Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Carbohydrate supramolecular chemistry: beyond the multivalent effect
Manuel
González-Cuesta
a,
Carmen
Ortiz Mellet
 *a and
José M.
García Fernández
*a and
José M.
García Fernández
 *b
*b
aDepartamento de Química Orgánica, Facultad de Química, Universidad de Sevilla, Profesor García González 1, Sevilla 41012, Spain. E-mail: mellet@us.es
bInstituto de Investigaciones Químicas (IIQ), CSIC – Universidad de Sevilla, Avda. Américo Vespucio 49, 41092 Sevilla, Spain. E-mail: jogarcia@iiq.csic.es
First published on 8th April 2020
Abstract
It has been amply constated that sugar ligand multivalency increases lectin-binding avidities dramatically, thereby modulating the capacity of carbohydrates to participate in supramolecular recognition processes involving transfer of biological information. The importance of this concept, the multivalent or glycoside cluster effect, in cell biology in general and in the glycosciences in particular is reflected in the ever-growing number of papers in the field. An impressive range of glycoarchitectures has been conceived to imitate the glycan coating of cells (the glycocalyx) in order to target complementary lectin receptors. However, these models rarely address the heterogeneity and the fluidity of the densely glycosylated cell membrane. They also disregard the impact that high-density nanosized arrangements could have in their interactions with the whole spectrum of carbohydrate-interacting proteins, among which glycosidases are notable representatives. For many years it has been tacitly assumed that: (i) efficient recognition by lectins generally requires high densities of the putative primary ligand and (ii) the mechanisms governing binding of a carbohydrate motif by a lectin or a glycosidase are totally disparate. Notwithstanding, an increasing amount of evidence seriously questions this paradigm. First, it was shown that secondary “innocent” ligands can play important roles in the recognition of heteroglycocluster constructs by lectins through synergistic or antagonistic contributions, a phenomenon termed the heterocluster effect. Second, the existence of multivalent effects in the inhibition of certain glycosidases by glycomimetic- and, even more disturbing, glyco-coated architectures (multivalent enzyme inhibition) was demonstrated. These observations call for a generalized multivalent effect governing the supramolecular chemistry of carbohydrate or glycomimetic structures in a biological context, with (hetero)multivalency acting as a multimodal switcher to drive the encoded information through different pathways. In this Feature Article we review the advancements made in the last few years in our understanding of the mechanisms underpinning the generalized multivalent effect, with an emphasis on the potential risks and opportunities derived from (hetero)multivalency-elicited promiscuity.
Introduction
The mutual recognition of different chemical entities is a fascinating phenomenon at the core of fundamental processes in life. The co-assembly of viral RNA and protein capsids to bring about functional viruses, the infection of a host cell, the immune response and the metastasis of tumors are examples of critical events determined by the reciprocal identification of specific biomolecular partners in a highly complex dynamic environment. Exquisite matching relationships are achieved through the complementary 3D disposition of functional hotspots in the intervening species, enabling the establishment of a network of noncovalent interactions that stabilizes a transient supramolecular body. The whole sequence can be seen as a decrypting process whereby precisely encoded information is transferred, leading to the activation or deactivation of specific signaling pathways. For the communication flow to occur, a minimum time of contact between the participating ligand and receptor units is necessary, which in turn requires reaching a “biologically useful” binding affinity threshold. In some cases, this is warranted by the one-to-one complementation of the parties leading to a “lock-and-key” fitting mode. A typical example is the formation of antibody–antigen complexes. In other cases, however, the interactions engaged in the pairing episode are intrinsically too weak to reach effective ligand–host dissociation constants and require an amplification contrivance. In nature this is achieved by deploying several replicates of one or the two components so that they can interrelate in a cooperative manner. The system as a whole then behaves differently from anticipations based on the individual interactions acting in isolation, resulting in a net increase in the lifetime of the bound state. The accumulated strength resulting from individual affinities performing concurrently is known as avidity, whereas the general strategy is called multivalency.1–4The recognition of carbohydrates by protein receptors (lectins) relies on multivalency much more intensely than processes implying any other type of biomolecule. Sets of glycan recognition motifs (glycotopes) and cognate lectins possessing several carbohydrate recognition domains (CRDs) have typically to play concertedly in order to afford productive associations.5–8 In this manner, individually weak affinities are enhanced by orders of magnitude, an observation termed the multivalent or glycoside cluster effect. Since its enunciation in 1995,9 the multivalent effect has dominated carbohydrate supramolecular chemistry, delivering not only a general principle that has led to decisive advances in our understanding of glycobiology but also a versatile tactic to interfere in carbohydrate-mediated processes for fundamental studies or biomedical applications.10–13 The advent of “click” chemistry and the implementation of “precision macromolecular synthesis” concepts, together with the developments in nanotechnology and self-assembly, have further provided a fantastic toolbox to sculpt the topology of glycoobjects and, eventually, incorporate refined functional properties.14–32 Collectively, this vast work has contributed to settle the assumption that presenting a given glycotope in multiple copies on a suitable scaffold is a safe and efficacious way to target a complementary lectin, preserving or even enhancing binding selectivity.
