Open Access Article
Open Access ArticleDynamic modulation of proximity-induced enzyme activity using supramolecular polymers†
Eva
Magdalena Estirado‡
a,
Bas J. H. M.
Rosier‡
 a,
Tom F. A.
de Greef
a,
Tom F. A.
de Greef
 abc and
Luc
Brunsveld
abc and
Luc
Brunsveld
 *a
*a
aLaboratory of Chemical Biology and Institute for Complex Molecular Systems, Eindhoven University of Technology, P.O. Box 513, 5600 MB Eindhoven, The Netherlands. E-mail: L.brunsveld@tue.nl
bComputational Biology Group, Department of Biomedical Engineering, Eindhoven University of Technology, P.O. Box 513, 5600 MB Eindhoven, The Netherlands
cInstitute for Molecules and Materials, Radboud University, Nijmegen, The Netherlands
First published on 14th April 2020
Abstract
Synthetic supramolecular polymers are used as dynamic nanoscaffolds for the activation of the apoptotic signalling enzyme caspase-9. Recruitment of caspase-9 to the nanoscaffold results in an increase in enzymatic activity due to enhanced proximity, with a bell-shaped response as a function of nanoscaffold concentration. The modularity of the system allows for dynamic regulation of enzyme activity through variation of the recruitment-motif density along the supramolecular polymer.
Spatial and temporal coordination of biomolecular components are of key importance for efficient intracellular communication and faithful signal transduction.1,2 Stable organization of functionally related components occurs on the microscale through vesicle-based compartmentalization,3 or on the nanoscale using dedicated scaffold proteins that assemble enzymes into signalling cascades.4 Transient organization of components based on non-covalent interactions allows the living cell to finely tune regulatory behaviours with examples including rapidly exchanging membraneless organelles,5,6 open-ended receptor clustering on lipid membranes,7,8 and dynamic rearrangement of cytoskeletal polymers.9
Synthetic supramolecular polymers are similarly based on non-covalent interactions and could therefore be used to mimic specific features of transient cellular organization in a controlled setting.10–12 Examples include the assembly of quasi-one-dimensional fibrillar aggregates with tunable kinetic characteristics, hierarchical assemblies, and branched networks such as hydrogels and coacervates.13–15 In addition, supramolecular polymers can be used as modular templates for the assembly of various biomolecules, including nanoparticles,16,17 nucleic acids,18,19 and proteins.20–23
Recent work has revealed that intracellular signalling pathways heavily rely on the dynamic assembly of higher-order fibril-like aggregates that are structurally well-defined but open-ended and sometimes quasi-infinite.24,25 As such, these assemblies function as platforms for the transient recruitment of signalling proteins that undergo proximity-induced activation,26,27 and subsequently elicit immune or inflammatory responses, or apoptosis. The nature of the non-covalent interactions involved in these assemblies allows for nucleated growth resulting in signal amplification,26 the appearance of threshold concentrations and cooperative incorporation of other enzymes.28 Pathway inhibition can be dynamically regulated by co-assembly of non-functional fiber monomers.29 Bottom-up construction of synthetic systems mimicking these natural systems has remained very difficult, especially because of the combined need for synthetically accessible systems with physical chemical properties, regarding assembly and dynamics, matching the biological systems at hand.
