T m filtering by 1H-methyl labeling in a deuterated protein for pulsed double electron–electron resonance EPR†
Thomas
Schmidt
and
G. Marius
Clore
 *
*
Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892-0520, USA. E-mail: mariusc@mail.nih.gov
First published on 12th August 2020
Abstract
Modulating the phase-memory relaxation time (Tm) of a spin label by introducing 1H-methyl groups in a perdeuterated protein background is used in DEER experiments to assign interactions in multimodal P(r) distributions. Proof of principle is demonstrated using Protein A where one nitroxide label occupies two distinct regions of conformational space. The presence of a single protonated methyl group in close proximity (4–8 Å) to only one of the two nitroxide rotamer ensembles results in a selective and substantial decrease in Tm, manifested by differential decay of the peak intensities in the bimodal P(r) distance distribution as a function of the total dipolar evolution time. This form of Tm filtering will facilitate DEER structural analysis of biomolecular systems with three spin labels, including complexes involving multimeric proteins.
Pulsed double electron–electron resonance (DEER) EPR spectroscopy provides a means of accurately measuring long-range distances between pairs of spin labels and, as such, is an invaluable tool for conformational analysis of proteins and other biological macromolecules.1,2 With complete deuteration of both protein and solvent, distances up to 170 Å can potentially be measured.3–5 When more than two spin labels are present, analysis can prove challenging, not only due to artefacts arising from multi-spin effects1,6,7 but because assignment of distances to specific interactions between paramagnetic centres may be difficult a priori. One approach to resolve this issue makes use of orthogonal spin-labeling to probe interactions between different spin labels (e.g., a nitroxide and a paramagnetic metal centre such as Gd3+ or Cu2+) separately from those between like spin labels.8–11 Here we propose another strategy that makes use of specific 1H methyl labeling in a perdeuterated protein background,12 thereby permitting phase-memory relaxation time (Tm) filtering in DEER, analogous to T2 filtering in solution NMR.13 We demonstrate this approach on a model system, Protein A, in which one of the nitroxide spin labels occupies two distinct regions of conformational space giving rise to two separate distances in the DEER-derived P(r) probability distance distribution.
The model system employed is AviTag-Protein A with the nitroxide spin label (R1) attached to engineered cysteine residues close to the N- (Q39C) and C- (K88C) termini of the ordered Protein A domain (residue numbering according to the complete construct which includes the AviTag; the Protein A domain extends from residues 30 to 90, and residues 1–38 are disordered).14 Previous Q-band DEER has shown the presence of two clearly resolved distances in the P(r) distribution, at 33 and 38 Å,14 arising from the Q39C-R1 label occupying two distinct regions of conformational space (labeled a and b in Fig. 1A), as judged by the predicted P(r) distribution generated from the atomic coordinates using the spin-label rotamer library program MMM.15,16
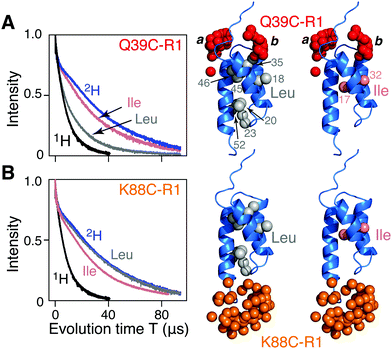 | ||
| Fig. 1 Two-pulse Hahn spin-echo decay curves for nitroxide spin-labeled AviTag-Protein A. The left hand panels show the spin echo curves for fully deuterated (blue), fully protonated (black), and Leu (grey) and Ile (pink) methyl protonated in a deuterated background samples of AviTag-Protein A nitroxide spin labeled at either (A) Q39C-R1 or (B) K88C-R1. The pulse scheme used is shown in Fig. S1A (ESI†). The left hand panels show the corresponding backbone ribbon diagram of Protein A (blue; PDB code 1BDD)22 with the methyl groups of Leu or Ile (CδH3) shown as grey and pink spheres, respectively, and the oxygen atoms of the calculated Q39C-R1 and K88C-R1 ensembles in red and orange, respectively. The nitroxide label ensembles were calculated from the atomic coordinates using the nitroxide rotamer library program MMM. The Q39C-R1 spin label samples two distinct regions of conformational spaces indicated by the labels a and b; only b is close to a methyl group. | ||
Hahn spin echo decay curves recorded with a two-pulse Hahn echo sequence (ESI,† Fig. S1A) with the nitroxide label either at the N- (Q39C-R1; Fig. 1A) or C (K88C-R1; Fig. 1B) terminal ends of the ordered Protein A domain were recorded on four samples of AviTag-Protein A in fully deuterated solvent (70% D2O/30% d8-glyerol v/v): fully deuterated, fully protonated, and either Leu (Cδ1H3 and Cδ2H3) or Ile (CδH3) methyl-protonated on a perdeuterated background (see ESI† for details of protein expression, purification, nitroxide labeling, deuteration, and incorporation of protonated Leu or Ile methyl groups). There are 7 leucines (Fig. 1A, right) and 2 isoleucines (Fig. 1B, right). The apparent Tm (Tappm) values for the fully deuterated and fully protonated samples are ∼35 and 9 μs, respectively (Fig. 1A and B). For Q39C-R1, the introduction of Leu 1H-methyl groups in a perdeuterated background results in a two-fold reduction in the Tappm value to ∼17 μs (Fig. 1A); no reduction in Tappm, however, is observed for the corresponding K88C-R1 sample (Fig. 1B). Introduction of CδH3 methyl-protonated Ile in a perdeuterated background results in only a minimal reduction in the Tappm value (to ∼32 μs) for both the Q39C-R1 and K88C-R1 samples (Fig. 1A and B), and it seems likely that this small reduction may in fact be due to the presence of a small amount of residual H2O owing to incomplete exchange with the D2O buffer. It has previously been shown that protons, and especially methyl groups in close proximity (4–8 Å) to the unpaired electron of a nitroxide spin label are the largest contributor to transverse electron relaxation,17–21 and examination of the Protein A structure (Fig. 1, right panels) shows that our results are fully consistent with this phenomenon, since only the Q39C-R1 spin label in conformer b is sufficiently close to a methyl group (specifically the two methyls of Leu35; Fig. 1A and also Fig. S2, ESI†) to induce a significant reduction in Tappm.
Fig. 2 presents the results for Leu methyl-protonated, otherwise deuterated [Leu-CH3/2H]-AviTag-Protein A spin-labeled at both Q39C-R1 and K88C-R1 (Fig. 2A). As expected, the Tappm for the Hahn spin-echo decay curve of the Q39C-R1/K88C-R1 doubly-spin labeled sample is intermediate between that for the Q39C-R1 and K88C-R1 single spin-labeled samples (Fig. 1B). DEER data were recorded with a refocussed four-pulse scheme (Fig. S1B, ESI†)23 varying the length of the second echo period T (= 2τ2), keeping the length of the acquisition time tmax constant at 4 μs, with the exception of the data at τ2 = 4 μs, where tmax was set to 3 μs (Fig. S3 and ESI† Methods). For tmax = 3 and 4 μs, the upper limits for an accurate mean distance determination are 50(tmax/2)1/3 = 57 and 63 Å, respectively; the upper limits for an accurate determination of the width of the P(r) distribution are 40(tmax/2)1/3 = 46 and 50 Å, respectively.1 The DEER echo curves were analysed to generate P(r) distributions using five different methods: model free Tikhonov regularization with the program DeerAnalysis without (Fig. 2C) and with (Fig. S4A, ESI†) validation;24 the program DD in which the DEER data are modelled as originating from the sum of two Gaussians (Fig. S5A, ESI†);25 the program WavPDS (wavelets for pulse dipolar signals) to filter out noise using a wavelet denoising method,26 followed by Tikhonov regularization24 (Fig. S6A, ESI†); and WavPDS followed by singular value decomposition (SVD) (Fig. S7A, ESI†).27,28 Since model-independent Tikhonov regularization, model-dependent Gaussian modelling and model-independent SVD represent three completely different approaches for extracting P(r) distributions from DEER echo curves, a comparison of the results provides an independent means of assessing the accuracy of the resulting P(r) distributions.29,30 All five methods of analysis yield quantitatively very similar results, in which the integrated intensity of the b component of the bimodal P(r) distribution centred at ∼33 Å decays more rapidly than that of the a component centred at ∼38 Å. Thus, one can immediately conclude that the Tm of the b component is shorter than that of the a component as a consequence of increased transverse electron relaxation arising from close proximity of the 1H-methyl groups of Leu35 to the unpaired electron in the b ensemble of the Q39C-R1 spin label (Fig. 2A).
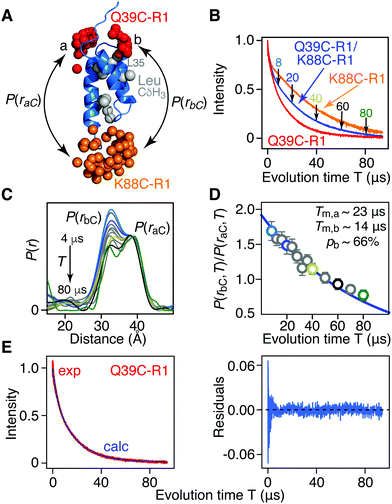 | ||
| Fig. 2 Phase memory relation time (Tm) filtering for [Leu-CH3/2H] AviTag-Protein A (Q39C-R1/K88C-R1) in an otherwise perdeuterated background. (A) Ribbon diagram of the protein A core domain22 (blue) with the oxygen atoms of the nitroxide spin label ensembles (generated using the rotamer library program MMM) colored in red (Q39C-R1) and orange (K88C-R1), and the leucine methyl groups in grey. The two distances between K88C-(R1) and the two conformer populations (a and b) of Q39C-R1 are indicated. (B) Hahn spin-echo curves for [Leu-CH3/2H] AviTag-Protein A, spin-labeled at Q39C-R1/K88C-r1 (blue), Q39C-R1 (red) and K88C-R1 (orange). (C) P(r) distributions for [Leu-CH3/2H] AviTag-Protein A (Q39C-R1/K88C-R1) obtained from the DEER echo curves using DeerAnalysis24 with Tikhonov regularization at evolution times T (= 2τ2) ranging from 8 to 80 μs. The color coding corresponds to that of the experimental points in panel D. The P(r) distributions are normalized with regard to the a peak at 38 Å. The DEER pulse scheme is shown in Fig. S1B (ESI†), and the raw and background-corrected DEER echo curves are shown in Fig. S3 (ESI†). Results of global best-fitting eqn (1) and (2) to the experimental evolution time T (= 2τ2) dependence of (D) the peak intensity ratio,P(rbC,T)/P(raC,T), in the P(r) distributions for [Leu-CH3/2H] AviTag-Protein A (Q39C-R1/K88C-R1) and (E) the Hahn echo curve for [Leu-CH3/2H] AviTag-Protein A (Q39C-R1) (left panel). The experimental ratios of the b to a peak intensities, P(rbC,T)/P(raC,T), are obtained from the integrated intensities of two Gaussians fitted to the bimodal P(r) distributions. The experimental data and best-fits are displayed by the circles and blue line, respectively in (D) and by the red and blue curves, respectively, in (E). Normalization in (E) was carried out by dividing the experimental and calculated curves by the optimized value of the sum of the two scale factors (λ1 + λ2). The right panel in (E) is a plot of the distribution of residuals between experimental and calculated Hahn echo curves. The corresponding P(r) distributions and best fits obtained using DeerAnalysis with validated Tikhonov regularization,24 the program DD with a sum of two Gaussians,25 the program WavPDS (to filter out noise)26 followed by Tikhonov regularization,24 and WavPDS followed by SVD26–28 are shown in Fig. S4–S7 (ESI†), respectively. | ||
The ratio of the integrated intensity of the b to a components of the P(r) distribution, P(rbC,T)/P(raC,T), for [Leu-CH3/2H]-AviTag-Protein A (Q39C-R1/K88C-R1) shown in Fig. 2D, is expected to decay as:
| P(rbC,T)/P(raC,T) = pb/(1 − pb)exp[−T(Rbm − Ram)/2] | (1) |
S(T) = λ1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−RESEEMT) + λ2[(1 − pb)exp(−TRam) + pb exp(−RESEEMT) + λ2[(1 − pb)exp(−TRam) + pb![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−TRbm)] exp(−TRbm)] | (2) |
| Methodab | R am (ms−1) | R bm (ms−1) | p B | R ESEEM (ms−1) |
|---|---|---|---|---|
| a Method used to derive the P(r) distributions from the DEER echo curves. The values of the two scale factors, λ1 and λ2, describing the amplitudes of the contributions of the ESEEM effect and phase-memory relaxation, respectively, to the Hahn spin echo curves are given in Table S1 (ESI). b The reduced χ2 of the global fits are as follows: DA (DeerAnalysis), 1.09; DA with validation, 1.09; DD, 1.10; WavPDS + DA, 1.10; WavPDS + SVD, 1.13. c DA with Tikhonov regularization. d Validated Tikhonov regularization was carried out varying the modulation depth (11 steps from 0.25 to 0.5), background density (11 steps from 0.04 to 0.15) and background start (6 steps from 500 to 1800 ns) for a total of 726 permutations. | ||||
| DAc | 44.3 ± 0.4 | 70.1 ± 0.5 | 0.66 ± 0.01 | 352 ± 7 |
| DA with validationd | 47.5 ± 0.4 | 67.2 ± 0.5 | 0.63 ± 0.01 | 321 ± 7 |
| DD | 46.8 ± 0.7 | 67.0 ± 0.7 | 0.66 ± 0.01 | 326 ± 9 |
| WavPDS + DA | 45.8 ± 0.4 | 69.3 ± 0.4 | 0.64 ± 0.01 | 322 ± 7 |
| WavPDS + SVD | 46.9 ± 0.4 | 69.3 ± 0.4 | 0.60 ± 0.01 | 329 ± 8 |
For [Ile-CδH3/2H] AviTag-Protein A (Fig. 3A), on the other hand, no difference in apparent Tm is observed between singly spin labeled (Q39C-R1 or K88C-R1) or doubly spin labeled (Q39C-R1/K88C-R1) samples (Fig. 3B), and no change in the ratio of peak intensities of the a and b components is observed in the DEER derived P(r) distributions obtained with evolution times T (= 2τ2) ranging from 10 to 60 μs (Fig. 3C and D). This is as expected, as the two Ile CδH3 methyl groups are not in close proximity to either the Q39C-R1 or K88C-R1 spin label (Fig. 3A and Fig. S2, ESI†), and provides a control for the results obtained with the [Leu-CH3/2H] labeling.
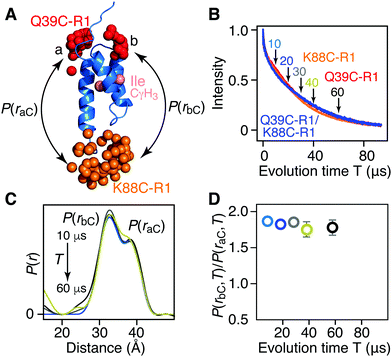 | ||
| Fig. 3 Introduction of protonated Ile CδH3 methyl groups in a deuterated background has no effect on Tm or DEER-derived P(r) distributions for AviTag-Protein A spin labeled at Q39C-R1 and/or K88C-R1. (A) Ribbon diagram of the protein A core domain (blue), with the oxygen atoms of the nitroxide spin label ensembles (generated using the rotamer library program MMM) colored in red (Q39C-R1) and orange (K88C-R1), and the Ile CδH3 methyl groups in pink. The two distances between K88C-(R1) and the two conformer populations (a and b) of Q39C-R1 are indicated. (B) Hahn spin-echo curves for [Ile-CγH3/2H] AviTag-Protein A, spin-labeled at Q39C-R1/K88C-r1 (blue), Q39C-R1 (red) and K88C-R1 (orange). (C) P(r) distributions for [Ile-CγH3/2H] AviTag-Protein A (Q39C-R1/K88C-R1) obtained from the DEER echo curves using DeerAnalysis24 with Tikhonov regularization at evolution times T (= 2τ2) ranging from 10 to 60 μs. The color coding corresponds to that of the experimental points in panel D. The P(r) distributions are normalized with regard to the a peak at 38 Å. The raw and background-corrected DEER echo curves are shown in Fig. S8 (ESI†). (D) Dependence of the peak intensity ratio,P(rbC,T)/P(raC,T), in the P(r) distributions for [Ile-CγH3/2H] AviTag-Protein A (Q39C-R1/K88C-R1) on evolution time T (= 2τ2). The ratios of b to a peak intensities, P(rbC,T)/P(raC,T), are obtained from the integrated intensities of two Gaussians fitted to the bimodal P(r) distributions. The same results are obtained when the DEER echo curves are processed using the programs DeerAnalysis with validated Tikhonov regularization (Fig. S9, ESI†),24 DD with two Gaussians,25 WavPDS26 followed by Tikhonov regularization,24 and WavPDS followed by SVD26,28 (Fig. S10, ESI†). | ||
In summary, we have shown that the introduction of a protonated methyl group(s) in an otherwise fully deuterated background, located in close proximity (less than 8 Å) to a nitroxide spin label can be used as a Tm filter in DEER spectroscopy. The example presented here of AviTag-Protein A provides proof of principle as the presence of a bimodal P(r) distribution is due to two distinctly different regions of conformational space sampled by one of the spin labels (specifically Q39C-R1). However, one can readily envision applications involving complexes between multimeric proteins. For example, in a complex between monomeric and homodimeric proteins the presence of three spin labels cannot be avoided if one wishes to probe distances between the two partner proteins in the complex. When the methyl group(s) is close to the nitroxide spin label in each subunit of the dimer, the relative contribution of the intersubunit distance can be selectively reduced by Tm filtering in the DEER-derived P(r) distribution relative to those of the intermolecular distances between monomer and dimer; conversely, a methyl group close to the nitroxide spin label on the monomer can be used to selectively reduce the contribution of the distances between monomer and dimer relative to the intersubunit distance within the dimer. Similar applications can be envisaged in the context of assigning interactions in monomeric proteins spin-labeled at three sites simultaneously.
We thank Drs. James Baber and John Louis for technical support. The work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Disease, National Institutes of Health, DK-029023 (to G. M. C.).
Conflicts of interest
There are no conflicts to declare.Notes and references
- G. Jeschke, Annu. Rev. Phys. Chem., 2012, 63, 419–446 CrossRef CAS PubMed.
- O. Duss, M. Yulikov, F. H. T. Allain and G. Jeschke, Methods Enzymol., 2015, 558, 279–331 CAS.
- R. Ward, A. Bowman, E. Sozudogru, H. El-Mkami, T. Owen-Hughes and D. G. Norman, J. Magn. Reson., 2010, 207, 164–167 CrossRef CAS PubMed.
- H. El Mkami and D. G. Norman, Methods Enzymol., 2015, 564, 125–152 CAS.
- T. Schmidt, M. A. Walti, J. L. Baber, E. J. Hustedt and G. M. Clore, Angew. Chem., Int. Ed., 2016, 55, 15905–15909 CrossRef CAS PubMed.
- G. Jeschke, M. Sajid, M. Schulte and A. Godt, Phys. Chem. Chem. Phys., 2009, 11, 6580–6591 RSC.
- T. von Hagens, Y. Polyhach, M. Sajid, A. Godt and G. Jeschke, Phys. Chem. Chem. Phys., 2013, 15, 5854–5866 RSC.
- E. Narr, A. Godt and G. Jeschke, Angew. Chem., Int. Ed., 2002, 41, 3907–3910 CrossRef CAS PubMed.
- P. Lueders, G. Jeschke and M. Yulikov, J. Phys. Chem. Lett., 2011, 2, 604–609 CrossRef CAS.
- A. Meyer, D. Abdullin, G. Schnakenburg and O. Schiemann, Phys. Chem. Chem. Phys., 2016, 18, 9262–9271 RSC.
- A. Meyer and O. Schiemann, J. Phys. Chem. A, 2016, 120, 3463–3472 CrossRef CAS PubMed.
- V. Tugarinov, V. Kanelis and L. E. Kay, Nat. Protoc., 2006, 1, 749–754 CrossRef CAS PubMed.
- F. Rastrelli, S. Jha and F. Mancin, J. Am. Chem. Soc., 2009, 131, 14222–14224 CrossRef CAS PubMed.
- J. L. Baber, J. M. Louis and G. M. Clore, Angew. Chem., Int. Ed., 2015, 54, 5336–5339 CrossRef CAS PubMed.
- Y. Polyhach, E. Bordignon and G. Jeschke, Phys. Chem. Chem. Phys., 2011, 13, 2356–2366 RSC.
- G. Jeschke, Protein Sci., 2018, 27, 76–85 CrossRef CAS PubMed.
- M. Lindgren, G. R. Eaton, S. S. Eaton, B. H. Jonson, P. Hammarström, M. Svensson and J. Carlsson, J. Chem. Soc., Perkin Trans. 2, 1997, 2549–2554 RSC.
- M. Persson, J. R. Harbridge, P. Hammarstrom, R. Mitri, L. G. Martensson, U. Carlsson, G. R. Eaton and S. S. Eaton, Biophys. J., 2001, 80, 2886–2897 CrossRef CAS PubMed.
- M. Huber, M. Lindgren, P. Hammarstrom, L. G. Martensson, U. Carlsson, G. R. Eaton and S. S. Eaton, Biophys. Chem., 2001, 94, 245–256 CrossRef CAS PubMed.
- H. El Mkami, R. Ward, A. Bowman, T. Owen-Hughes and D. G. Norman, J. Magn. Reson., 2014, 248, 36–41 CrossRef CAS PubMed.
- E. R. Canarie, S. M. Jahn and S. Stoll, J. Phys. Chem. Lett., 2020, 11, 3396–3400 CrossRef CAS PubMed.
- H. Gouda, H. Torigoe, A. Saito, M. Sato, Y. Arata and I. Shimada, Biochemistry, 1992, 31, 9665–9672 CrossRef CAS PubMed.
- M. Pannier, S. Veit, A. Godt, G. Jeschke and H. W. Spiess, J. Magn. Reson., 2000, 142, 331–340 CrossRef CAS PubMed.
- G. Jeschke, V. Chechik, P. Ionita, A. Godt, H. Zimmerman, J. E. Banham, C. R. Timmel, D. Hilger and H. Jung, Appl. Magn. Reson., 2006, 30, 473–498 CrossRef CAS.
- S. Brandon, A. H. Beth and E. J. Hustedt, J. Magn. Reson., 2012, 218, 93–104 CrossRef CAS PubMed.
- M. Srivastava, E. R. Georgieva and J. H. Freed, J. Phys. Chem. A, 2017, 121, 2452–2465 CrossRef CAS PubMed.
- M. Srivastava and J. H. Freed, J. Phys. Chem. Lett., 2017, 8, 5648–5655 CrossRef CAS PubMed.
- M. Srivastava and J. H. Freed, J. Phys. Chem. A, 2019, 123, 359–370 CrossRef PubMed.
- T. Schmidt, J. Jeon, Y. Okuno, S. C. Chiliveri and G. M. Clore, ChemPhysChem, 2020, 21, 1224–1229 CrossRef CAS PubMed.
- T. Schmidt, J. M. Louis and G. M. Clore, ChemBioChem, 2020 DOI:10.1002/cbic.202000263.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methods, DEER echo curves and DEER-derived P(r) distributions. See DOI: 10.1039/d0cc04369a |
| This journal is © The Royal Society of Chemistry 2020 |