While the success of the above notion is attested by many studies, a lot of evidence questions this one-dimensional picture. For instance, the recognition of glycans by complementary lectins has been found to be strongly dependent on density and architectural parameters.33–35 The biological information encoded by carbohydrates, the “glycocode”,36,37 does not seem to be written in the monosaccharide sequences forming the oligosaccharide chains, but in much more complex glycan patterns that are read by lectins behaving as pattern recognition proteins.38,39 The heterogeneity and the fluidity of the densely glycosylated cell membrane, the glycocalyx, contribute decisively to achieve the right arrangement for a perfect multi-spot match in a precise context.40 Importantly, a change in the glycocalyx configuration, e.g. in response to a different cell state, can promote dissimilar recognition processes while involving essentially the same glycotopes.41 Heterogeneity and promiscuity in multivalent carbohydrate recognition are indeed implicit in the glycome and play essential roles in adapting cellular responsiveness to environmental conditions.42 The complexity of developing suitable models and analytical tools to investigate these phenomena and exploit them in drug design represents a daunting challenge. Nevertheless, the last few years have witnessed a series of discoveries that have the potential to provoke a drastic revolution in our conceptualization of multivalency and convey the necessary instruments to move ahead. First, the synthesis of topologically defined high-density heteroglycoclusters and their evaluation as lectin ligands revealed unsuspected synergies between putative and non-cognate glycotopes. Next, a number of carbohydrate processing enzymes, namely glycosidases and glycosyl transferases, were found to be responsive to multivalent presentations of carbohydrate analogues (glycomimetics) in a way somehow reminiscent of carbohydrate–lectin recognition. Then, (hetero)multivalent glycomimetic or glycoligand and hybrid glycomimetic/glycoligand systems capable of triggering simultaneous recognition/inhibition of lectin/glycosidase pools were reported. This new scenario prompted the advancement of the “generalized multivalent effect”,43 a shift in paradigm whereby (hetero)multivalency is perceived as an action principle with the capability to induce, deter or enhance specific recognition phenomena towards a spectrum of lectins and carbohydrate processing enzymes in a multimodal manner. In this Feature Article we track the latest developments in this effervescent field, contextualizing our own results. We purposely focus on contributions that shed some light on the molecular basis behind the experimental observations, supply evidence of the relevance of the generalized multivalent effect in a biological context and explore the new channels that it opens in glycobiology and biomedicine. For a broader view of the state-of-the-art in carbohydrate supramolecular chemistry, including biomolecular recognition, self-assembly and host–guest carbohydrate chemistry, the reader is advised to consult more comprehensive or specialized reviews and monographs.44–51
Carbohydrate–lectin interactions in heterogeneous environments: the heteromultivalent effect
Imitating the densely glycosylated cell membrane to correctly evaluate glycan interactions and their biological consequences has been a constant motivation for research in the field of multivalency.52 Intuitively, the presence of a diverse ensemble of glycotopes in heteromultivalent glycosystems has the potential to enable simultaneous interactions with distant areas in the same lectin, e.g. subsites hosting different monosaccharide residues of an oligosaccharide ligand in the corresponding supramolecular complex, or with an arrangement of lectins with assorted specificities. The first vision was initially explored by Kobayashi and coworkers and systematized as the “carbohydrate module method”.53–55 Basically, it consists of deconstructing the glycan ligand whose lectin recognition properties are to be emulated into elemental motifs that are copolymerized together. Provided that the fragments in the side chain of the resulting synthetic heteroglycopolymers can act cooperatively with each other, enhanced affinities against the target lectin can be achieved (Fig. 1).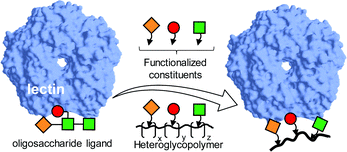 | ||
| Fig. 1 Schematic representation of the carbohydrate module method for the preparation of heteroglycopolymers mimicking oligosaccharide lectin ligands devised by Kobayashi and coworkers. The elemental oligosaccharide motifs are represented by geometrical (square, circle, diamond) forms.53–55 | ||