Here, we present a supramolecular polymeric nanoscaffold as a synthetic model system for the dynamic recruitment, organization and activation of the signalling enzyme caspase-9 (cysteine-dependent aspartic protease) as model system. The synthetic recruitment nanoscaffold consists of amphiphilic monomers made of bis-pyridine-based C3-symmetrical molecules decorated with single-stranded DNA handles (DNA-Disc, Fig. 1a).19 The hybrid DNA-decorated monomers self-assemble in water into columnar assemblies, displaying multiple copies of DNA recruitment handles (Fig. 1b).30 The handles are accessible for interacting through DNA-duplex formation with complementary DNA anti-handle strands located at the enzyme of interest, in our case caspase-9 (Fig. 1b). Full-length caspase-9 consists of a catalytic domain responsible for its protease activity, and a caspase recruitment domain (CARD) that, by interacting with the apoptosome, brings multiple caspases together to initiate proximity-induced activation.31–33 In the present system, the CARD domain has been replaced by a DNA strand, that recruits caspase-9 onto the nanoscaffold through DNA-duplex formation.34 The catalytic domain was expressed with an N-terminal non-canonical amino acid (p-azidophenylalanine), introduced by using amber codon suppression in E. coli, in order to site-specifically decorate the protein with the DNA anti-handle (ESI†).34 The DNA contains a 5′-end nucleotide linker between the enzyme and the recruitment recognition sequence (anti-handle) in order to avoid undesired interactions with the nanoscaffold. A detailed quantitative analysis has previously demonstrated that the kinetic characteristics of the caspase-9 enzyme–DNA conjugate is similar to wildtype caspase-9.34
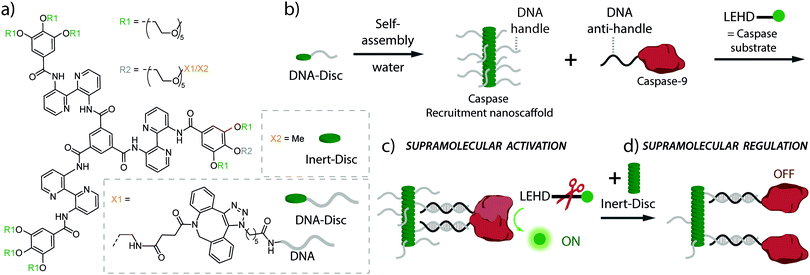 | ||
| Fig. 1 (a) Chemical structure of DNA-decorated discotic monomers (DNA-Disc) and unmodified discotics (Inert-Disc). (b) Self-assembly of the monomers in water results in columnar stacks, effectively displaying DNA handle strands. In the cartoon, DNA handles were partially omitted for clarity. (c) Hybridization between the DNA handles on the nanoscaffold and complementary DNA anti-handle strands located at the caspase-9, result in the recruitment of multiple caspases on the nanoscaffold, enhancing caspase-9 activity due to increased inter-enzyme proximity. While the natural substrate for caspase-9 is caspase-3, enzymatic activity is typically examined by use of the synthetic fluorogenic substrate Ac-LEHD-AFC (N-acetyl-Leu-Glu-His-Asp-7-amino-4-(trifluoromethyl)coumarin) that becomes fluorescent upon enzymatic cleavage.31 (d) The modularity of the system allows easy tuning of the nanoscaffold composition by the addition of Inert-Disc that intermixes with the DNA-Disc, resulting in stacks with varying DNA handle densities. This variation in handle density allows the supramolecular regulation of enzymatic activity by the increase in distance between enzymes. | ||
The modularity and dynamic nature of monomer intermixing in the supramolecular nanoscaffold allows straightforward tuning of the nanoscaffold characteristics, such as the systematic variation of the recruiter handle density in the nanoscaffold (Fig. 1c and d), which potentially regulates enzymatic activity through the variation of inter-enzyme distances, as seen in natural systems.
First, the ability of the supramolecular nanoscaffold to recruit caspase-9 was characterized. A series of control experiments were carried out to rule out possible unspecific interactions between the components in the system. A constant concentration of caspase-9 (25 nM, well below its dimerization affinity34) was incubated with non-DNA-functionalized monomers (Inert-Disc) at different concentrations (Fig. S1, ESI†). Upon the addition of Ac-LEHD-AFC substrate, the fluorescence intensity of the samples was monitored over time and the enzymatic activity was calculated using the initial slope of the fluorescence curves (monitored between the first 20 to 60 minutes after addition of the fluorogenic substrate, unless stated otherwise). The enzymatic activity (Fig. S1, ESI†) exhibited similar background levels when the experiment was performed in the presence and absence of the Inert-Disc, indicating the lack of interference between the supramolecular nanoscaffold and the substrate or caspase-9.
Next, the active assembly to the supramolecular nanoscaffold and resultant proximity-induced caspase-9 activation was analysed. Therefore, nanoscaffolds were assembled using a series of concentrations of DNA-Disc ranging from 0 to 500 nM, and subsequently incubating with caspase-9 for 2 hours at 4 °C, after which substrate was added to the sample. Fluorescence intensity traces at the different DNA-Disc concentrations are shown in Fig. S2 (ESI†), revealing that the presence of supramolecular recruitment nanoscaffold effectively enhances proteolytic cleavage, in comparison with the background activity of non-scaffolded enzymes. The enzymatic activities derived from the intensity traces for the entire concentration range (see Fig. 2a) showed a step-wise enhancement in enzyme activity with increasing concentration of DNA-Disc. Optimal enzyme activity was reached at equimolar amounts of both caspase-9 and DNA-Disc. Importantly, the nearly 5-fold increase in enzyme activity seen at 20 nM of DNA-Disc agrees with the enhancement seen in previous studies where caspase-9 was dimerized on a DNA origami-based nanoscaffold at roughly the same caspase-9 concentration,34 or with other in vitro systems that typically do not reach the activity of apoptosome activated caspase-9.31–33,35
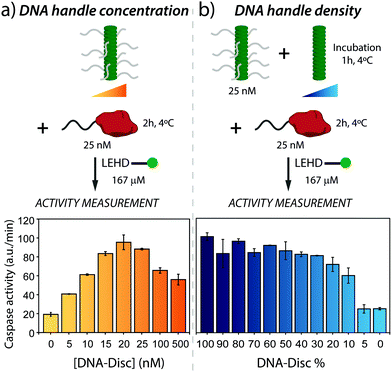 | ||
| Fig. 2 (a) Effect of nanoscaffold concentration on the enzymatic activity of caspase-9. Enzymatic activity increases with DNA-Disc concentration up to a 5-fold at equimolar concentrations of enzyme and DNA-Disc. At high nanoscaffold densities, combinatorial inhibition leads to a decrease in enzymatic activity. Experiments were conducted with supramolecular nanoscaffolds containing 100% DNA-Disc. In the cartoon, DNA handles were partially omitted for clarity. (b) Effect on caspase-9 activity of the variation in DNA handle density displayed by the nanoscaffold. Varying DNA handle densities is achieved by keeping the total concentration of DNA-Disc at 25 nM, and adding varying amounts of Inert-Disc, resulting in nanoscaffolds in which the DNA handle density varies between 100 and 0%. In both graphs, the enzyme activity is based on the initial slope of the kinetic traces (Fig. S2 and S3, ESI†). The error bars represent the standard deviation of the measurements performed in triplicates. | ||
A further increase in DNA-Disc concentration, beyond the optimum, diminished enzyme activity (Fig. 2a), which can be attributed to combinatorial inhibition.36,37 That is, suprastoichiometric levels of DNA-decorated supramolecular monomers promote sequestration of caspase-9 on the nanoscaffold in the inactive monomeric state, and therefore reduce proximity-induced formation of enzymatically active dimers. Note that full suppression of the activity to background levels was not observed, even at high DNA-Disc concentrations (500 nM). The intrinsic affinity between caspase-9 monomers (KD of dimerization of around 50 μM)38 in combination with a high effective concentration on the nanoscaffold could lead to a stabilization of the active caspase-9 dimer complex and therefore explain the enhanced enzymatic activity at high DNA-Disc concentrations.
Next, we evaluated the effect of changing the DNA handle density on the nanoscaffold on enzymatic activity. This can be achieved in a straightforward fashion by co-assembly of Inert-Disc and DNA-Disc, which results in homogeneous intermixing of both monomers along the supramolecular polymer, as previously shown.19 To this end, DNA-Disc at a constant concentration of 25 nM was co-assembled with varying concentrations of Inert-Disc (from 0 to 500 nM), leading to nanoscaffolds with handle densities ranging from 100 to 5% (Fig. 2b). The samples were incubated for one hour to ensure full co-assembly, followed by incubation with 25 nM of caspase-9 at 4 °C for 2 hours. After addition of enzyme substrate, the fluorescence was monitored over time (Fig. S3, ESI†) and the activity was determined. The control experiment with non-scaffolded caspase-9 showed typical values for background activity (0% DNA-Disc, Fig. 2b).
