Open Access Article
Open Access ArticleSynthetic lectins for selective binding of glycoproteins in water†
Likun
Duan
,
Milad
Zangiabadi
and
Yan
Zhao
 *
*
Department of Chemistry, Iowa State University, Ames, Iowa 50011-3111, USA. E-mail: zhaoy@iastate.edu; Fax: +1-515-294-0105; Tel: +1-515-294-5845
First published on 24th July 2020
Abstract
Although synthetic mimics of lectins can be extremely useful in biological and biomedical research, molecular recognition of carbohydrates has been hampered by their strong solvation in water and subtle structural differences among analogues. Molecularly imprinted nanoparticle receptors were prepared with glycans directly cleaved from glycoproteins. Functionalized with boroxole groups in the binding sites, these water-soluble synthetic lectins bound the parent glycoproteins selectively in water with an association constant of Ka = 104–105 M−1. The strong binding enabled the receptors to protect the targeted glycans from enzymatic cleavage. When clicked onto magnetic nanoparticles, the receptors enabled facile isolation of glycoproteins from a mixture.
Lectins are nonimmunological proteins that bind specific glycans (or carbohydrates) on glycoproteins, glycolipids, or oligosaccharides.1,2 Since glycosylation is the most common post-translational modification of proteins and glycans are involved in numerous biological processes including cell adhesion, bacterial and viral infection, inflammation, and cancer development,3–7 it is difficult to overstate the importance of lectins in biological and biomedical research.
The significance of lectin–glycan interactions and poor availability of many lectins have motivated chemists to develop their synthetic mimics.6–9 The molecular recognition, especially in aqueous media, however, represents a formidable challenge because inverting a single hydroxyl in a glycan, connecting the monosaccharide building blocks by different hydroxyls, or altering the (α/β) glycosidic linkages all bring profound changes to the biological properties of the glycan. Not only so, the main functional group of carbohydrates (i.e., hydroxyl) is found in the solvent molecule. Thus, in order for a lectin mimic to carry out its intended task competently, it needs to not only overcome a tremendous competition from the solvent water but also differentiate the complex and fine structural differences among analogous glycans.
Many boronic acid-based carbohydrate-binding materials have been prepared10–16 through molecular imprinting and found applications in chemistry and biology since Wulff's pioneering work.17,18 The advantage of molecular imprinting is the facile construction of complex binding pockets with template-complementary binding groups. Nonetheless, most molecularly imprinted polymers and materials are macroscopic in nature, which limits their application as lectin mimics that are much smaller in dimension and soluble in water.
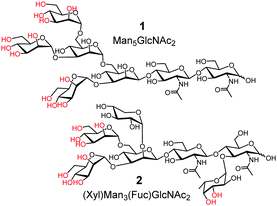
The challenge in glycoprotein recognition is exemplified by two common glycoproteins. Avian ovalbumin (OVA) has high-mannose glycans such as Man5GlcNAc2 (1) and Man6GlcNAc2 (Fig. 1, Man = D-mannose; GlcNAc = N-acetyl-D-glucosamine).19 A variety of other structures are also present with varying numbers of Man and additional GlcNAc. A major glycan of horseradish peroxidase (HRP) is 2.20 It shares the same Man3GlcNAc2 core structure as 1, but contains a xylose (Xyl) and a fucose (Fuc) at specific locations.
Structural complexity of these glycans and their microheterogeneity make it inconceivable to construct effective molecular receptors based on traditional supramolecular principles (i.e., preorganization and complementarity). Even though boronic acid9,17,21–24 and benzoboroxole derivatives25–27 can bind specific 1,2- or 1,3-diols of sugars (highlighted in red in 1 and 2), preorganizing these binding groups on an organic framework to match the proper hydroxyls is still too difficult.
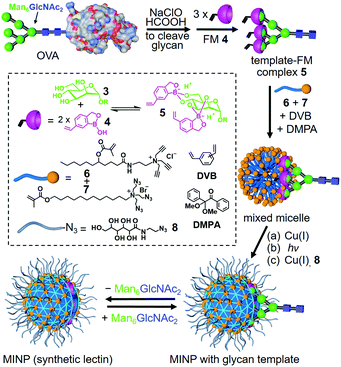
Our strategy to deal with the complexity and microheterogeneity of the glycans is shown in Scheme 1, through molecular imprinting within cross-linked micelles, a method developed by our group recently.28 The key is to employ glycans cleaved directly from the glycoprotein as the templates, following a reported oxidative protocol.29 The glycan template (shown as a generic structure 3) is then combined with functional monomer (FM) 4 to form negatively charged (amphiphilic) boronates (5), which are stabilized by the hydrophobic microenvironment of cationic mixed micelles of 6 and 7.30 The highly efficient click reaction between 6 and 7 is facilitated by the intimate mixing of alkyne and azide groups on the micellar surface. With divinylbenzene (DVB) and 2,2-dimethoxy-2-phenylacetophenone (DMPA, a photolytic radical initiator) inside, the micelles are core-cross-linked by free radical polymerization upon UV irradiation, with the boroxole FMs polymerized into the micellar core in the meantime. Another round of click reaction with 8 decorates the surface of the doubly cross-linked micelle with a layer of hydrophilic ligand, which enables the MINP to precipitate readily from acetone and be purified by simple washing (with acetone/water and methanol).
Table 1 shows the binding properties of MINP(OVA) and MINP(HRP), prepared from glycans cleaved from the corresponding glycoproteins. Consistent with the importance of boronate formation, MINP(OVA) displayed stronger binding for the templating glycans as the FM/template ratio increased from 4 to 8 (entries 1–3). Molecular imprinting was clearly responsible for the recognition, since negligible binding was observed for nonimprinted nanoparticles (NINPs) prepared with the same 8 equiv FM 4 but without the glycan templates (entry 4). Concanavalin A (Con A) binds ovalbumin glycans with Ka ≈ 30 × 104 M−1 in aqueous buffer.31 It is encouraging that the binding constant of our MINP (16.3 × 104 M−1) was more than half of the biological value. The imprinting factor (IF), defined as Ka(MINP)/Ka(NINP), was exceedingly large (16.3/0.02 ≈ 800). Molecular imprinting is known to be highly effective when carried out in the confined nanospace of micelles.32 The much larger IF value in comparison to what was achieved (>100) for peptides and related compounds in micellar imprinting33,34 was likely caused by the hydrophilicity of the glycan templates that reduced nonspecific binding.
| Entry | Templating glycan source | FM/template ratio | Guestb | K a (×104 M−1) | −ΔG (kcal mol−1) |
|---|---|---|---|---|---|
| a The titrations were performed in duplicates with the indicated errors in HEPES buffer (10 mM, pH 7.4) at 298 K. b Glycan indicates the guest was the same templating glycan mixture from the corresponding glycoprotein. c Binding was very weak and the binding constant was estimated from ITC titration. | |||||
| 1 | OVA | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Glycan | 8.45 ± 0.07 | 6.72 |
| 2 | OVA | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Glycan | 10.5 ± 0.1 | 6.85 |
| 3 | OVA | 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Glycan | 16.3 ± 0.2 | 7.11 |
| 4 | None | 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
Glycan | 0.02c | 3.19 |
| 5 | OVA | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
OVA | 1.75 ± 0.04 | 5.79 |
| 6 | OVA | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
HRP | 0.59 ± 0.03 | 5.14 |
| 7 | OVA | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
OVA | 4.28 ± 0.05 | 6.31 |
| 8 | OVA | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
HRP | 0.79 ± 0.03 | 5.31 |
| 9 | OVA | 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
OVA | 9.41 ± 0.21 | 6.78 |
| 10 | OVA | 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
HRP | 0.73 ± 0.02 | 5.27 |
| 11 | None | 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
OVA | 0.02c | 3.17 |
| 12 | HRP | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Glycan | 0.51 ± 0.03 | 5.06 |
| 13 | HRP | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Glycan | 1.31 ± 0.07 | 5.61 |
| 14 | HRP | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Glycan | 1.09 ± 0.03 | 5.51 |
| 15 | HRP | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
HRP | 0.30 ± 0.02 | 4.74 |
| 16 | HRP | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
OVA | 0.12 ± 0.02 | 4.20 |
| 17 | HRP | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
HRP | 1.09 ± 0.06 | 5.50 |
| 18 | HRP | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
OVA | 0.21 ± 0.02 | 4.53 |
| 19 | HRP | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
HRP | 1.01 ± 0.04 | 5.46 |
| 20 | HRP | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
OVA | 0.32 ± 0.02 | 4.78 |
Importantly, all the MINP(OVA)s showed substantial binding affinities for the parent glycoprotein, up to Ka = 9.41 × 104 M−1 in HEPES buffer (entry 9). The binding constants for OVA generally decreased by 2–4-fold in comparison to those for the glycans. The amide-containing surfactant 6 is known to help the binding of oligosaccharides by micelle-stabilized hydrogen bonds with those hydroxyls not engaged in boronate formation.30 Thus, other parts of the glycan—in addition to the red hydroxyls shown in 1—should also play some roles in the binding.35 Under such a situation, binding of the same glycan on the parent protein is anticipated to encounter some steric hindrance, especially for the part of the glycan close to the protein surface. Reduced binding for the glycoprotein is thus a reasonable result, in comparison to that for the glycan.
Equally important was the selectivity of MINP(OVA), which bound HRP, a glycoprotein with some similarity in the glycan structure, much more weakly. As the FM/template ratio increased from 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 all the way to 8
1 all the way to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the ratio of binding constants for OVA/HRP increased from 3 to 5 to 13
1, the ratio of binding constants for OVA/HRP increased from 3 to 5 to 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (entries 5–10). Not surprisingly, the binding for the protein by the nonimprinted particles was nearly undetectable, highlighting the importance of molecular imprinting in the recognition (entry 11).
1 (entries 5–10). Not surprisingly, the binding for the protein by the nonimprinted particles was nearly undetectable, highlighting the importance of molecular imprinting in the recognition (entry 11).
According to the preferred binding motifs of boroxole (i.e., cis-3,4-diol and trans-4,6-diol for mannose),30 Man5GlcNAc2 (1) can react with up to 6 equiv. FM 4, and Man6GlcNAc2 with 8 equiv. The strongest binding observed at the 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, as well as the highest binding selectivity, suggests that the proposed boronate formation was in operation. Because the template–FM was formed in situ in the aqueous micellar solution, it is remarkable that nearly strict stoichiometry appeared to be maintained between the glycan and the FM.
1 ratio, as well as the highest binding selectivity, suggests that the proposed boronate formation was in operation. Because the template–FM was formed in situ in the aqueous micellar solution, it is remarkable that nearly strict stoichiometry appeared to be maintained between the glycan and the FM.
The binding of MINP(HRP) showed similar behavior. The maximum binding for the templating glycan was observed in this case at FM/template = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (entry 13). This is very close to the maximum number (i.e., 5) of boroxoles that could react with (Xyl)Man3(Fuc)GlcNAc2 (2), the most abundant glycan on the protein. Selectivity between HRP and OVA was also the highest at FM/template = 4
1 (entry 13). This is very close to the maximum number (i.e., 5) of boroxoles that could react with (Xyl)Man3(Fuc)GlcNAc2 (2), the most abundant glycan on the protein. Selectivity between HRP and OVA was also the highest at FM/template = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (entries 17 and 18), suggesting the proposed boronate binding motif was general. Because fewer boronate bonds are expected for 2 (with MINP(HRP)) than 1 (with MINP(OVA)), the overall weaker binding of MINP(HRP) was reasonable.
1 (entries 17 and 18), suggesting the proposed boronate binding motif was general. Because fewer boronate bonds are expected for 2 (with MINP(HRP)) than 1 (with MINP(OVA)), the overall weaker binding of MINP(HRP) was reasonable.
To further confirm the proposed mannose–boroxole interactions, we compared the ITC titrations of OVA before and after the treatment with a broad-specificity exoglycosidase, α1-2,3,6 mannosidase (Fig. S28, ESI†). The much weaker interactions for the enzyme-treated sample indicate that the strong binding for the glycoprotein was derived from simultaneous interactions of multiple mannose units on the glycan. After enzymatic cleavage, even though the mannose would stay in the same solution, these molecules could only bind to the MINP individually, apparently much less effectively than when they were clustered on the parent structure.
Fig. 2 shows the binding selectivities of the two MINPs, when they were titrated with a number of glyco- and nonglycoproteins. For MINP(OVA), only HRP and IgG showed noticeable cross-reactivity, likely because (some of the) glycans on these proteins bear certain degrees of resemblance to the OVA glycans (Fig. 1 and Fig. S11, ESI†). For MINP(HRP), OVA and IgG showed higher cross-reactivities than alpha-1-acid glycoprotein (AGP) and transferrin (TF). None of the nonglycosylated proteins, including bovine serum albumin (BSA), lysozyme, trypsin, and cytochrome complex (Cyt C), showed any significant binding toward either MINP.
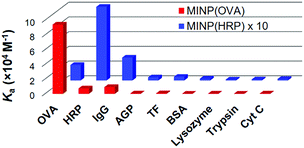 | ||
| Fig. 2 Binding constants of MINP(OVA) and MINP(HRP) for different proteins in 10 mM HEPES buffer (pH 7.4). See Table S1 (ESI†) for the actual binding data. | ||
The strong binding obtained for the glycoprotein allowed us to use the MINP as a “protective group” for the glycan in enzymatic reactions. As shown in Fig. S28 (ESI†), α1-2,3,6 mannosidase degraded the OVA glycans readily. Fig. 3 shows the amounts of mannose released from the glycoprotein when treated with the enzyme. In the presence of 1 equiv. NINP, mannose release slowed down slightly in comparison to the reaction in buffer. The addition of 1 or 2 equiv. MINP(OVA), on the other hand, inhibited the enzymatic cleavage much more strongly over a period of 10 h.
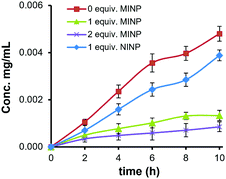 | ||
| Fig. 3 Concentration of D-mannose released from OVA by α1-2,3,6 mannosidase in the presence of different amounts of MINP(OVA) and NINP. | ||
After surface–core double cross-linking, the alkyne-excessive mixed micelle was typically terminated with monoazide 8 for enhanced hydrophilicity and facile purification of the final MINP (Scheme 1). Without the termination, the alkyne-containing intermediate MINPs could be easily clicked onto azide-functionalized magnetic nanoparticles (MNPs), prepared from modified procedures from the literature (Scheme S1, ESI†).36 The resulting MINP(OVA)-MNP composite was then evaluated for selective extraction of the target protein (OVA) from a mixture of four glycoproteins (OVA, HRP, TF, and IgG). The structures of representative glycans for these proteins are shown in Fig. 1 and Fig. S11 (ESI†).
MALDI MS analysis showed the presence of all four proteins in the mixture prior to extraction (Fig. 4a). Note that OVA (MW 45 kDa) and HRP (MW 44 kDa) overlapped due to their similar molecular weights. When MINP(OVA)-MNP was incubated with the protein mixture for 2 h, the remaining solution showed three proteins, with OVA missing (Fig. 4b). When the MINP(OVA)-MNP was rinsed with buffer multiple times, and treated with 100 mM acetic acid to release the bound glycoprotein, the resulting solution showed a dominant OVA peak in MALDI, indicative of successful extraction of the targeted protein (Fig. 4c). In a separate control experiment, we showed that NINP-derived materials could not extract any proteins (Fig. S11, ESI†), consistent with a negligible degree of nonspecific adsorption (Table 1).
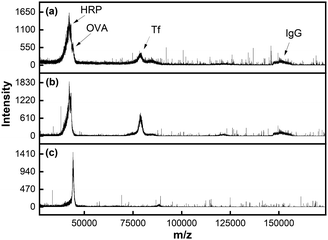 | ||
| Fig. 4 MALDI-TOF MS spectraof (a) a glycoprotein mixture (OVA/HRP/TF/IgG), (b) the remaining solution after MINP(OVA)-MNP extraction, and (c) the released proteins from MINP(OVA)-MNP. | ||
Micellar imprinting is a powerful way to make protein-sized receptors for a variety of biologically interesting molecules including drugs,37 peptides,38 and carbohydrates.30 In this work, imprinting was performed with glycans directly cleaved from glycoprotein to overcome the complexity and microheterogeneity of glycans on glycoproteins. Water-solubility, nanodimension (5 nm), and facile preparation are additional highlights of these synthetic lectins, and can enable applications not possible with macroscopic sugar-binding materials. As highly cross-linked polymeric nanoparticles, MINP tolerate high temperature,28,39 organic solvent,39 and extreme pH.40 In view of the importance of carbohydrates in biomolecular recognition, poor availability of many lectins, and the fragile nature of natural glycan-binding proteins, readily available, robust synthetic lectins can open up new applications in biological and biomedical research.
We thank NIGMS (R01GM113883) for financial support.
Conflicts of interest
There are no conflicts to declare.Notes and references
- M. E. Taylor, K. Drickamer, R. L. Schnaar, M. E. Etzler and A. Varki, in Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar and P. H. Seeberger, Cold Spring Harbor Laboratory Press, Cold Spring Harbor (NY), 2015, pp. 361–372 Search PubMed.
- J. P. Ribeiro and L. K. Mahal, Curr. Opin. Chem. Biol., 2013, 17, 827–831 CrossRef CAS PubMed.
- C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357–2364 CrossRef CAS PubMed.
- D. H. Dube and C. R. Bertozzi, Nat. Rev. Drug Discovery, 2005, 4, 477 CrossRef CAS PubMed.
- J. P. Kamerling and G.-J. Boons, Comprehensive glycoscience: from chemistry to systems biology, Elsevier, Amsterdam, Boston, 1st edn, 2007 Search PubMed.
- B. Wang and G.-J. Boons, Carbohydrate recognition: biological problems, methods, and applications, Wiley, Hoboken, NJ, 2011 Search PubMed.
- S. Jin, Y. Cheng, S. Reid, M. Li and B. Wang, Med. Res. Rev., 2010, 30, 171–257 CAS.
- A. P. Davis and T. D. James, Carbohydrate Receptors, Wiley-VCH, Weinheim, 2005 Search PubMed.
- T. D. James, M. D. Phillips and S. Shinkai, Boronic acids in saccharide recognition, RSC Publishing, Cambridge, 2006 Search PubMed.
- S. Shinde, Z. El-Schich, A. Malakpour, W. Wan, N. Dizeyi, R. Mohammadi, K. Rurack, A. Gjörloff Wingren and B. Sellergren, J. Am. Chem. Soc., 2015, 137, 13908–13912 CrossRef CAS PubMed.
- S. Kunath, M. Panagiotopoulou, J. Maximilien, N. Marchyk, J. Sänger and K. Haupt, Adv. Healthcare Mater., 2015, 4, 1322–1326 CrossRef CAS PubMed.
- Z. Bie, Y. Chen, J. Ye, S. Wang and Z. Liu, Angew. Chem., Int. Ed., 2015, 54, 10211–10215 CrossRef CAS PubMed.
- J. K. Awino, R. W. Gunasekara and Y. Zhao, J. Am. Chem. Soc., 2016, 138, 9759–9762 CrossRef CAS PubMed.
- R. Xing, S. Wang, Z. Bie, H. He and Z. Liu, Nat. Protoc., 2017, 12, 964 CrossRef CAS PubMed.
- L. Jiang, M. E. Messing and L. Ye, ACS Appl. Mater. Interfaces, 2017, 9, 8985–8995 CrossRef CAS PubMed.
- Z. Liu and H. He, Acc. Chem. Res., 2017, 50, 2185–2193 CrossRef CAS PubMed.
- G. Wulff and W. Vesper, J. Chromatogr., 1978, 167, 171–186 CrossRef CAS.
- G. Wulff, Angew. Chem., Int. Ed. Engl., 1995, 34, 1812–1832 CrossRef CAS.
- D. J. Harvey, D. R. Wing, B. Kuster and I. B. Wilson, J. Am. Soc. Mass Spectrom, 2000, 11, 564–571 CrossRef CAS PubMed.
- B. Y. Yang, J. S. Gray and R. Montgomery, Carbohydr. Res., 1996, 287, 203–212 CrossRef CAS PubMed.
- K. T. Kim, J. J. L. M. Cornelissen, R. J. M. Nolte and J. C. M. van Hest, J. Am. Chem. Soc., 2009, 131, 13908–13909 CrossRef CAS PubMed.
- A. Pal, M. Bérubé and D. G. Hall, Angew. Chem., Int. Ed., 2010, 49, 1492–1495 CrossRef CAS PubMed.
- X. Wu, Z. Li, X.-X. Chen, J. S. Fossey, T. D. James and Y.-B. Jiang, Chem. Soc. Rev., 2013, 42, 8032–8048 RSC.
- S. D. Bull, M. G. Davidson, J. M. H. Van den Elsen, J. S. Fossey, A. T. A. Jenkins, Y. B. Jiang, Y. Kubo, F. Marken, K. Sakurai, J. Z. Zhao and T. D. James, Acc. Chem. Res., 2013, 46, 312–326 CrossRef CAS PubMed.
- H. Kim, Y. J. Kang, S. Kang and K. T. Kim, J. Am. Chem. Soc., 2012, 134, 4030–4033 CrossRef CAS PubMed.
- M. Dowlut and D. G. Hall, J. Am. Chem. Soc., 2006, 128, 4226–4227 CrossRef CAS PubMed.
- M. Bérubé, M. Dowlut and D. G. Hall, J. Org. Chem., 2008, 73, 6471–6479 CrossRef.
- J. K. Awino and Y. Zhao, J. Am. Chem. Soc., 2013, 135, 12552–12555 CrossRef CAS PubMed.
- X. Z. Song, H. Ju, Y. Lasanajak, M. R. Kudelka, D. F. Smith and R. D. Cummings, Nat. Methods, 2016, 13, 528–534 CrossRef CAS PubMed.
- R. W. Gunasekara and Y. Zhao, J. Am. Chem. Soc., 2017, 139, 829–835 CrossRef CAS PubMed.
- D. K. Mandal, N. Kishore and C. F. Brewer, Biochemistry, 1994, 33, 1149–1156 CrossRef CAS PubMed.
- K. Chen and Y. Zhao, Org. Biomol. Chem., 2019, 17, 8611–8617 RSC.
- S. Fa and Y. Zhao, Chem. Mater., 2019, 31, 4889–4896 CrossRef CAS.
- L. Duan and Y. Zhao, J. Org. Chem., 2019, 84, 13457–13464 CrossRef CAS PubMed.
- Because the MINP mainly relies on the boroxole functional monomer for binding, most of the binding is expected to be derived from the interactions with the terminal sugar residues. Even though hydrogen bonds between the sugar and MINP (ref. 30) could contribute to the binding, uncomplexed hydroxyls are strongly solvated by water and, in turn, poorly imprinted.
- X. Liu, Z. Ma, J. Xing and H. Liu, J. Magn. Magn. Mater., 2004, 270, 1–6 CrossRef CAS.
- J. K. Awino and Y. Zhao, ACS Biomater. Sci. Eng., 2015, 1, 425–430 CrossRef CAS.
- J. K. Awino, R. W. Gunasekara and Y. Zhao, J. Am. Chem. Soc., 2017, 139, 2188–2191 CrossRef CAS PubMed.
- X. Xing and Y. Zhao, Org. Biomol. Chem., 2018, 16, 2855–2859 RSC.
- X. Xing and Y. Zhao, New J. Chem., 2018, 42, 9377–9380 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, ITC titration curves, and spectroscopic data. See DOI: 10.1039/d0cc02892d |
| This journal is © The Royal Society of Chemistry 2020 |