 Open Access Article
Open Access ArticleA novel, rationally designed lanthanoid chelating tag delivers large paramagnetic structural restraints for biomolecular NMR†
Daniel
Joss
 ,
Florine
Winter
and
Daniel
Häussinger
,
Florine
Winter
and
Daniel
Häussinger
 *
*
Department of Chemistry, University of Basel, St. Johanns-Ring 19, Basel 4056, Switzerland. E-mail: daniel.haeussinger@unibas.ch
First published on 14th September 2020
Abstract
Herein, a novel and rationally designed ortho-substituted pyridine activator is reported that reacts rapidly and selectively with cysteine thiols. It forms reduction-stable conjugates and induces large pseudocontact shifts, residual dipolar couplings and paramagnetic relaxation enhancement on both ubiquitin S57C and human carbonic anhydrase II S50C constructs under physiological conditions.
Paramagnetic effects induced by metal centres with anisotropic distribution of unpaired electrons on biomacromolecules,1,2e.g. lanthanoid ions complexed with suitable chelators for attachment, provide a unique and precise tool for the study of the properties of biomacromolecules by solution NMR.3–6 More specifically, lanthanoid chelating tags (LCT) induce long-range restraints that provide the unique opportunity to elucidate the structure of complexes, dynamics and interactions of biomacromolecules and ligands.7–16 In order to induce large paramagnetic effects, i.e. large pseudocontact shifts (PCS), residual dipolar couplings (RDC) and paramagnetic relaxation enhancement (PRE), rigid immobilization of the lanthanoid chelator on the surface of the biomacromolecule of interest is crucial.5,6 Conformational and positional invariant LCTs are ideally suited for monitoring protein interactions, as they can be placed far away from the interface and nevertheless provide accurate spatial information up to 200 Å.
Earlier approaches to suppress the translational and rotational freedom of the chelator on the surface of the protein, and hence to induce long-range restraints in solution, include: (1) two linker moieties attached to a double cysteine mutant,17–19 (2) restriction of the translational and rotational mobility,20–22 (3) placement of bulky groups on the macrocyclic ring scaffold.22,23
In order to restrict the remaining rotational averaging by para-substituted linker moieties,20,21 we rationally designed a novel, ortho-substituted pyridine activator that leads to a hampered rotation around the Scysteine–Cpyridine bond and to a short, reduction-stable linkage between the LCT and the biomacromolecule (Fig. 1). As carved out during the design process, cysteine-selective and rapid conjugation in ortho-position requires a very delicate tuning of an electron-withdrawing π-acceptor type functional group and a suitable leaving group in order to balance high reactivity, potential hydrolysis and steric demands of the incoming cysteine thiol as nucleophile. During the nucleophilic aromatic substitution tagging reaction, a nitro substituent in meta-position stabilises the Meisenheimer complex intermediate24 and, in combination with a methylsulphone leaving group, allows for fast and selective ligation of the LCT with a cysteine residue of the targeted biomacromolecule.
 | ||
| Fig. 1 Structure of Ln-M7-Nitro in Λ(δδδδ) conformation and indication of the important parts of the chelator. | ||
The seven methyl groups on the cyclen-based ring scaffold of the proposed LCT (Ln-(2R,2′R,2′′R)-2,2′,2′′-((2S,5S,8S,11S)-2,5,8,11-tetramethyl-10-((6-(methylsulphonyl)-5-nitropyridin-2-yl)methyl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)tripropanoate = Ln-M7-Nitro), and on the pendant carboxylate side arms provide a significant rigidification of the chelator.25–28 Reduction-stable thioether linkages of LCT-protein were earlier shown to provide an attachment suitable for acquisition of paramagnetic restraints by in-cell NMR.12,13
In order to demonstrate the applicability of our proposed approach, we synthesized paramagnetic thulium, dysprosium, terbium, ytterbium and gadolinium complexes in combination with the lutetium complex as diamagnetic reference and investigated their potential for the generation of PCSs (position of amide groups in 3D space), RDCs (orientation of the amide N–H vectors in space) and PRE (distance of amide protons from the paramagnetic centre) on both ubiquitin S57C (9 kDa) and human carbonic anhydrase II S50C (hCA II, 30 kDa) constructs.
With the idea of the installation of a leaving group in the ortho-position of the pyridine moiety of the envisioned LCT in mind, a suitable bromopyridine linker precursor 1 was synthesized starting from the commercially available 2-chloro-6-methyl-3-nitropyridine (ESI,† pp. 3–4). A sevenfold methyl-substituted cyclen precursor with three side arms already installed (2) was then reacted with the brominated linker precursor 1 in an alkylation reaction (Fig. 2). The precursor of the LCT with all four side arms installed (3) was then deprotected by heating of the compound with hydrochloric acid in acetonitrile to give the free ligand 4. Subsequently, the chelator was loaded with lanthanoid ions suitable for the investigation of paramagnetic effects on biomacromolecules (5) and the leaving group was activated by oxidation of the thioether to the methylsulphone (6).
 | ||
| Fig. 2 Synthetic route to Ln-M7-Nitro starting from sevenfold methyl-substituted cyclen precursor 2via alkylation, deprotection, metalation and oxidation. | ||
In order to test the magnitude of the induced PCS and RDC effects of Ln-M7-Nitro on ubiquitin S57C and selectively 15N leucine labelled hCA II S50C protein constructs, paramagnetic Tm-, Dy-, Tb- and Yb-loaded complexes were conjugated to the solvent exposed single cysteine residues on the protein constructs, whereas the Lu-complex was used as diamagnetic reference when conjugated to the protein. Subsequently, 1H–15N HSQC (acquisition of PCSs) and 1H–15N HSQC IPAP (acquisition of RDCs, no decoupling in the 15N dimension) experiments were acquired. Ln-M7-Nitro was thereby shown to induce anisotropy parameters on ubiquitin S57C of up to Δχax = 94.3 × 10−32 m3 for Dy-, Δχax = 65.9 × 10−32 m3 for Tb- and Δχax = 60.9 × 10−32 m3 for Tm-M7-Nitro (Table 1). When compared to other LCTs, the anisotropy parameters induced by Ln-M7-Nitro are exceedingly large and the value for the Dy-loaded chelator attached to ubiquitin S57C constitutes an unprecedented value of Δχax for an LCT-protein construct.17,19,20,22,29 The obtained Q-factors and the distance of the fitted lanthanoid metal centre to the Cbeta–carbon of the cysteine residue (6.9 Å) indicate excellent fits. Notably, RDCs up to 41.3 Hz were obtained for the Dy-loaded chelator, while RDCs of up to 35.9 Hz were obtained using the Tb complex (Tables S11 and S12, ESI†). In order to confirm the results obtained on ubiquitin S57C (9 kDa) and to extend our methodology to a larger protein, selectively 15N leucine labelled hCA II S50C (30 kDa) with a single solvent exposed cysteine was chosen as suitable target. Similar anisotropy parameters as for ubiquitin S57C were found (Table 1) and the Ln–Cbeta,cys distance of 7.1 Å indicated an excellent quality of the obtained fits. As can be seen from the anisotropy parameters depicted as isosurfaces (Fig. 3), the properties of the induced effects match for the pairs Dy and Tb as well as for Tm and Yb – intrinsic properties of the lanthanoids that were described in literature.5 When compared to seven-fold methyl substituted, pyridine derived LCTs, as e.g. Tm-M7FPy-DOTA21 attached to ubiquitin S57C, Tm-M7-Nitro shows a superior performance, which can most likely be attributed to the switch of the ligation site on the pyridine linker moiety from the para- to the ortho-position and therefore to an increased rotational and translational rigidity of the LCT relative to the protein surface. Since PCSs and RDCs are susceptible to different motional averaging mechanisms, a joint fit of PCSs and RDCs should result in the same tensors as the individual fits in case of a non-mobile LCT.30,31 We were delighted to find that Δχax for the combined PCS/RDC fit agreed on average with the PCS fit within 5 and 9% for ubiquitin and hCA II, respectively, whereas the agreement with the RDC fit was even better (0.3 and 1%, see Table S18, ESI†). Ln-M7-Nitro might, therefore, be also useful to detect dynamics. The moderate Curie line broadening of approx. only 20 Hz in the case of hCA II, observed at 600 MHz in 24 Å distance from the lanthanoid, holds great promise also for applications at ultra-high fields (Fig. S40, ESI†).
| Protein mutant | PDB | N° PCS | Ln3+ | Δχax (10−32 m3) | Δχrh (10−32 m3) | X metal (Å) | Y metal (Å) | Z metal (Å) | α (°) | β (°) | γ (°) | Q (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ubiquitin S57C | 1UBI 34 | 32 | Tm | 60.9 ± 1.9 | 12.3 ± 1.3 | 16.3 | 16.9 | 12.3 | 110.9 | 70.4 | 101.4 | 5.1 |
| 32 | Dy | 94.3 ± 2.2 | 10.7 ± 0.7 | 133.7 | 161.5 | 128.4 | 2.4 | |||||
| 44 | Tb | 65.9 ± 1.6 | 27.9 ± 0.8 | 128.0 | 162.4 | 110.4 | 7.0 | |||||
| hCA II S50C | 3KS3 35 | 10 | Tm | 71.0 ± 1.8 | 6.2 ± 0.9 | −31.0 | 4.4 | 14.9 | 174.7 | 17.4 | 139.0 | 4.3 |
| 22 | Dy | 95.9 ± 1.1 | 13.1 ± 2.5 | 179.8 | 112.5 | 64.5 | 2.1 | |||||
| 38 | Tb | 63.6 ± 0.9 | 22.5 ± 1.4 | 4.9 | 62.5 | 109.5 | 2.0 | |||||
| 22 | Yb | 13.8 ± 0.6 | 7.5 ± 0.4 | 36.7 | 163.0 | 115.4 | 3.5 |
| Protein mutant | PDB | N° RDC | Ln3+ | Δχax (10−32 m3) | Δχrh (10−32 m3) | X metal (Å) | Y metal (Å) | Z metal (Å) | α (°) | β (°) | γ (°) | Q (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ubiquitin S57C | 1UBI 34 | 17 | Tm | 66.3 | −13.1 | 16.3 | 16.9 | 12.3 | 164.3 | −103.2 | 107.4 | 18.0 |
| 15 | Dy | 106.7 | −3.5 | −131.4 | −14.7 | 137.1 | 18.2 | |||||
| 21 | Tb | 79.2 | −32.5 | 143.2 | −15.7 | 146.2 | 20.5 | |||||
| hCA II S50C | 3KS3 35 | 18 | Tb | 74.9 | −23.3 | −31.0 | 4.4 | 14.9 | −23.2 | −111.9 | −3.3 | 18.3 |
| 11 | Yb | 13.9 | −6.0 | −126.4 | −151.1 | −123.3 | 50.1 |
 | ||
| Fig. 3 Anisotropy parameters induced by Ln-M7-Nitro on ubiquitin S57C (top) and selectively 15N leucine labelled hCA II S50C (bottom) depicted as isosurfaces with a given shift in ppm (generated using Numbat32). Top (from left to right): Tm-, Dy- and Tb-M7-Nitro-UbS57C (inner layer: 4.0 ppm, outer layer: 1.5 ppm). Bottom (from left to right): Tm-, Dy-, Tb- and Yb-M7-Nitro attached to selectively 15N leucine labelled hCA II S50C (inner layer: 3.0 ppm, outer layer: 0.8 ppm). | ||
Due to the stabilization of the intermediate Meisenheimer complex24 during the ligation reaction by the newly developed activator, Ln-M7-Nitro provides a fast, quantitative and therefore clean ligation to the protein. As confirmed by ESI-HRMS, a ligation yield of >95% is already observed after 30 min at room temperature (Fig. 4). The 3-nitro-substituted 2-(methylsulphonyl)pyridine constitutes a novel, highly reactive and selective moiety for tagging of cysteines in biomacromolecules under close to physiological conditions (highly diluted (∼100 μM), aqueous buffer, pH 7.0). While the methylsulphonyl leaving group shows a suitable balance in terms of reactivity and selectivity, a fluorine leaving group led to hydrolysis and chlorine showed no reaction.
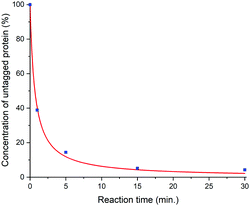 | ||
| Fig. 4 Kinetics of the tagging reaction of monomeric ubiquitin S57C (90 μM) with Lu-M7-Nitro (540 μM) in phosphate buffer (10 mM, pH 7.0, 0.2 mM TCEP) followed by ESI-HRMS. The experimental data points were fitted with the equation for second order kinetics [Ubi] = (k·t + 1/[Ubi]t=0)−1 to give k = 274.5 M−1 s−1. A comparison of Ln-M7-Nitro and previously reported LCTs is shown in Fig. S41 (ESI†). | ||
Besides the favourable properties of Ln-M7-Nitro for the generation of anisotropic paramagnetic effects, we were curious about the properties of Ln-M7-Nitro as PRE tag. PRE is an isotropic effect that enhances the relaxation of spins in vicinity to the paramagnetic metal centre and causes thereby line broadening, which can be quantified by plotting the intensity ratio of paramagnetic and diamagnetic peaks vs. the distance of the amide proton to the metal centre. Upon attachment of the Gd complex of the newly developed LCT to both ubiquitin S57C and hCA II S50C, strong PREs were detected in 1H–15N HSQC experiments and evaluated with respect to their distance dependence, the amino acid sequence of the protein and the theoretically predicted values for an LCT-protein construct with the same molecular weight (Fig. S9–S12, ESI†).36 PRE was previously used to resolve structures and interactions of biomacromolecules in solution.37
To conclude, a novel, rationally designed ortho-substituted pyridine activator was developed that delivers an LCT with unprecedented paramagnetic properties and, thereby, allows for acquisition of experimental structural restraints for the investigation of structure and dynamics of biomacromolecules and their complexes via PCSs, RDCs and PREs. The fast and clean ligation reaction of the new activator in combination with the strong, induced paramagnetic effects render Ln-M7-Nitro as an ideal LCT for challenging applications.
The Chemistry Department of the University of Basel and the SNSF grant 200021_130263 are acknowledged for financial support. Biological structures were generated using the open source software PyMOL. C. E. Housecroft, E. C. Constable, T. Müntener, R. Vogel and P. Rieder are acknowledged for helpful discussions and S. Fischer for involvement in synthetic side projects.
Conflicts of interest
There are no conflicts to declare.Notes and references
- I. Bertini, C. Luchinat, G. Parigi and R. Pierattelli, ChemBioChem, 2005, 6, 1536–1549 Search PubMed.
- J. H. Prestegard, H. M. Al-Hashimi and J. R. Tolman, Q. Rev. Biophys., 2000, 33, 371–424 Search PubMed.
- J. Koehler and J. Meiler, Prog. Nucl. Magn. Reson. Spectrosc., 2011, 59, 360–389 Search PubMed.
- W.-M. Liu, M. Overhand and M. Ubbink, Coord. Chem. Rev., 2014, 273–274, 2–12 Search PubMed.
- C. Nitsche and G. Otting, Prog. Nucl. Magn. Reson. Spectrosc., 2017, 98–99, 20–49 Search PubMed.
- D. Joss and D. Häussinger, Prog. Nucl. Magn. Reson. Spectrosc., 2019, 114–115, 284–312 Search PubMed.
- G. Pintacuda, M. John, X.-C. Su and G. Otting, Acc. Chem. Res., 2007, 40, 206–212 Search PubMed.
- J.-Y. Guan, P. H. J. Keizers, W.-M. Liu, F. Löhr, S. P. Skinner, E. A. Heeneman, H. Schwalbe, M. Ubbink and G. Siegal, J. Am. Chem. Soc., 2013, 135, 5859–5868 Search PubMed.
- C. Göbl, T. Madl, B. Simon and M. Sattler, Prog. Nucl. Magn. Reson. Spectrosc., 2014, 80, 26–63 Search PubMed.
- K. D. Brewer, T. Bacaj, A. Cavalli, C. Camilloni, J. D. Swarbrick, J. Liu, A. Zhou, P. Zhou, N. Barlow, J. Xu, A. B. Seven, E. A. Prinslow, R. Voleti, D. Häussinger, A. M. J. J. Bonvin, D. R. Tomchick, M. Vendruscolo, B. Graham, T. C. Südhof and J. Rizo, Nat. Struct. Mol. Biol., 2015, 22, 555–564 Search PubMed.
- T. Saio, K. Ogura, H. Kumeta, Y. Kobashigawa, K. Shimizu, M. Yokochi, K. Kodama, H. Yamaguchi, H. Tsujishita and F. Inagaki, Sci. Rep., 2015, 5, 16685 Search PubMed.
- T. Müntener, D. Häussinger, P. Selenko and F.-X. Theillet, J. Phys. Chem. Lett., 2016, 7, 2821–2825 Search PubMed.
- B.-B. Pan, F. Yang, Y. Ye, Q. Wu, C. Li, T. Huber and X.-C. Su, Chem. Commun., 2016, 52, 10237–10240 Search PubMed.
- K. Zimmermann, D. Joss, T. Müntener, E. S. Nogueira, M. Schäfer, L. Knörr, F. W. Monnard and D. Häussinger, Chem. Sci., 2019, 10, 5064–5072 Search PubMed.
- D. Joss, R. Vogel and D. Häussinger, Ref. Mod. in Chem, Mol. Sci. and Chem. Eng., 2020, DOI: 10.1016/B978-0-12-409547-2.14848-6.
- C. A. Barnes, Y. Shen, J. Ying, Y. Takagi, D. A. Torchia, J. R. Sellers and A. Bax, J. Am. Chem. Soc., 2019, 141, 9004–9017 Search PubMed.
- P. H. Keizers, A. Saragliadis, Y. Hiruma, M. Overhand and M. Ubbink, J. Am. Chem. Soc., 2008, 130, 14802–14812 Search PubMed.
- W.-M. Liu, P. H. J. Keizers, M. A. S. Hass, A. Blok, M. Timmer, A. J. C. Sarris, M. Overhand and M. Ubbink, J. Am. Chem. Soc., 2012, 134, 17306–17313 Search PubMed.
- M. D. Lee, M. L. Dennis, B. Graham and J. D. Swarbrick, Chem. Commun., 2017, 53, 13205–13208 Search PubMed.
- F. Yang, X. Wang, B.-B. Pan and X.-C. Su, Chem. Commun., 2016, 52, 11535–11538 Search PubMed.
- T. Müntener, J. Kottelat, A. Huber and D. Häussinger, Bioconjugate Chem., 2018, 29, 3344–3351 Search PubMed.
- D. Joss and D. Häussinger, Chem. Commun., 2019, 55, 10543–10546 Search PubMed.
- D. Joss, M.-S. Bertrams and D. Häussinger, Chem. – Eur. J., 2019, 25, 11910–11917 Search PubMed.
- C. N. Neumann, J. M. Hooker and T. Ritter, Nature, 2016, 534, 369–373 Search PubMed.
- D. Häussinger, J.-R. Huang and S. Grzesiek, J. Am. Chem. Soc., 2009, 131, 14761–14767 Search PubMed.
- A. C. L. Opina, M. Strickland, Y.-S. Lee, N. Tjandra, R. A. Byrd, R. E. Swenson and O. Vasalatiy, Dalton Trans., 2016, 45, 4673–4687 Search PubMed.
- D. Joss, R. M. Walliser, K. Zimmermann and D. Häussinger, J. Biomol. NMR, 2018, 29–38 Search PubMed.
- D. Parker, R. S. Dickins, H. Puschmann, C. Crossland and J. A. K. Howard, Chem. Rev., 2002, 102, 1977–2010 Search PubMed.
- F. Peters, M. Maestre-Martinez, A. Leonov, L. Kovacic, S. Becker, R. Boelens and C. Griesinger, J. Biomol. NMR, 2011, 51, 329–337 Search PubMed.
- D. Shishmarev and G. Otting, J. Biomol. NMR, 2013, 56, 203–216 Search PubMed.
- I. Bertini, C. Luchinat, G. Parigi and E. Ravera, NMR of Paramagnetic Molecules, Elsevier, Boston, 2nd edn, 2017, pp. 277–312 Search PubMed.
- C. Schmitz, M. J. Stanton-Cook, X.-C. Su, G. Otting and T. Huber, J. Biomol. NMR, 2008, 41, 179 Search PubMed.
- M. Rinaldelli, A. Carlon, E. Ravera, G. Parigi and C. Luchinat, J. Biomol. NMR, 2015, 61, 21–34 Search PubMed.
- R. Ramage, J. Green, T. W. Muir, O. M. Ogunjobi, S. Love and K. Shaw, Biochem. J., 1994, 299(Pt 1), 151–158 Search PubMed.
- B. S. Avvaru, C. U. Kim, K. H. Sippel, S. M. Gruner, M. Agbandje-McKenna, D. N. Silverman and R. McKenna, Biochemistry, 2010, 49, 249–251 Search PubMed.
- J. L. Battiste and G. Wagner, Biochemistry, 2000, 39, 5355–5365 Search PubMed.
- G. M. Clore and J. Iwahara, Chem. Rev., 2009, 109, 4108–4139 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc04337k |
| This journal is © The Royal Society of Chemistry 2020 |