The samples featuring DNA handles at different densities showed a non-linear variation of enzymatic activity. A decrease from 100 to 50% in handle density did not have a significant effect on activity, however, activity gradually diminished between 50 to 10%, and dropped sharply to background activity levels after 10% (Fig. 2b). To gain insights into these results, it is useful to estimate the expected distance between DNA handles for a given DNA handle density. The distance between the monomer cores forming the supramolecular nanoscaffold has been estimated to be 0.34 nm.39 Assuming homogeneous co-assembly of Inert-Disc and DNA-Disc, the distance between DNA handles is ≤3.4 nm for nanoscaffolds containing ≥10% DNA-Disc. Given previous work that demonstrated markedly enhanced activity at a DNA handle separation of 6 nm,34 we conclude that the handle density is sufficiently high to enable efficient caspase-9 dimerization across the range of nanoscaffolds containing ≥20% DNA-Disc, explaining the constant enzyme activity in the first part of the graph in Fig. 2b. In nanoscaffolds with 5% DNA-Disc the average inter-handle distance is 6.8 nm, which does not result in enzymatic activity. An explanation can be that, compared to the DNA origami-based system which employs a fixed two-dimensional scaffold, the columnar supramolecular nanoscaffold displays DNA handles in three dimensions, and therefore might lower effective caspase-9 dimerization at this DNA-Disc concentration. These experiments demonstrate that co-assembly with high concentrations of Inert-Disc affect enzymatic activity by lowering the DNA handle density on the supramolecular nanoscaffold.
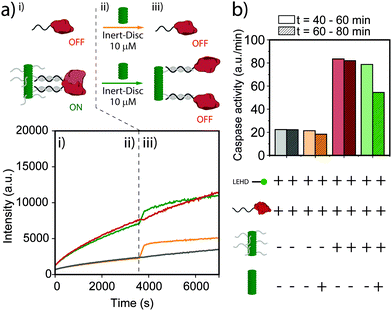
Finally, we explored dynamic regulation of enzymatic activity by addition of Inert-Disc after equilibration of DNA-functionalized nanoscaffolds. The enzyme activity of an equimolar solution at 25 nM of DNA-Disc and caspase-9 was followed over time in the presence of substrate (Fig. 3a). After approximately 60 minutes, a solution of Inert-Disc was added to a final concentration of 10 μM. Note that the instant increase of the fluorescence for both the non-scaffolded and scaffolded caspase-9 (Fig. 3a, green and orange curves) is a consequence of the characteristic absorption peak at 350 nm and emission peak at 520 nm of the nanoscaffold in the aggregated state, which does not affect the slope of the curve (enzyme activity). The histogram in Fig. 3b represents the enzymatic activity extracted from Fig. 3a before and after the addition of the Inert-Disc. The addition of Inert-Disc to the non-scaffolded caspase-9 showed a negligible effect on the enzyme activity. In contrast, the scaffolded version displayed a decrease in enzyme activity of roughly 35%. The intrinsic affinity between caspase-9 monomers could explain the residual enzymatic activity after the addition of Inert-Disc, similar to the observations in Fig. 2. These results indicate the potential of dynamic supramolecular polymers to regulate enzymatic activity by simple addition of a non-functional monomer, which results in an increased distance between the individual enzymes and a concomitant decrease in proximity-induced dimerization.
In this communication, we mimic proximity-induced activation of enzymes with a fully synthetic and programmable nanoscaffold, making use of DNA-decorated supramolecular polymers and engineered signalling proteins (caspase-9). Tethering of enzymes on the nanoscaffold via hybridization between their complementary DNA handle strands resulted in their activation due to proximity-induced dimerization. The extent of enzyme activity could be modulated by varying the concentration of DNA-functionalized monomers, resulting in a bell-shaped activity response with a 500% increase in activity at optimal equimolar concentrations of enzyme and supramolecular monomer. Moreover, the enzymatic activity could be inhibited by decreasing the average density of DNA handles displayed on the nanoscaffold, achieved by co-assembly of non-functionalized monomers either a priori or dynamically after equilibration.
Taken together, these results provide insights into the potential of the implementation of supramolecular polymers as general recruitment nanoscaffolds for the distance-dependent activation of signalling proteins. The modularity of the supramolecular polymer and the DNA element, together with their befitting assembly characteristics, opens many possibilities for the recruitment of other proteins of interest and the development of programmable enzyme assembling systems.
This work was supported by The Netherlands Organization for Scientific Research (NWO) through Gravity Program 024.001.035, and through the European Union's program Horizon 2020 Marie Curie Innovative Training Network MULTI APP (2015-2019).
Conflicts of interest
There are no conflicts of interest to declare.Notes and references
- R. P. Bhattacharyya, A. Reményi, B. J. Yeh and W. A. Lim, Annu. Rev. Biochem., 2006, 75, 655–680 CrossRef CAS PubMed.
- B. N. Kholodenko, J. F. Hancock and W. Kolch, Nat. Rev. Mol. Cell Biol., 2010, 11, 414–426 CrossRef CAS PubMed.
- A. H. Chen and P. A. Silver, Trends Cell Biol., 2012, 22, 662–670 CrossRef CAS PubMed.
- M. C. Good, J. G. Zalatan and W. A. Lim, Science, 2011, 332, 680–686 CrossRef CAS PubMed.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS PubMed.
- Y. Shin and C. P. Brangwynne, Science, 2017, 357, eaaf4382 CrossRef PubMed.
- N. C. Hartman and J. T. Groves, Curr. Opin. Cell Biol., 2011, 23, 370–376 CrossRef CAS PubMed.
- J. B. Casaletto and A. I. McClatchey, Nat. Rev. Cancer, 2012, 12, 387–400 CrossRef CAS PubMed.
- D. A. Fletcher and R. D. Mullins, Nature, 2010, 463, 485–492 CrossRef CAS PubMed.
- K. Petkau-Milroy, M. H. Sonntag and L. Brunsveld, Chem. – Eur. J., 2013, 19, 10786–10793 CrossRef CAS PubMed.
- R. Freeman, J. Boekhoven, M. B. Dickerson, R. R. Naik and S. I. Stupp, MRS Bull., 2015, 40, 1089–1100 CrossRef CAS PubMed.
- O. J. G. M. Goor, S. I. S. Hendrikse, P. Y. W. Dankers and E. W. Meijer, Chem. Soc. Rev., 2017, 46, 6621–6637 RSC.
- S. Van Dun, C. Ottmann, L. G. Milroy and L. Brunsveld, J. Am. Chem. Soc., 2017, 139, 13960–13968 CrossRef CAS PubMed.
- C. Facciotti, V. Saggiomo, A. Bunschoten, R. Fokkink, J. B. Ten Hove, J. Wang and A. H. Velders, Soft Matter, 2018, 14, 9542–9549 RSC.
- S. Hirayama, K. Oohora, T. Uchihashi and T. Hayashi, J. Am. Chem. Soc., 2020, 142, 1822–1831 CrossRef CAS PubMed.
- S. H. Jung, J. Jeon, H. Kim, J. Jaworski and J. H. Jung, J. Am. Chem. Soc., 2014, 136, 6446–6452 CrossRef CAS PubMed.
- Y. Vyborna, M. Vybornyi and R. Häner, Chem. Commun., 2017, 53, 5179–5181 RSC.
- S. P. W. Wijnands, E. W. Meijer and M. Merkx, Bioconjugate Chem., 2019, 30, 1905–1914 CrossRef CAS PubMed.
- M. Á. Alemán García, E. Magdalena Estirado, L. G. Milroy and L. Brunsveld, Angew. Chem., Int. Ed., 2018, 57, 4976–4980 CrossRef PubMed.
- Q. Luo, Z. Dong, C. Hou and J. Liu, Chem. Commun., 2014, 50, 9997–10007 RSC.
- M. K. Müller, K. Petkau and L. Brunsveld, Chem. Commun., 2011, 47, 310–312 RSC.
- K. Petkau-Milroy, D. A. Uhlenheuer, A. J. H. Spiering, J. A. J. M. Vekemans and L. Brunsveld, Chem. Sci., 2013, 4, 2886–2891 RSC.
- S. P. W. Wijnands, W. Engelen, R. P. M. Lafleur, E. W. Meijer and M. Merkx, Nat. Commun., 2018, 9, 65 CrossRef PubMed.
- J. C. Kagan, V. G. Magupalli and H. Wu, Nat. Rev. Immunol., 2014, 14, 821–826 CrossRef CAS PubMed.
- M. Bienz, Trends Biochem. Sci., 2014, 39, 487–495 CrossRef CAS PubMed.
- Q. Qiao, C. Yang, C. Zheng, L. Fontán, L. David, X. Yu, C. Bracken, M. Rosen, A. Melnick, E. H. Egelman and H. Wu, Mol. Cell, 2013, 51, 766–779 CrossRef CAS PubMed.
- Y. Tan and J. C. Kagan, Cell, 2019, 177, 384–398 CrossRef CAS PubMed .e11.
- L. David, Y. Li, J. Ma, E. Garner, X. Zhang and H. Wu, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 1499–1504 CrossRef CAS PubMed.
- T.-M. Fu, Y. Li, A. Lu, Z. Li, P. R. Vajjhala, A. C. Cruz, D. B. Srivastava, F. DiMaio, P. A. Penczek, R. M. Siegel, K. J. Stacey, E. H. Egelman and H. Wu, Mol. Cell, 2016, 64, 236–250 CrossRef CAS PubMed.
- E. Magdalena Estirado, M. A. Aleman Garcia, J. Schill and L. Brunsveld, J. Am. Chem. Soc., 2019, 141, 18030–18037 CrossRef CAS PubMed.
- M. Renatus, H. R. Stennicke, F. L. Scott, R. C. Liddington and G. S. Salvesen, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 14250–14255 CrossRef CAS PubMed.
- Y. Chao, E. N. Shiozaki, S. M. Srinivasula, D. J. Rigotti, R. Fairman and Y. Shi, PLoS Biol., 2005, 3, e183 CrossRef PubMed.
- Y. Li, M. Zhou, Q. Hu, X. Bai, W. Huang, S. H. W. Scheres and Y. Shi, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 1542–1547 CrossRef CAS PubMed.
- B. J. H. M. Rosier, A. J. Markvoort, B. Gumí Audenis, J. A. L. Roodhuizen, A. den Hamer, L. Brunsveld and T. F. A. de Greef, Nat. Catal., 2020, 3, 295–306 CrossRef CAS PubMed.
- D. T. Dang, H. D. Nguyen, M. Merkx and L. Brunsveld, Angew. Chem., Int. Ed., 2013, 52, 2915–2919 CrossRef CAS PubMed.
- A. Levchenko, J. Bruck and P. W. Sternberg, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 5818–5823 CrossRef CAS PubMed.
- J. J. McCann, U. B. Choi and M. E. Bowen, Structure, 2014, 22, 1458–1466 CrossRef CAS PubMed.
- S. J. Riedl and G. S. Salvesen, Nat. Rev. Mol. Cell Biol., 2007, 8, 405–413 CrossRef CAS PubMed.
- T. Metzroth, A. Hoffmann, R. Martín-Rapún, M. M. J. Smulders, K. Pieterse, A. R. A. Palmans, J. A. J. M. Vekemans, E. W. Meijer, H. W. Spiess and J. Gauss, Chem. Sci., 2011, 2, 69–76 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details and supporting figures. See DOI: 10.1039/d0cc02120b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |