Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biosynthesis of oxygenated brasilane terpene glycosides involves a promiscuous N-acetylglucosamine transferase†
Jin
Feng
 a,
Frank
Surup
a,
Frank
Surup
 b,
Maurice
Hauser
a,
Anna
Miller
a,
Jan-Peer
Wennrich
b,
Maurice
Hauser
a,
Anna
Miller
a,
Jan-Peer
Wennrich
 b,
Marc
Stadler
b,
Marc
Stadler
 b,
Russell J.
Cox
b,
Russell J.
Cox
 a and
Eric
Kuhnert
a and
Eric
Kuhnert
 *a
*a
aInstitute for Organic Chemistry and Centre for Biomolecular Drug Research (BMWZ), Leibniz University Hannover, Schneiderberg 38, Hannover 30167, Germany. E-mail: eric.kuhnert@oci.uni-hannover.de
bDepartment Microbial Drugs, Helmholtz Centre for Infection Research (HZI), Inhoffenstraße 7, 38124 Braunschweig, Germany
First published on 16th September 2020
Abstract
Investigation of the metabolome of the ascomycete Annulohypoxylon truncatum led to the identification of novel oxygenated brasilane glycosides and the revision of the stereochemistry of the brasilane A octahydro-1H-indene core scaffold to trans. The bra biosynthetic gene cluster containing five genes (braA–braE) was identified and verified by heterologous expression experiments in Aspergillus oryzae demonstrating that BraC is a multifunctional P450 monooxygenase. In vitro studies of BraB revealed it to be a very rare fungal UDP-GlcNAc dependent N-acetylglucosamine transferase. UDP-glucose is also accepted as a donor, and a broad acceptor substrate tolerance for various primary and secondary alcohols was observed.
The fungal family Hypoxylaceae is a hotspot for the identification of novel secondary metabolites.1 Genome sequencing of representative species enabled us to assess the biosynthetic capabilities of these fungi and correlate produced compounds with their respective biosynthetic genes.2 During a screening for new metabolites we found various species capable of producing brasilane-type sesquiterpenoid glycosides with an unusual N-acetylglucosamine moiety. The brasilane core structure is widespread in nature and has been isolated from various sources including basidiomycetes, various ascomycetes, algae and sea hare.3–8 Recently, two groups have independently reported the characterisation of the terpene cyclase responsible for brasilane biosynthesis in Trichoderma spp.9,10 These reports facilitated the search for the underlying biosynthetic gene clusters in the Hypoxylaceae which led us to investigate the participating tailoring enzymes.
Fermentation of the Annulohypoxylon truncatum strain CBS 140778 in 2 L YMG media led to the production of various glycosylated metabolites. Subsequent chromatographic purification yielded three pure compounds that were analyzed by NMR spectroscopy. Structure elucidation revealed the known sesquiterpenoid brasilane A 16 and two new oxygenated congeners brasilanes D 2 and E 3 [Scheme 1, see ESI,† Sections S2 and S6 for additional information]. All three terpenes feature an unusual N-acetylglucosamine moiety, with 2 being hydroxylated at C-12 and 3 possessing an epoxide functionality between C-5 and C-10 and an aldehyde at C-12. Careful analysis of coupling constants and ROESY data of 1 revealed that the previously assigned relative configuration of brasilane A6 was incorrect. Since overlapping signals hampered the interpretation of spectra measured in chloroform-d, methanol-d4, acetonitrile-d3, DMSO-d6 and acetone-d6, we selected pyridine-d5 and benzene-d6 for the measurement of ROESY and J-resolved NMR spectra (see ESI,† Chapter S2 and S6). On the β-face of the molecule, strong 1,3-diaxial ROESY correlations were observed between 2-Ha/4-Hb/6-H, in addition to correlations between 15-H3 and 4-Hb, 6-H, 8a-H. On the α side of the molecule, there are strong ROESY correlations between 1-H/14-H3 and 8-Hb/7-Hb. Taken together, these ROESY correlations indicated a trans fusion of the central octahydro-1H-indene scaffold, which was confirmed by the large coupling constant JH1,H6 = 12.5 Hz, observed in the signal pattern of 6-H. All compounds were evaluated for biological activity in antimicrobial and cytotoxicity assays, but no significant activity was observed. Nevertheless, oxygenated brasilane glycosides have so far not been reported in the literature, suggesting the involvement of a specific enzymatic machinery in the producer organism.
