Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers†
Liyun
Zhang‡
 *a,
Xiaona
Fang‡
b,
Xingbo
Liu‡
c,
Huichao
Ou
b,
Haiyan
Zhang
b,
Jinjun
Wang
a,
Qian
Li
a,
Huanyi
Cheng
c,
Wenyi
Zhang
c and
Zhaofeng
Luo
*b
*a,
Xiaona
Fang‡
b,
Xingbo
Liu‡
c,
Huichao
Ou
b,
Haiyan
Zhang
b,
Jinjun
Wang
a,
Qian
Li
a,
Huanyi
Cheng
c,
Wenyi
Zhang
c and
Zhaofeng
Luo
*b
aState Key Laboratory of Medicinal Chemical Biology, College of Life Science, Nankai University, Tianjin, 300350, P. R. China. E-mail: liyunzhang@nankai.edu.cn
bSchool of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, P. R. China. E-mail: lzf@ustc.edu.cn
cAbace Biotechnology Co. LTD, Beijing, 100176, P. R. China
First published on 23rd July 2020
Abstract
Here, we report for the first time DNA aptamers targeted toward the COVID-19 nucleocapsid protein (Np). Np is one of the most abundant structural proteins and it serves as a diagnostic marker for the accurate and sensitive detection of COVID-19. After five rounds of selection, we obtained four DNA sequences with an affinity below 5 nM. The best one displayed a superb binding performance toward Np with a Kd value of 0.49 nM. Interestingly, we found that the four pairs of aptamers could bind to Np successively, suggesting a sandwich-type interaction. Using these sandwiched aptamers in ELISA and colloidal gold immunochromatographic strips, we were able to detect Np at the tens of pM level. The results demonstrate that aptamers are powerful molecular tools for virus detection, diagnosis, and antiviral therapy.
Novel coronavirus pneumonia has now become a global pandemic. Millions of cases have been confirmed, and hundreds of thousands have died. It has enormous implications for global economic and social development. One of the key factors in tackling the spread of COVID-19 across the globe is testing. Currently, the most common testing methods are based on the reverse transcription fluorescence quantitative polymerase chain reaction (RT-PCR), which is designed according to the COVID-19 sequence.1 RT-PCR tests need laboratories, specialized instruments, and technicians. They also take several hours. Due to the false-negative rate of RT-PCR, computed tomography (CT) scans are used for clinical diagnosis of COVID-19.2 However, CT systems are expensive, require technical expertise, and cannot accurately diagnose COVID-19. The third type of test measures the antibody responses to the virus in the blood serum.3 Immunoglobulin G and M (IgG and IgM) typically take a few weeks to develop against a new infection and last much longer in the bloodstream than the virus itself, providing a historical picture of past infections. However, they are not suitable for early detection and diagnosis. Therefore, it is imperative to develop new strategies to realize the early, sensitive, and specific detection of COVID-19.
Aptamers are a class of nucleic acid sequences that have high-affinity recognition targets similar to antibodies.4 Aptamers can be easily chemically modified to enhance their stability, affinity, and detection sensitivity. Moreover, the process of aptamer-sensor detection is much safer, conveniently operated, and time-saving. Synthetic aptamers are much cheaper than antibodies with no batch difference. Therefore, aptamers are employed in different fields and are most attractive in applications involving therapeutics and diagnoses.5–7 Importantly, aptamers play a vital role in the detection of viruses and antiviral therapy,8 including influenza,9–11 dengue12 Ebola,13 and SARS.14–16 In this work, we have used the SELEX procedure to select DNA aptamers binding specifically to the COVID-19 nucleocapsid protein (Np).
A 76mer ssDNA library containing 36-nucleotide long random sequences was constructed and screened by SELEX to discover ssDNA aptamers that specifically bound to the COVID-19 N protein. After five rounds of selection, the enriched pool presented a considerable increase in affinity for the target protein (Fig. S1, ESI†). The quantitative polymerase chain reaction was performed to confirm the convergence of the aptamer species in the enriched pool (Fig. S2, ESI†).17 Then, the profoundly enriched DNA pool was sequenced using a high-throughput method. Finally, four aptamer candidates were selected for optimization and characterization. After truncation, a 58-nucleotide sequence, named Np-A48, was identified as the strongest binding aptamer to Np with an affinity of 0.49 nM (Fig. 1A and Table 1). All aptamers showed a strong binding ability to Np with their affinity below 5 nM. The secondary structure of these aptamers was proposed by mfold, as shown in Fig. S3 (ESI†).
| Entry Np- | Sequence (5′…3′) | k a (M−1 s−1) | k d (s−1) | K d (nM) |
|---|---|---|---|---|
| The association rate constant (ka), dissociation rate constant (kd), and equilibrium dissociation constant (Kd, Kd = ka/kd) were determined by SPR analysis. | ||||
| A48 | GCTGGATGTCGCTTACGACAATATTCCTTAGGGGCACCGCTACATTGACACATCCAGC | 8.80 × 105 | 3.85 × 10−4 | 0.49 ± 0.05 |
| A58 | GCTGGATGTCACCGGATTGTCGGACATCGGATTGTCTGAGTCATATGACACATCCAGC | 1.92 × 104 | 1.34 × 10−4 | 0.70 ± 0.06 |
| A61 | GCTGGATGTTGACCTTTACAGATCGGATTCTGTGGGGCGTTAAACTGACACATCCAGC | 4.58 × 104 | 1.18 × 10−4 | 2.74 ± 0.08 |
| A15 | GCTGGATGTTCATGCTGGCAAAATTCCTTAGGGGCACCGTTACTTTGACACATCCAGC | 2.84 × 105 | 1.16 × 10−3 | 4.38 ± 0.06 |
COVID-19 Np (pI 10.07) is positively charged at physiological pH. Therefore, non-specific electrostatic interactions will interfere with the specific binding of the aptamers. The nature of this binding is also expected since encapsidation of the entire viral genome would require the N protein to bind to diverse sequences with reasonable affinity.18 To obtain a high-affinity aptamer, we used several strategies during the selection process: first, measurements of the binding of random sequences from pool 0 were performed. As shown in Fig. S1 (ESI†), the isolated aptamers bind more tightly than any arbitrary DNA sequence. Second, we added Salmon extract double-stranded DNA (10 μM) into the 0.5 μM enriched DNA pool for equilibration of non-specific DNA–Np interactions. Under these conditions, only the high-affinity single-stranded DNA aptamer could be captured by Np. Third, a high ionic strength was used to balance the electronegativity of the buffer system. We found that 500 mM sodium chloride was the optimized concentration in the selection system. Fourth, a small peptide consisting of eight histidine residues (His8) was used as the counter selection during our SELEX process. This strategy was used to avoid screening the aptamer to target the His-tag of Np. Finally, a high proportion of serum (40% v:v) was added to the selection system to improve the specificity, stability, and adaptability of the aptamers.
Next, we investigated the specificity of the aptamers. As shown in Fig. 1B, the four aptamers displayed distinct selectivity toward Np. We then evaluated their application in sensing Np in the presence of sputum (50% v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v PBS buffer), urine, and human serum. Most encouragingly, the binding dynamic curves showed that these aptamers were able to bind to the target rapidly and stably in the complex biological sample systems (Fig. S4–S6, ESI†). The difference of the binding kinetics in the different matrices may be related to the aptamer binding sites of the target, as discussed further below. The other components of the biomatrix did not cause significant interference or suppression of the interaction between aptamer and Np. By a simple comparison, the aptamer–Np complexes were found to be more stable in sputum and serum than in urine. These results demonstrate the potential of the new aptamers for the development of point-of-care testing (POCT) products to detect COVID-19 Np in samples obtained from suspected infected persons.
v PBS buffer), urine, and human serum. Most encouragingly, the binding dynamic curves showed that these aptamers were able to bind to the target rapidly and stably in the complex biological sample systems (Fig. S4–S6, ESI†). The difference of the binding kinetics in the different matrices may be related to the aptamer binding sites of the target, as discussed further below. The other components of the biomatrix did not cause significant interference or suppression of the interaction between aptamer and Np. By a simple comparison, the aptamer–Np complexes were found to be more stable in sputum and serum than in urine. These results demonstrate the potential of the new aptamers for the development of point-of-care testing (POCT) products to detect COVID-19 Np in samples obtained from suspected infected persons.
