edn, 2017, vol. 82, ch. 2, pp. 35–62 Search PubMed.
- I. V. Korendovych and W. F. DeGrado, Q. Rev. Biophys., 2020, 53, 1 CrossRef.
- L. Pal and G. Basu, Protein Eng., 1999, 12, 811 CrossRef CAS.
- R. Hummel, C. Toniolo and G. Jung, Angew. Chem., Int. Ed. Engl., 1987, 26, 1150 CrossRef.
- J. Clayden, A. Castellanos, J. Solà and G. A. Morris, Angew. Chem., Int. Ed., 2009, 48, 5962 CrossRef CAS.
- (a) T. S. Yokum, T. J. Gauthier, R. P. Hammer and M. L. McLaughlin, J. Am. Chem. Soc., 1997, 119, 1167 CrossRef CAS; (b) P. Pengo, L. Pasquato, S. Moro, A. Brigo, F. Fogolari, Q. B. Broxterman, B. Kaptein and P. Scrimin, Angew. Chem., Int. Ed., 2003, 42, 3388 CrossRef CAS.
- (a) R. A. Brown, T. Marcelli, M. De Poli, J. Solà and J. Clayden, Angew. Chem., Int. Ed., 2012, 51, 1395 CrossRef CAS; (b) M. De Poli, L. Byrne, R. A. Brown, J. Solà, A. Castellanos, T. Boddaert, R. Wechsel, J. D. Beadle and J. Clayden, J. Org. Chem., 2014, 79, 4659 CrossRef CAS; (c) B. A. F. Le Bailly and J. Clayden, Chem. Commun., 2014, 50, 7949 RSC; (d) M. Tomsett, I. Maffucci, B. A. F. Le Bailly, L. Byrne, S. M. Bijvoets, M. G. Lizio, J. Raftery, C. P. Butts, S. J. Webb, A. Contini and J. Clayden, Chem. Sci., 2017, 8, 3007 RSC.
- (a) R. A. G. D. Silva, S. C. Yasui, J. Kubelka, F. Formaggio, M. Crisma, C. Toniolo and T. A. Keiderling, Biopolymers, 2002, 65, 229 CrossRef CAS; (b) A. Moretto, M. Crisma, F. Formaggio, B. Kaptein, Q. B. Broxterman, T. A. Keiderling and C. Toniolo, Pept. Sci., 2007, 88, 233 CrossRef CAS.
- (a) S. J. Pike, J. E. Jones, J. Raftery, J. Clayden and S. J. Webb, Org. Biomol. Chem., 2015, 13, 9580 RSC; (b) J. E. Jones, V. Diemer, V. C. Adam, J. Raftery, R. E. Ruscoe, J. T. Sengel, M. I. Wallace, A. Bader, A. S. L. Cockroft, J. Clayden and S. J. Webb, J. Am. Chem. Soc., 2016, 138, 688 CrossRef CAS; (c) C. Adam, A. D. Peters, M. G. Lizio, G. F. S. Whitehead, V. Diemer, J. A. Cooper, S. L. Cockroft, J. Clayden and S. J. Webb, Chem. – Eur. J., 2018, 24, 2249 CrossRef CAS.
- Z. Biron, S. Khare, A. O. Samson, Y. Hayek, F. Naider and J. Agister, Biochemistry, 2002, 41, 12687 CrossRef CAS.
- F. Formaggio, M. Crisma, P. Rossi, P. Scrimin, B. Kaptein, Q. B. Broxterman, J. Kamphuis and C. Toniolo, Chem. – Eur. J., 2000, 6, 4498 CrossRef CAS.
- R. B. Merrifield, J. Am. Chem. Soc., 1963, 85, 2149 CrossRef CAS.
- L. Carpino and G. Han, J. Org. Chem., 1972, 37, 3404 CrossRef CAS.
- (a) J. M. Collins and N. E. Leadbeater, Org. Biomol. Chem., 2007, 5, 1141 RSC; (b) J. Murray, J. Aral and L. Miranda, Methods Mol. Biol., 2011, 716, 73 CrossRef CAS; (c) S. L. Pedersen, A. P. Tofteng, L. Malik and K. J. Kensen, Chem. Soc. Rev., 2012, 41, 1826 RSC.
- J. M. Collins, K. A. Porter, S. K. Singh and G. S. Vanier, Org. Lett., 2014, 16, 940 CrossRef CAS.
- R. I. Hara, Y. Mitsuhasi, K. Saito, Y. Maeda and T. Wada, ACS Comb. Sci., 2018, 20, 13 CrossRef.
- (a) M. Itoh, Bull. Chem. Soc. Jpn., 1973, 46, 2219 CrossRef CAS; (b) J. Izdebski, Pol. J. Chem., 1979, 53, 1049 CAS.
- R. Subirós-Funosas, R. Prohens, R. Barbas, A. El-Faham and F. Albericio, Chem. – Eur. J., 2009, 15, 9394 CrossRef.
- (a) A. Kumar, Y. E. Jad, B. G. de la Torre, A. El-Faham and F. Albericio, J. Pept. Sci., 2017, 23, 763 CrossRef CAS; (b) A. El-Faham and F. Albericio, Chem. Rev., 2011, 111, 6557 CrossRef CAS.
- K. B. H. Salah and N. Inguimbert, Org. Lett., 2014, 16, 1783 CrossRef.
- J. M. Collins and S. K. Singh, US Pat., 0066031 A1, 2018 Search PubMed.
- A. El-Faham and F. Albericio, Org. Process Res. Dev., 2018, 22, 760 CrossRef.
- M. Schiell, J. Hofmann, M. Kurz, F. R. Schmidt, L. Vértesy, M. Vogel, J. Wink and G. Seibert, J. Antibiot., 2001, 54, 220 CrossRef CAS.
- U. Orcel, M. De Poli, M. De Zotti and J. Clayden, Chem. – Eur. J., 2013, 19, 16357 CrossRef CAS.
- T. Boddaert, J. Solà, M. Helliwell and J. Clayden, Chem. Commun., 2012, 48, 3397 RSC.
- J. Solà, G. A. Morris and J. Clayden, J. Am. Chem. Soc., 2011, 133, 3712 CrossRef.
- B. A. F. Le Bailly, L. Byrne, V. Diemer, M. Foroozandeh, G. A. Morris and J. Clayden, Chem. Sci., 2015, 6, 2313 RSC.
- Toniolo and coworkers report that the sign of the signal at 222 nm in Aib-containing helices is slightly negative, rather than positive. The origin of this consistent discrepancy is still unclear. See E. Longo, A. Moretto, F. Formaggio and C. Toniolo, Chirality, 2011, 23, 756–760 CrossRef CAS.
- I. L. Karle, J. L. Flippen-Anderson, R. Gurunath and P. Balaram, Protein Sci., 1994, 3, 1547 CrossRef CAS.
- (a) S. C. Yasui, T. A. Kiederling, F. Formaggio, G. M. Bnora and C. Toniolo, J. Am. Chem. Soc., 1986, 108, 4988 CrossRef CAS; (b) K. Otoda, Y. Kitagawa, S. Kimura and Y. Imanishi, Biopolymers, 1993, 33, 1337 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Peptide synthesis, peptide purification, circular dichroism spectroscopy and analytical data. See DOI: 10.1039/d0cc04698a |
| This journal is © The Royal Society of Chemistry 2020 |