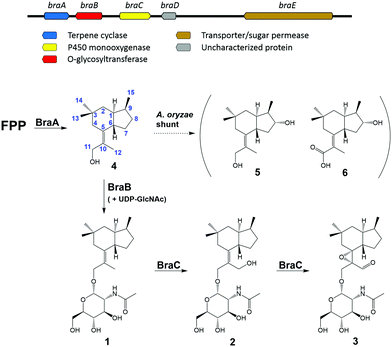 | ||
| Scheme 1 Brasilane biosynthetic gene cluster (bra) from A. truncatum and biosynthetic pathway for the production of 1–6 based on experimental evidence. | ||
Recently, two independent studies found and characterized the terpene cyclases (TaTC6, Tvi09626) responsible for bra-silane backbone assembly in Trichoderma spp.9,10 Using BLASTp homology searches with Tvi09626 as a template, we found a candidate biosynthetic gene cluster (BGC, bra) in the A. truncatum genome containing five genes termed braA–E (Scheme 1). BraA was identified as a terpene cyclase showing 70.9% similarity to Tvi09626, while BraB was annotated as a putative O-glycosyltransferase (O-GT) with homology to the O-GT PaGT involved in fusicoccin biosynthesis in Diaporthe (Phomopsis) amygdali.11 Except for its homology to the latter enzyme, BraB (and also PaGT) shows no conserved motifs recognized by CAZyme analysis.12 InterPro scan identified a partial capsular polysaccharide synthesis protein family domain,13 which is part of the nucleotide-diphospho-sugar transferase superfamily, suggesting UDP-sugars as donor substrates. BraC exhibited high identity to various known cytochrome P450 monooxygenases and BraE likely constitutes an ABC transporter. No functionality could be assigned to BraD which lacks any currently recognizable functional motifs.
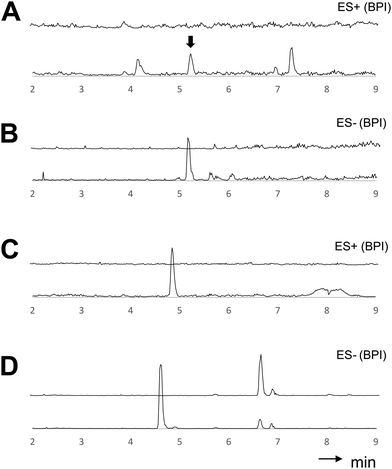
To verify whether the bra BGC is responsible for the bio-synthesis of 1–3, heterologous expression experiments in Aspergillus oryzae NSAR1 were conducted. For this purpose, the braA–C genes were directly cloned from gDNA into the vector pTYGS-arg using the arginine deficiency as auxotrophic marker.14 Transformation of A. oryzae with the constructed vector resulted in well-growing transformants (A. oryzae braABC), which were screened for the production of new metabolites in DPY media. Media extracts of six transformants showed the presence of two new conspicuous peaks (Fig. 1C), which correlated well with 2 and 3 by UV, retention time and mass spectra. Isolation and structure elucidation of the respective compounds confirmed their identity as brasilane D 2 and E 3. In contrast, all extracts were absent of 1, implying that the compound was fully converted. As 2 and 3 show different levels of oxygenation, BraC is likely responsible for two consecutive oxygenations. Cytochrome P450 monooxygenases catalyzing multiple steps within a biosynthetic pathway are commonly observed and this can explain the occurrence of two main compounds.15,16
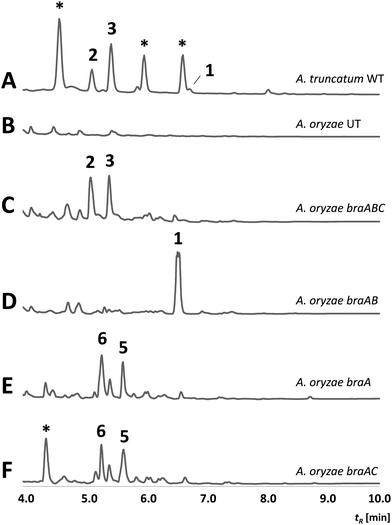 | ||
| Fig. 1 HPLC-DAD chromatograms (200–600 nm) of supernatant extracts from; (A) wild type of A. truncatum; (B) untransformed A. oryzae (negative control); (C–F) A. oryzae transformants expressing different combinations of braA, braB and braC. Numbers refer to structures depicted in Scheme 1. Unrelated compounds are indicated with *. | ||
To reconstitute the biosynthesis of 1, an additional vector containing only braA and braB was constructed and trans-formed into A. oryzae to yield a dominant compound, which as expected was identified as brasilane A 1 (Fig. 1D). In order to determine the sequence of the biosynthetic steps two further vectors were assembled and transformed. One vector contained braA and braC and the other contained only braA. Analysis of the resulting A. oryzae transformants (A. oryzae braAC; A. oryzae braA) showed production profiles with various new compounds, which were identical between A. oryzae braAC and A. oryzae braA (Fig. 1E and F). The two major products were purified, and elucidation of their structures showed them to be two new oxygenated brasilanes designated as brasilane F 5 and xylarenic acid B 6 (Scheme 1). Against our expectation neither 5 nor 6 were oxygenated at C-12, C-5 or C-10. Instead, both molecules displayed a hydroxyl group at C-8, and 6 had an additional carboxylic acid functionality at C-10. While ROESY correlations and a series of 1D NOE NMR experiments (Fig. S60–S64, ESI†) confirmed the relative configuration of the new molecules including the trans configuration of 1-H/6-H, their absolute stereochemistry was determined by derivatisation of 8-OH with Mosher's acid (MTPA, see ESI,† Chapter S2). As the chemical profiles of A. oryzae braAC and A. oryzae braA transformants were identical, we concluded that BraC does not accept brasilane aglycones. Instead, unknown native A. oryzae enzymes process the terpene backbone leading to the formation of 5 and 6. The influence of the heterologous host on shunt formation or even complete pathway disruption has been observed during the heterologous expression of various biosynthetic pathways, such as solanapyrone, cytochalasans or betaenone,17 and therefore was not surprising. To confirm that BraC was functional in the A. oryzae braAC transformants, 1 was fed to the fermentation broth of A. oryzae braAC. After one day of incubation 2 and 3 were detected in the crude extracts, demonstrating that glycosylation occurs prior to oxygenation (Fig. S4, ESI†).
The product of BraA in A. oryzae NSAR1 could neither be unambiguously detected by HPLC-MS nor GCMS in our experimental setup. As all isolated brasilanes show the same absolute configuration as reported by Murai et al.9 and because BraA is highly similar to TaTC6 (Fig. S3, ESI†), it can be assumed that the terpene alcohol trichobrasilenol 4 is the most likely product of BraA. To further confirm this prediction, BraA was overexpressed in E. coli and purified for subsequent in vitro studies. For this purpose, the braA gene was obtained from cDNA of the A. oryzae braA transformant and cloned into the pET28a+ vector. The purified protein was tested in vitro in the presence of farnesyl pyrophosphate (FPP) and the solution was extracted with pentane. Analysis of the crude extract by GCMS revealed a single peak with a mass of 222.1 (Fig. S5, ESI†). The respective fragmentation pattern corresponds well with those previously reported for trichobrasilenol.9
Based on these results, the full biosynthetic pathway towards 2 and 3 can be deduced (Fig. 1A), starting by the activity of BraA, which converts farnesyl pyrophosphate into the sesquiterpene alcohol 4. Subsequently, 4 is glycosylated by BraB putatively using UDP-GlcNAc as sugar donor to yield 1. The latter then undergoes two rounds of oxidation performed by the P450 monooxygenase BraC. In the first round BraC hydroxylates C-12 forming 2, which serves as substrate in the second round to establish the epoxide at the bond between C-5 and C-10 and oxidize the alcohol at C-12 to an aldehyde leading to the final product 3.
Reports of fungal secondary metabolites containing N-acetylglucosamine units are scarce in the literature.18 The few known cases besides brasilanes A, D and E comprise the glycopeptide supprescins from Mycosphaerella pinodes, isopimarane glycosides from Paraconiothyrium sp. and a series of glycosylated compounds from Malbranchea filamentosa.19–23 This data is especially surprising in light of the fact that GlcNAc is one of the most abundant sugars on earth used for protein regulation and to form chitin, which constitutes the major part of the fungal cell walls and the exoskeleton of invertebrates, and murein as structural component of bacterial cell walls.24 We were therefore interested in investigating the promiscuity of BraB in vitro. For this purpose, the codon optimized intron-free gene braB was synthesized and cloned into the pET28a+ vector for protein overexpression in E. coli. Protein isolation resulted in high concentrations of soluble protein. As 4 could not be directly obtained for in vitro characterization (note that deglycosylation under acidic conditions of 1 did result in very low amounts of 4 being insufficient for further reactions), the terpene cyclase BraA was used for in vitro generation of 4.
To test the UDP-sugar selectivity of BraB, the enzyme was co-incubated with BraA, FPP, dithiothreitol (DTT) and different UDP-sugars (UDP-GlcNAc, UDP-glucose, UDP-galactose) at 30 °C overnight. In the presence of UDP-GlcNAc biosynthesis of 1 was observed by LCMS (Fig. S6, ESI†). In addition, a clear reaction product was also found when UDP-glucose was used as alternative sugar donor (Fig. S7, ESI†), which is likely to be identical with the known compound hypoxyside isolated from the related fungus Hypoxylon fuscum.3 In the latter organism an O-GT with different selectivity may be active, but lack of genomic information currently precludes further investigations. In contrast, UDP-galactose was not accepted as substrate (Fig. S8, ESI†). To further assess the selectivity of BraB, a competition assay with a UDP-Glc/UDP-GlcNAc mixture was conducted, which resulted in 1 as predominant product (Fig. S9, ESI†) and thus showing that BraB has a strong preference for UDP-GlcNAc. This also explains why brasilane glycosides with GlcNAc moiety are the predominant pathway products in A. truncatum and the heterologous host.
Due to the strong affinity of BraB for UDP-GlcNAc and to set it apart from other known fungal glycosyl transferases, we define the protein as an N-acetylglucosamine transferase rendering it the first of its kind associated with fungal secondary metabolism.
After the evaluation of the donor substrate selectivity, we studied the tolerance of BraB for various acceptor substrates in vitro. Therefore, a set of secondary and tertiary alcohols was tested in the presence of UDP-GlcNAc, the majority of which were converted into their respective glycosides (Table 1, Fig. 2 and Fig. S10–S14, ESI†). The highest product yield observed by LCMS after 12 h of incubation was achieved for perillyl alcohol (PA) and 3,4-dichlorophenol. Based on the obtained data, it can be seen that BraB is able to glycosylate a broad range of substrates.
| Acceptor substrate | Conversion |
|---|---|
| + Glycosylated product detected, (+) product detected in trace amounts, − no conversion. | |
| Geraniol | + |
| Linalool | + |
| Perillyl alcohol | + |
| Menthol | − |
| Benzyl alcohol | (+) |
| Serine | − |
| Serotonin | − |
| 3,4-Dichlorophenol | + |
To demonstrate that the observed in vitro products are in agreement with the prediction and that BraB can be used for biotechnological applications, in vivo preparative scale reactions were conducted. Therefore, 1 L of E. coli BraB transformants (same strain as used for protein overexpression) were incubated overnight with 75 mg of PA in total resulting in the formation of a new compound 7 (Fig. S16, ESI†). Subsequent product isolation resulted in 10 mg of pure compound. HMBC correlations from H2-7 to C-1′ and 1′-H to C-7, respectively, confirmed the metabolite as a perillyl sugar with a GlcNAc moiety (see ESI,† Chapter S2 and S6) and emphasising the function of BraB as a UDP-GlcNAc transferase.
N-Acetyl glucosamine transferases are well known in primary metabolism, but their substrate selectivities are normally very narrow. In contrast, N-acetyl glucosamine transferases are very rarely found in secondary metabolism. Two prominent examples are ORF1 and ORF10* from the bacterial glycopeptide teicoplanin biosynthetic pathway, which also appear to have tight substrate selectivity.25 BraB is the first N-acetyl glucosamine transferase to have been identified from a fungal biosynthetic pathway, and its broad substrate selectivity should offer significant opportunities for use in pathway engineering and other biotechnology applications. In addition, the biotransformation can be easily scaled-up in vivo using bacterial hosts, which provides a cost-efficient method to generate GlcNAc-glycosylated products.
In summary, we have elucidated the structures of four new brasilane congeners and characterized the complete biosynthetic pathway for brasilane glycoside formation in A. truncatum. Furthermore, a new multi-catalytic cytochrome P450 monooxygenase as well as the first biosynthetic N-acetylglucosamine transferase in fungi was identified. In vitro characterization of the latter showed a broad substrate tolerance for acceptor compounds enabling biotechnological and chemical applications.
This work was funded by the DFG (Deutsche Forschungsge-meinschaft) priority program “Taxon-Omics: New Approaches for Discovering and Naming Biodiversity” (SPP 1991). DFG is also thanked for 600 MHz NMR equipment (INST 187/686-1). JF was funded by the China Scholarship Council (201709110130) and AM was funded by Erasmus+ and the Carnegie Trust. We would also like to thank Klaus-Peter Conrad for the measurement of HRESIMS data, and Christel Kakoschke for the measurement of NMR spectra (HZI, Braunschweig). Dr Daniel Wibberg (CeBiTec, Bielefeld) is acknowledged for assistance in genome annotation.
Conflicts of interest
There are no conflicts to declare.References
- S. E. Helaly, B. Thongbai and M. Stadler, Nat. Prod. Rep., 2018, 35, 992–1014 RSC.
- D. Wibberg, M. Stadler, C. Lambert, B. Bunk, C. Spröer, C. Rückert, J. Kalinowski, R. J. Cox and E. Kuhnert, Fungal Divers., 2020 DOI:10.1007/s13225-020-00447-5.
- B. B. Basnet, B. Chen, Y. M. Suleimen, K. Ma, S. Guo, L. Bao, Y. Huang and H. Liu, Planta Med., 2019, 85, 1088–1097 CrossRef CAS.
- S. Caccamese, V. Amico and P. Neri, J. Nat. Prod., 1990, 53, 1287–1296 CrossRef CAS.
- D. Iliopoulou, C. Vagias, D. Galanakis, D. Argyropoulos and V. Roussis, Org. Lett., 2002, 4, 3263–3266 CrossRef CAS.
- D.-B. Hu, S. Zhang, J.-B. He, Z.-J. Dong, Z.-H. Li, T. Feng and J.-K. Liu, Fitoterapia, 2015, 104, 50–54 CrossRef CAS.
- M. O. Stallard, W. Fenical and J. S. Kittredge, Tetrahedron, 1978, 34, 2077–2081 CrossRef CAS.
- Z. Wu, D. Li, F. Zeng, Q. Tong, Y. Zheng, J. Liu, Q. Zhou, X.-N. Li, C. Chen, Y. Lai, H. Zhu and Y. Zhang, Phytochemistry, 2018, 156, 159–166 CrossRef CAS.
- K. Murai, L. Lauterbach, K. Teramoto, Z. Quan, L. Barra, T. Yamamoto, K. Nonaka, K. Shiomi, M. Nishiyama, T. Kuzuyama and J. S. Dickschat, Angew. Chem., Int. Ed., 2019, 58, 15046–15050 CrossRef CAS.
- X. Sun, Y.-S. Cai, Y. Yuan, G. Bian, Z. Ye, Z. Deng and T. Liu, Beilstein J. Org. Chem., 2019, 15, 2052–2058 CrossRef CAS.
- M. Noike, Y. Ono, Y. Araki, R. Tanio, Y. Higuchi, H. Nitta, Y. Hamano, T. Toyomasu, T. Sassa, N. Kato and T. Dairi, PLoS One, 2012, 7, e42090 CrossRef CAS.
- V. Lombard, H. Golaconda Ramulu, E. Drula, P. M. Coutinho and B. Henrissat, Nucleic Acids Res., 2014, 42, D490–D495 CrossRef CAS.
- S. Hunter, R. Apweiler and T. K. Attwood, et al. , Nucleic Acids Res., 2009, 37, D211–D215 CrossRef CAS.
- L. Kahlert, E. F. Bassiony, R. J. Cox and E. J. Skellam, Angew. Chem., 2020, 132, 5865–5871 CrossRef.
- Y. Matsuda and I. Abe, Nat. Prod. Rep., 2016, 33, 26–53 RSC.
- S. Saikia, E. J. Parker, A. Koulman and B. Scott, J. Biol. Chem., 2007, 282, 16829–16837 CrossRef CAS.
- Y. He, B. Wang, W. Chen, R. J. Cox, J. He and F. Chen, Biotechnol. Adv., 2018, 36, 739–783 CrossRef CAS.
- S. I. Elshahawi, K. A. Shaaban, M. K. Kharel and J. S. Thorson, Chem. Soc. Rev., 2015, 44, 7591–7697 RSC.
- K. Kawai, D. Wakana, T. Hosoe, T. Itabashi, H. Okada and K. Fukushima, Heterocycles, 2008, 75, 1109 CrossRef.
- T. Shiraishi, K. Saitoh, H. M. Kim, T. Kato, M. Tahara, H. Oku, T. Yamada and Y. Ichinose, Plant Cell Physiol., 1992, 33, 663–667 CAS.
- D. Wakana, T. Itabashi, K. Kawai, T. Yaguchi, K. Fukushima, Y. Goda and T. Hosoe, J. Antibiot., 2014, 67, 585–588 CrossRef CAS.
- D. Wakana, T. Hosoe, T. Itabashi, K. Fukushima and K. Kawai, Mycotoxins, 2008, 58, 1–6 CrossRef CAS.
- Y. Shiono, M. Kikuchi, T. Koseki, T. Murayama, E. Kwon, N. Aburai and K. Kimura, Phytochemistry, 2011, 72, 1400–1405 CrossRef CAS.
- J.-K. Chen, C.-R. Shen and C.-L. Liu, Mar. Drugs, 2010, 8, 2493–2516 CrossRef CAS.
- T. Li, F. Huang, S. Haydock, T. Mironenko, P. Leadlay and J. Spencer, Chem. Biol., 2004, 11, 107–119 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, structure elucidation, supplementary tables and figures, NMR spectra. See DOI: 10.1039/d0cc03950k |
| This journal is © The Royal Society of Chemistry 2020 |