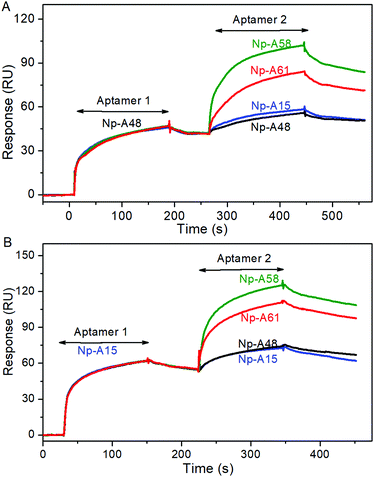
By adopting appropriate three-dimensional stem and loop structures, the aptamers selectively bind to their targets. Differences in the sequence often lead to differences in the structure. In the following experiments, we explored the possibility of sandwich-type binding between different aptamers and Np. The Np was coupled on the surface of a CM5 chip by using a typical EDC/NHS protocol. In the first run, the aptamer Np-A48 flowed into the channel. Subsequently, a response signal up to 47 RU was observed. Then, the second aptamer flowed through the same channel; if the second aptamer bound at different sites, the response signal should jump up to a second plateau. As shown in Fig. 2A, the green line represented Np-A58 and the red line represented Np-A61. In contrast, when Np-A15 flowed through the same channel, there were no significant response signal changes. When we first ran Np-A15 instead of Np-A48 (Fig. 2B), we had similar results that further confirmed that Np-A15 and Np-A48 share the same binding site. The four pairs of aptamers (Np-A48 or Np-A15 with Np-A58 and Np-A61) display sandwich-type interactive features.
The correlation between COVID-19 and SARS-CoV was about 79%, suggesting that COVID-19 might use the same molecular doorway to enter cells as SARS.19 The N protein serves multiple purposes, such as packaging the RNA genome into helical ribonucleoproteins, modulating host cell metabolism, and regulating viral RNA synthesis during replication and transcription.20 On the basis of the SPR assay, the N-terminal RNA-binding domain (NTD) and the C-terminal dimerization domain (CTD) of COVID-19 Np should capture aptamer 1 and aptamer 2, respectively. As shown in the SPR sensorgram, Np-A48 and Np-A58 bind to Np successively, resulting in the formation of a ternary complex, aptamer 1|Np|aptamer 2. No matter whether we first ran Np-A48 or Np-A15, they both showed competition, proving the fact that these two aptamers occupy the same binding site. The mfold secondary structures of Np-A48 and Np-A15 share a virtually identical loop sequence at one end of a base-paired stem. Similarly, the aptamers NP-A58 and Np-A61 share identical loops that cap the base-paired stems. Since these identical regions were selected independently, it is highly likely that they correspond to the recognition domains of the aptamers for their N protein target. The performance of the aptamers binding at different sites on the same target is also shown in their effects on the microenvironment. Different biological matrices (sputum, serum, and urine) contain different components, which directly or indirectly interfere with the binding of the aptamers and the target, causing differences in the binding dynamics. Benefiting from the sandwich combination performance that the detection process could reduce the impact of interference factors as much as possible.
To prove that these aptamers can be applied in future clinical samples, enzyme-linked immunosorbent assay (ELISA) was performed by using a combination of Np aptamers and antibodies (Table S2, ESI†). The sensitivity of ELISA was evaluated by testing standard solutions of Np at different concentrations (400, 200, 100, 50, 25, 12.5, 6.25, and 0 ng mL−1) in PBS solution (pH 7.4). The limit of detection (LOD) for aptamer–hybrid ELISA reached 1 ng mL−1 (∼20 pM).21 The sandwiched aptamers were then fabricated into a gold nanoparticle immunochromatographic strip (GIS) to detect spiked Np in human serum or urine. Fig. 3 represents an illustration of GIS and the images of the strips. The test line in the presence of 1 ng mL−1 can be clearly seen by the naked eye. The intensity of the test lines in pixels was quantified by ImageJ software (Fig. S7, ESI†). Importantly, all of the detection process can be done in 15 minutes.
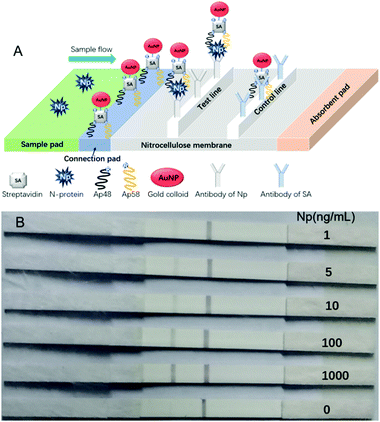 | ||
| Fig. 3 (A) Illustration and schematic diagram of the strip test results. (B) Visual assessment sensitivities of strips for spiked Np in urine. | ||
To fight the current global pandemic, we aim to provide useful aptamers as molecular tools. The application of these aptamers in ELISA and GLS assay shows that they should enable the rapid development of POCT and biosensors for personal or home use. The binding mechanism of aptamer and antibody to the same target is different. The smaller size of the aptamers (about 2–3 nm in diameter), as compared to antibodies (about 12–15 nm in diameter), subjects them to less steric hindrance on the surface of the target (herein, it is COVID-19 Np). In theory, the smaller size allows for the binding of more recognition motifs on the same surface area of the protein. Therefore, the antibody–aptamer hybrid showed good performance. For either ELISA or GLS, when only using aptamers or antibodies, the range of LOD was 50–100 ng mL−1 (Table S3, ESI†). The aptamer–antibody hybrid used in ELISA or GLS showed greatly improved performance (50-fold enhancement) compared to using one alone. The novelty of our biosensors is the use of a pair of sandwich-type aptamers. As shown in Fig. 3A, the formation of the Np-A48|Np|Np-A58 complex makes the detection of Np in a sophisticated system robust and sensitive.
With the spread of the epidemic, more and more analytical detection methods have been developed, as we have summarized in Table S4 (ESI†). Each method has its advantages and disadvantages. The detection limit for the PCR method is low, but there are false positives. The antibody method is not suitable for early diagnosis.
Recently, a field-effect transistor (FET)-based biosensing device was reported to achieve a limit of detection for COVID-19 spike protein at the level of 1.6 × 101 pfu mL−1 (clinical samples: 2.42 × 102 copies per mL).22 The limit of detection of our sensors (ELISA and GLS) for testing N-protein is better than that of the reported one.23 However, other studies based on our new aptamers may find that the analytical performance can be improved by orders of magnitude (new work will be submitted and published by our collaborators). These results indicated that aptamers are powerful molecular tools to combat COVID-19. Although many aptamers are used both in virus detection and antiviral therapy,8 there is still a lack of high-performance aptamer-based market products for the fight against the current outbreak of COVID-19. Future work will continue to advance the acquisition of high-performance aptamers and the development of possible application products and technologies.
In conclusion, we have developed four DNA aptamers, all of which have high affinity to bind with COVID-19 Np. Among them, four pairs (Np-A48/Np-A58, Np-A48/Np-A61, Np-A15/Np-A58, and Np-A15/Np-A61) can form ternary complexes with Np. Using these sandwich type aptamers in ELISA and GLS assay, the limit of detection has reduced to the tens of pM level. All the results suggest that the new aptamers hold great potential in the development of POCT and biosensors to address the current epidemic crisis.
We acknowledge the financial support from the National Natural Science Foundation of China (No. 31570755 and 5197021057) and the COVID-19 emergency science and technology special fund (YD2070002016). L. Zhang thanks Nankai university for the support of the hundred youth program (019157) and acknowledges the funding support from the Natural Science Foundation of Tianjin (19JCZDJC33400).
Conflicts of interest
There are no conflicts to declare.References
- F. Wu, S. Zhao, B. Yu, Y.-M. Chen, W. Wang, Z.-G. Song, Y. Hu, Z.-W. Tao, J.-H. Tian, Y.-Y. Pei, M.-L. Yuan, Y.-L. Zhang, F.-H. Dai, Y. Liu, Q.-M. Wang, J.-J. Zheng, L. Xu, E. C. Holmes and Y.-Z. Zhang, Nature, 2020, 579, 265–269 CrossRef CAS PubMed.
- Z. Y. Zu, M. D. Jiang, P. P. Xu, W. Chen, Q. Q. Ni, G. M. Lu and L. J. Zhang, Radiology, 2020, 200490 Search PubMed.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. C. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. W. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS PubMed.
- M. Famulok and G. Mayer, Chem. Biol., 2014, 21, 1055–1058 CrossRef CAS PubMed.
- G. Zhu and X. Chen, Adv. Drug Delivery Rev., 2018, 134, 65–78 CrossRef CAS PubMed.
- Y.-H. Lao, K. K. L. Phua and K. W. Leong, ACS Nano, 2015, 9, 2235–2254 CrossRef CAS PubMed.
- B. Santosh and P. K. Yadava, BioMed Res. Int., 2014, 2014, 540451 Search PubMed.
- X. Zou, J. Wu, J. Gu, L. Shen and L. Mao, Front. Microbiol., 2019, 10, 1462 CrossRef PubMed.
- J. Bhardwaj, N. Chaudhary, H. Kim and J. Jang, Anal. Chim. Acta, 2019, 1064, 94–103 CrossRef CAS PubMed.
- I. Hmila, M. Wongphatcharachai, N. Laamiri, R. Aouini, B. Marnissi, M. Arbi, S. Sreevatsan and A. Ghram, J. Virol. Methods, 2017, 243, 83–91 CrossRef CAS PubMed.
- Y. Pang, Z. Rong, J. Wang, R. Xiao and S. Wang, Biosens. Bioelectron., 2015, 66, 527–532 CrossRef CAS PubMed.
- C. R. Basso, B. P. Crulhas, M. Magro, F. Vianello and V. A. Pedrosa, Talanta, 2019, 197, 482–490 CrossRef CAS PubMed.
- S.-L. Hong, M.-Q. Xiang, M. Tang, D.-W. Pang and Z.-L. Zhang, Anal. Chem., 2019, 91, 3367–3373 CrossRef CAS PubMed.
- C. Roh and S. K. Jo, J. Chem. Technol. Biotechnol., 2011, 86, 1475–1479 CrossRef CAS PubMed.
- S. J. Cho, H. M. Woo, K. S. Kim, J. W. Oh and Y. J. Jeong, J. Biosci. Bioeng., 2011, 112, 535–540 CrossRef CAS PubMed.
- D. G. Ahn, I. J. Jeon, J. D. Kim, M. S. Song, S. R. Han, S. W. Lee, H. Jung and J. W. Oh, Analyst, 2009, 134, 1896–1901 RSC.
- Z. Luo, L. He, J. Wang, X. Fang and L. Zhang, Analyst, 2017, 142, 3136–3139 RSC.
- C.-K. Chang, M.-H. Hou, C.-F. Chang, C.-D. Hsiao and T.-H. Huang, Antiviral Res., 2014, 103, 39–50 CrossRef CAS PubMed.
- R. Lu, X. Zhao, J. Li, P. Niu, B. Yang, H. Wu, W. Wang, H. Song, B. Huang, N. Zhu, Y. Bi, X. Ma, F. Zhan, L. Wang, T. Hu, H. Zhou, Z. Hu, W. Zhou, L. Zhao, J. Chen, Y. Meng, J. Wang, Y. Lin, J. Yuan, Z. Xie, J. Ma, W. J. Liu, D. Wang, W. Xu, E. C. Holmes, G. F. Gao, G. Wu, W. Chen, W. Shi and W. Tan, Lancet, 2020, 395, 565–574 CrossRef CAS.
- S.-Y. Lin, C.-L. Liu, Y.-M. Chang, J. Zhao, S. Perlman and M.-H. Hou, J. Med. Chem., 2014, 57, 2247–2257 CrossRef CAS PubMed.
- S. Zhang, A. Garcia-D'Angeli, J. P. Brennan and Q. Huo, Analyst, 2014, 139, 439–445 RSC.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park, H. G. Kim, S.-J. Kim, J.-O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- Z. Chen, Q. Wu, J. Chen, X. Ni and J. Dai, Virol. Sin., 2020, 35, 351–354 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The details of experiments and supporting figures and tables are included. See DOI: 10.1039/d0cc03993d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |