Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
How trimerization of CTR1 N-terminal model peptides tunes Cu-binding and redox-chemistry†
Thibaut
Galler
a,
Vincent
Lebrun
a,
Laurent
Raibaut
 a,
Peter
Faller
a,
Peter
Faller
 *a and
Nina E.
Wezynfeld
*a and
Nina E.
Wezynfeld
 *ab
*ab
aInstitut de Chimie, UMR 7177, CNRS-Universitéde Strasbourg, 4 rue Blaise Pascal, Strasbourg 67000, France. E-mail: pfaller@unistra.fr
bChair of Medical Biotechnology, Faculty of Chemistry, Warsaw University of Technology, Warsaw 00-664, Poland. E-mail: nwezynfeld@ch.pw.edu.pl
First published on 7th September 2020
Abstract
Employing peptide-based models of copper transporter 1 (CTR1), we show that the trimeric arrangement of its N-terminus tunes its reactivity with Cu, promoting Cu(II) reduction and stabilizing Cu(I). Hence, the employed multimeric models of CTR1 provide an important contribution to studies on early steps of Cu uptake by cells.
In humans, copper uptake by cells is mostly provided by Cu-transporter CTR1 (copper-transporter 1),1 a homo-trimeric protein,2 consisting of an extracellular, a transmembrane, and a shorter intracellular domain. Recently, the crystal structure of the transmembrane domain was solved from salmon CTR1 and shows three transmembrane helices per monomer.3 However, structural insights into the initial steps of Cu uptake by CTR1, including capture of copper by the extracellular domain of CTR1 and Cu(II) reduction, could not be obtained from this work.
In the past, those initial steps of Cu uptake by CTR1 were mostly investigated using monomeric models of the CTR1 N-terminus. Those studies showed that CTR1 has a strong Cu(II) binding site, spanning the first three N-terminal CTR1 residues Met-Asp-His (MDH), a sequence which belongs to an ATCUN motif (Amino Terminal Cu(II) and Ni(II) binding motif) (Xxx-Zzz-His).4,5 Meanwhile, the adjacent His residues, His5–His6 (bis-His motif), were proposed as the first potential Cu(I) binding site.6 It should be noted that Cu(II) from the blood must be reduced extracellularly to Cu(I) before being transported into the cytosol. The reduction could occur (i) prior to the binding to CTR1, for example by STEAP4,7 or (ii) while Cu is bound to CTR1. In the latter, Cu(II) is captured first by the ATCUN motif of CTR1 and then reduced to Cu(I). This hypothesis is based on (i) high affinity of the ATCUN for Cu(II), (ii) susceptibility of Cu(II)–ATCUN to reduction, and (iii) the affinity of Cu(I) for the adjacent bis-His motif (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 10.2 ± 0.2 at pH 7.4).6
K = 10.2 ± 0.2 at pH 7.4).6
Although the studies on monomeric models of the N-terminal portion of CTR1 gave important insights into the Cu(II)-binding and its chemical reactivity, there is no knowledge of how the trimeric structure could affect the interaction of the CTR1 N-termini and Cu. More generally, very little is known about the impact of multimeric peptide models on metal binding. Such information could help in understanding why nature evolved CTR1 as a trimer. To address this question, we synthesized for the first-time trimeric models of the N-terminal domain of CTR1 (see Fig. 1). By comparing their reactivity with respect to monomeric analogues, we show here that indeed, trimerization significantly impacts Cu(II) reduction and Cu(I) coordination, hence suggesting an important role of this arrangement for copper transport in the native CTR1 transporter.
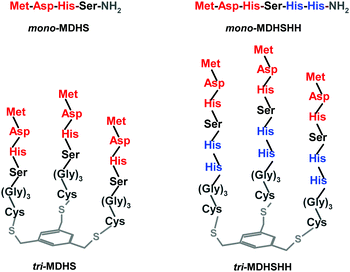 | ||
| Fig. 1 Schemes of synthesized monomers and trimers of the CTR1 N-terminus. The ATCUN motif is highlighted in red and the bis-His motif is highlighted in blue. The platform and the linker are grey. | ||
The description and schemes of the trimer synthesis are available in the Experimental section and in the ESI,† Fig. S1. The monomeric and trimeric models we elaborated are depicted in Fig. 1. The first couple (mono- and tri-MDHS) contains the first four residues of the CTR1 sequence, Met-Asp-His-Ser, which includes the ATCUN motif. In the second ones (mono- and tri-MDHSHH), the CTR1 sequence was elongated with two further His residues, a potential Cu(I) binding site. In trimers, a (Gly)3Cys linker was added to anchor the peptides on the mesitylene platform, to ensure flexibility of the trimeric structure and to avoid interferences from potential interactions between CTR1 sequences and the platform. The purity of the obtained peptide fractions was checked by HPLC (see the ESI,† Fig. S2), whereas their identity was confirmed by ESI-MS (see Fig. S3, S4 and Tables S1, S2, ESI†).
At the beginning, the Cu(II) binding to the trimeric models of the CTR1 N-terminus was compared to their monomeric analog at pH 7.4 using UV-vis spectroscopy (see Fig. 2). All spectra related to the first Cu(II) equivalent are very similar for both monomeric and trimeric models, with a band at 525 nm, characteristic for 4N coordination of the ATCUN-like Cu(II) complexes with the His3 imidazole, the N-terminal amine, and the first two amides involved in Cu(II) binding.5,8 The observed coordination mode is consistent with previous studies on Cu(II)–CTR1 monomeric models at pH 7.4.4,6,9 Further addition of Cu(II) to the monomeric models (above 1 equiv.) resulted in an increase of the baseline for mono-MDHS (Fig. 2 left, dashed lines) or an apparent red-shift and the further increase of the baseline for mono-MDHSHH (Fig. 2 right, dashed lines). The observed red-shift in the latter one likely corresponds to the binding of the second Cu(II) equivalent to the bis-His motif leading to the appearance of a band of the second Cu(II) binding site, while the baseline increase resulted from precipitation of unbound Cu(II).
In contrast, the addition of the second and the third equivalent of Cu(II) to the trimers caused a continuous increase of the intensity of the band at 525 nm, in line with the Cu(II) coordination to the two remaining apo-ATCUN sites in the trimer. We have not noticed any indication of a trimeric structure affecting Cu(II) coordination to the ATCUN motif under these conditions (λmax and ε not significantly different). Plus, addition of Cu(II) above 3 equiv. of the tri-MDHSHH led to the precipitation of unbound copper after the ATCUN saturation by Cu(II) (see Fig. S5, ESI†). Unlike what we described for mono-MDHSHH, here no signals indicating the Cu(II) binding to the further bis-His motif have been noticed. As such, the Cu(II)-ATCUN complexes could act as a steric hindrance for the excess of Cu(II), potentially limiting the amount of Cu(II) bound to the CTR1 N-terminus. In summary, all the data indicate that properties of the strongest Cu(II)-site (ATCUN) are very similar to monomers and trimers and suggest hence that there is no mutual influence of these three sites in the trimers.
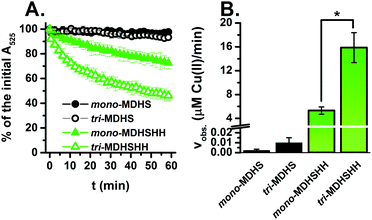
In the next step we investigated the susceptibility of Cu(II) bound to the CTR1 models (trimers and monomers) to reduction in the presence of ascorbate. Ascorbate (Asc) is a major low-molecular reductant in the extracellular environment of 10–100 μM concentration in plasma.10 In the presence of 0.2 mM Asc, the reduction of Cu(II) bound to our CTR1 N-terminus models was monitored following the characteristic d–d band at 525 nm (Fig. 3A). The whole UV-vis spectra of Cu(II) reduction are shown in the ESI,† Fig. S6. In accordance with the previous studies on other monomeric CTR1 models,6 the presence of the bis-His motif significantly accelerates the Cu(II) reduction as shown in Fig. 3A (green vs. black). Interestingly, this process was significantly faster for tri-MDHSHH than for mono-MDHSHH, with about 50% conversion after 60 min versus 25%. This trend was confirmed by the initial velocities calculated from the slope of the linear decrease in A525: Cu(II) reduction is three times faster when bound to tri-MDHSHH than bound to mono-MDHSHH (see Fig. 3B). It should be noted that those incubations were performed with the same Cu-load per CTR1 sequence, one Cu(II) per one trimer (Cu(II)1-tri-MDHSHH) and one Cu(II) per three monomers Cu(II)1-(mono-MDHSHH)3. On the other hand, Cu(II) bound to the MDHS models was much more resistant to reduction by Asc. In the presence of 0.2 mM Asc only about 2% and 6% of Cu(II) was reduced after 1 h for mono-MDHS and tri-MDHS, respectively. The difference between their velocities (calculated for the whole one-hour linear kinetic) was not significantly different at a significance level α = 0.05 (p = 0.17). Therefore, we decided to increase the Asc concentration to 5 mM. As shown in Fig. S7A (ESI†), it accelerated the reaction, leading to the conversion degree of 7% and 28% for mono-MDHS and tri-MDHS, respectively. Moreover, the difference between the reduction velocities for those models became more evident and significantly relevant (p < 0.001) (see Fig. S7B, ESI†). Overall, these results clearly show that the trimeric arrangement facilitates the reduction to Cu(I).
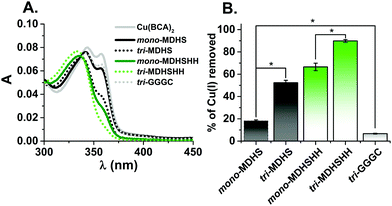
In order to assess the ability of the different models to bind Cu(I), we performed a ligand competition with bicinchoninic acid (BCA) that forms a stable Cu(I)–(BCA)2 complex.11 The latter was preformed, and we monitored its disappearance after addition of monomeric or trimeric CTR1 monomers to final concentrations of 25 μM and 8.3 μM, respectively, per 1.8 μM Cu(I) (see Fig. 4A). The addition of each model peptide caused a decrease in the intensity of the band at 360 nm, characteristic for Cu(I)–(BCA)2 (see Fig. S8, ESI†), suggesting that the peptides successfully compete for Cu(I) with BCA, where the Cu(I)–(BCA)2 binding constant is within the nanomolar range under the studied conditions.11 The amount of Cu(I) removed from Cu(I)–(BCA)2 after addition of each CTR1 model was calculated from the initial absorbance change at 360 nm and shown in Fig. 4B. These values suggest that the trimers had withdrawn significantly more Cu(I) ions from Cu(I)–(BCA)2 than the monomers (p < 0.001) and the trimeric organization could stabilize Cu(I) complexes. Under the applied conditions, this effect is more pronounced for the MDHS sequence with almost a 300% higher amount of removed Cu(I) between mono- vs. tri-MDHS, compared to 33% for mono- and tri-MDHSHH. The platform combined with the linker without CTR1 sequences (tri-GGGC) withdraws only very little Cu(I), suggesting that thioethers of the platform are not primary Cu(I) binding sites of tri-MDHS and tri-MDHSHH. Moreover, the longer peptides with the additional bis-His motif were much better competitors against Cu(I)–(BCA)2 than the shorter models, suggesting that the two adjacent His are the predominant Cu(I)-binding site.
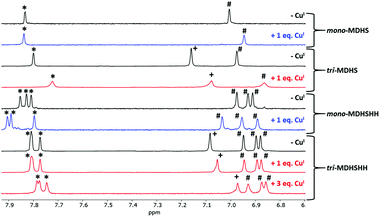
The Cu(I) binding to the CTR1 models was further investigated by NMR. Fig. 5 shows the aromatic region, where the CH of the imidazole as well as benzylic protons from the platform were observed. Mono-MDHS exhibits two resonances from CH(δ) and CH(ε) of the sole His. Upon Cu(I) addition, the resonance of CH(δ) shifts, suggesting the coordination to His3. In addition, the CH3 resonance of Met1 also significantly shifted (see Fig. S9, ESI†), in line with the coordination of the thioether as well. In the case of the tri-MDHS, both His peaks shift (CH(δ) and CH(ε)) and are broadened while CH3 of the Met1 shifted much less compared to mono-MDHS. This indicates that with 1 equiv. Cu(I) per trimer, His (not Met) residues are the main ligands.
For mono-MDHSHH, the resonances of CH(δ) and (ε) from the three His can be distinguished. Upon addition of 1 equiv. of Cu(I), all three CH(ε) resonances shift, but two more than the last. We assigned them, based on the competition with Cu(I)–(BCA)2 and the similar case in Aβ peptides and CTR11–14 peptides11,12 with reported preferential Cu(I) binding to the bis-His motif. For the tri-MDHSHH, CH(ε) signals of the two His residues from the bis-His motif overlapped, whereas CH(δ) from all His residues are distinctly resolved. Upon addition of 1 equiv. of Cu(I) per trimer, the shift of all His resonances for tri-MDHSHH was not so pronounced as for the monomeric analog. However, the further addition of Cu(I), up to 3 equiv., resulted in better resolution of the initially overlapped two CH(ε) resonances for tri-MDHSHH. Again, this can be assigned to the preferential binding of Cu(I) to the bis-His and, as above, the spectra are in line with a fast Cu(I) exchange between the sites. On the other hand, no significant changes in Cu(I) binding were noticed for the CH3 Met1 of mono-MDHSH and tri-MDHSHH (see Fig. S9, ESI†). In contrast, the signals assigned to benzylic protons from the platform drifted significantly upon Cu(I) addition (see Fig. 5) to the trimers that could result from the changes in the entire trimeric structure upon Cu(I) binding and from supramolecular interactions between the trimer units.
Overall, our study shows that trimeric models of CTR1 possess additional features compared to monomeric ones, which are of potential relevance to mimic Cu interaction with natural CTR1. While trimeric models mostly parallel the monomeric ones by the main Cu(II)- and Cu(I)-coordination sites, they change the behavior on the transition between them, i.e. the redox reaction. Indeed, in both models, Cu(II) is preferentially bound in the ATCUN motif, and Cu(I) is preferentially bound in the bis-His motif. Transition from Cu(II) to Cu(I) involves an important rearrangement of the Cu-peptide including at least one transition state influenced by different factors. First, as shown in the past for the monomeric models, the presence of a Cu(I)-binding site (e.g. the bis-His motif) promotes the reduction of Cu(II) by Cu(I) stabilization, a trend that is also visible with trimeric models. Second, prearrangement of the peptide chains arising from trimerization may explain the remarkably faster reduction of Cu(II) to Cu(I) with trimeric models than with monomeric ones. Indeed, trimeric organization may maintain in close vicinity several Cu(II) and Cu(I) sites in the trimers, and/or promote weak interactions between the residues, resulting in a change of the second coordination sphere and facilitating the transition. Two recent publications studied reduction reactions of Cu(II)–ATCUN motifs,13,14 and they suggested that Cu(II) bound to the 4N in ATCUN is not reduced, but a low populated state in equilibrium, where Cu(II) is bound to only two nitrogen donors (terminal NH2, Im).14 This state with non-saturated equatorial coordination by the peptide could bind external ligands (such Glu)13 or a reducing agent. We think that such type of low populated states (susceptible for reduction) could be stabilized by trimerization.
Finally, Cu(I) was generally bound tighter to the trimer than to the monomer. Competition with Cu(I)–(BCA)2 indicates that the main Cu(I)-binding site is the bis-His motif, a well-known digonal site in which Cu(I) is bound to two imidazole nitrogens, as observed also for β-amyloids.15–18 However, the His3 is likely to be involved as well, as Cu(I)-binding impacts its NMR resonances. Ligand exchange reactions occur faster than the NMR time-sale, i.e. less than seconds. Thus, the stronger Cu(I) affinity of the trimers could be due to the preorganization of the three chains towards each other and/or changes in the second coordination sphere.
Cu(I)-Transport by CTR1 is not coupled to energy conversion like hydrolysis of ATP. The driving force is likely the high Cu(I) affinity of the thiol-proteins in the cytosol, which are not or little present extracellularly. Hence, Cu can only be taken up in the Cu(I) oxidation state. But, Cu(I) can be oxidized by O2, and hence a scaffold that allows fast reduction and transfer to the Cu(I)-binding sites might be important to ensure fast transport and to avoid re-oxidation, which can lead to ROS production.
Overall, our data indicate that the trimeric form of CTR1 is not only crucial to build the Cu(I)-selective channel in the membrane-embedded portion of CTR1, but could also be of importance for extracellular Cu(I)-binding and in the reduction process from Cu(II)-bound to the CTR1 N-terminal sequence MDH to the first Cu(I) sites. Considering the variety of proteins that bind metals and adopt multimeric structures including metal-transporters19,20 and enzymes,21 the use of multimeric models could provide valuable information about the impact of multimerization in other metal-transporters or metalloenzymes.
This study was financed by the Frontier Research in Chemistry Foundation/Labex CSC, Strasbourg (CSC-PFA-17). N. E. W. was supported by the Mobility Plus Programme funded by the Polish Ministry of Science and Higher Education (1632/MOB/V/2017/0). We thank Dr. Jean-Louis Schmitt (ISIS, Strasbourg) for access to the LC-MS.
Conflicts of interest
There are no conflicts to declare.References
- H. Öhrvik and D. J. Thiele, J. Trace Elem. Med. Biol., 2015, 31, 178–182 CrossRef.
- C. J. De Feo, S. G. Aller and V. M. Unger, Biometals, 2007, 20, 705–716 CrossRef.
- F. Ren, B. L. Logeman, X. Zhang, Y. Liu, D. J. Thiele and P. Yuan, Nat. Commun., 2019, 10, 1386 CrossRef.
- K. Bossak, S. C. Drew, E. Stefaniak, D. Płonka, A. Bonna and W. Bal, J. Inorg. Biochem., 2018, 182, 230–237 CrossRef CAS.
- P. Gonzalez, K. Bossak, E. Stefaniak, C. Hureau, L. Raibaut, W. Bal and P. Faller, Chem. – Eur. J., 2018, 24, 8029–8041 CrossRef CAS.
- K. L. Haas, A. B. Putterman, D. R. White, D. J. Thiele and K. J. Franz, J. Am. Chem. Soc., 2011, 133, 4427–4437 CrossRef CAS.
- R. S. Ohgami, D. R. Campagna, A. McDonald and M. D. Fleming, Blood, 2006, 108, 1388–1394 CrossRef CAS.
- C. Harford and B. Sarkar, Acc. Chem. Res., 1997, 30, 123–130 CrossRef CAS.
- E. Stefaniak, D. Płonka, S. C. Drew, K. Bossak-Ahmad, K. L. Haas, M. J. Pushie, P. Faller, N. E. Wezynfeld and W. Bal, Metallomics, 2018, 10, 1723–1727 RSC.
- C. Kuiper, M. C. M. Vissers and K. O. Hicks, Free Radical Biol. Med., 2014, 77, 340–352 CrossRef CAS.
- R. De Ricco, D. Valensin, S. Dell’Acqua, L. Casella, C. Hureau and P. Faller, ChemBioChem, 2015, 16, 2319–2329 CrossRef CAS.
- M. J. Pushie, K. Shaw, K. J. Franz, J. Shearer and K. L. Haas, Inorg. Chem., 2015, 54, 8544–8551 CrossRef CAS.
- A. Santoro, N. E. Wezynfeld, E. Stefaniak, A. Pomorski, D. Płonka, A. Krężel, W. Bal and P. Faller, Chem. Commun., 2018, 54, 12634–12637 RSC.
- R. Kotuniak, M. J. F. Strampraad, K. Bossak-Ahmad, U. E. Wawrzyniak, I. Ufnalska, P.-L. Hagedoorn and W. Bal, Angew. Chem., 2020, 59, 11234–11239 CrossRef CAS.
- C. Hureau, V. Balland, Y. Coppel, P. L. Solari, E. Fonda and P. Faller, J. Biol. Inorg. Chem., 2009, 14, 995–1000 CrossRef CAS.
- A. Santoro, G. Walke, B. Vileno, P. P. Kulkarni, L. Raibaut and P. Faller, Chem. Commun., 2018, 54, 11945–11948 RSC.
- R. A. Himes, G. Y. Park, G. S. Siluvai, N. J. Blackburn and K. D. Karlin, Angew. Chem., Int. Ed., 2008, 47, 9084–9087 CrossRef CAS.
- M. J. Pushie, E. Stefaniak, M. R. Sendzik, D. Sokaras, T. Kroll and K. L. Haas, Inorg. Chem., 2019, 58, 15138–15154 CrossRef CAS.
- J. M. Argüello, D. Raimunda and M. González-Guerrero, J. Biol. Chem., 2012, 287, 13510–13517 CrossRef.
- I. De Domenico, D. M. V. Ward, G. Musci and J. Kaplan, Blood, 2007, 109, 2205–2209 CrossRef CAS.
- R. W. Strange, F. E. Dodd, Z. H. L. Abraham, J. G. Grossmann, T. Brüser, R. R. Eady, B. E. Smith and S. S. Hasnain, Nat. Struct. Biol., 1995, 2, 287–292 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, ESI-MS spectra, UV-vis titration with Cu(II), UV-vis spectra of the reduction of Cu(II)–CTR1 models with ascorbate, kinetics for the BCA competitions, and 1H-NMR spectra of the models in the aliphatic region. See DOI: 10.1039/d0cc04693k |
| This journal is © The Royal Society of Chemistry 2020 |