The simplicity of the carbohydrate module method makes it very appealing, but the successful implementation remains challenging. It presupposes that the individual sugar moieties in the heteroglycopolymer will bind at the sites in the target lectin where they are found in the complex between the lectin and the full oligosaccharide ligand that inspired the design, giving rise to allosteric cooperativity.56 Yet, the effectiveness of this mechanism is sometimes questionable.57 Moreover, it is not always clear which modules in a natural glycan are essential for protein recognition: some are directly involved in binding and others are required for determining the right conformation. Underestimating the latter can totally offset the enthalpic benefit of cooperativity due to the entropic penalty associated with linker flexibility.58,59 Recently, Zentel and coworkers60 and Tacke and coworkers61 conveyed compelling evidence of the potential of mimicking the selectin recognition properties of the sialyl Lewisx (SLex) tetrasaccharide (NeuNAc-α(2–3)-Gal-β(1–4)-[Fuc-α(1–3)]-GlcNAc) by the carbohydrate module approach for biomedical applications. Selectins are a family of cell adhesion molecules with an extracellular lectin domain that bind to fucosylated and sialylated glycoproteins and play a key role in the innate immune response.62–64 SLex is the common structure required for binding in several natural glycoproteins that behave as ligands of the selectins in the inflammatory cascade, but the monovalent tetrasaccharide itself has rather low affinities to all selectin members. The authors used a biocompatible poly(2-hydroxypropyl)methacrylamide (PHMA) backbone to multivalently and randomly present the fucose (Fuc), galactose (Gal) and neuraminic acid (NeuAc) substructures, bearing or not a sulfated tyramide sidechain to account for sulfation of some natural selectin glycoprotein ligands at tyrosine residues (Fig. 2). Both heteroglycopolymers behaved as selectin binders in vitro in different cells.60 Additionally, the latter heteroglycopolymer strongly bound to resident liver macrophages in vivo (mice) and meaningfully inhibited toxic liver injury and reduced the injury in a model of immune-mediated hepatitis.61
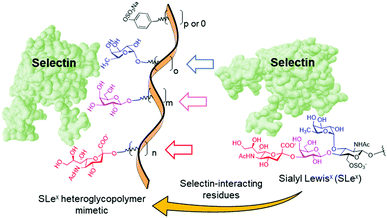 | ||
| Fig. 2 Structure of the sialyl Lewisx (SLex) tetrasaccharide and schematic representation of the heteroglycopolymers conceived by Zentel and coworkers60 and Tacke and coworkers61 to emulate the SLex selectin binding capabilities. | ||
Heteromultivalent prototypes having the crucial glycotopes prearranged in segregated domains are expected to be better suited to simultaneously target lectins differing in their sugar selectivities. This concept has been probed by the groups of Roy and coworkers and Renaudet and coworkers with Janus65 (Fig. 3A) and heterolayered hybrid glycodendrimers66 (Fig. 3B) or regioselectively addressable functionalized template (RAFT) cyclopeptide scaffolded heteroglycoclusters (Fig. 3C),67,68 respectively. Multiconjugates combining sugar head groups optimized for the Pseudomonas aeruginosa (P. aeruginosa) lectins LecA and LecB (namely Gal and Fuc) or LecA and concanavalin A (ConA), a model mannose (Man)-binding lectin used throughout many fundamental studies, were synthesized and evaluated. Differently from the carbohydrate module approach, multidomain heterogeneous prototypes are intended to achieve multispecificity, which may be delicate if the target lectins have different requirements in terms of the optimal valency or architecture of their ligands. For instance, the formation of a complex between the Janus heterovalent glycodendrimer in Fig. 3A and LecB was much more complete than that formed with LecA, likely due to the higher affinity of the fucoside residues towards LecB in comparison to that of the galactoside residues towards LecA.65 Increasing the Gal glycotope valency or density, as in the heteromultivalent systems depicted in Fig. 3B and C,66,67 led to increased LecB affinities. Considering that LecB is much less sensitive to multivalency than LecA, Nierengarten and coworkers proposed a more sophisticated design where ten Gal units are attached at the two rims of a pillar[5]ene platform and two Fuc residues are located as stoppers in a rotaxanated axle.69 The authors demonstrated that such a “supermolecule” can indeed achieve high binding affinities towards the two lectins (Fig. 3D).
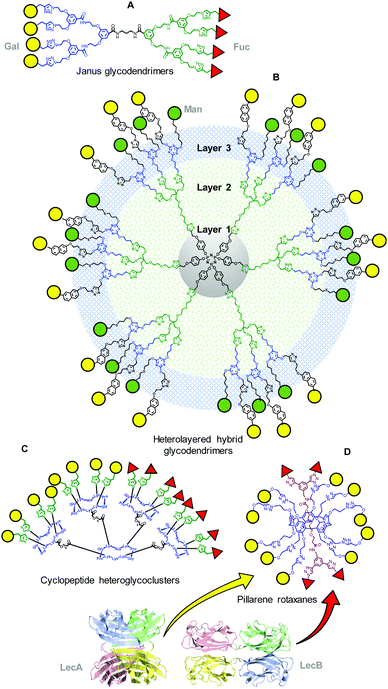 | ||
| Fig. 3 Representative structures of the different multidomain-type heteromultivalent prototypes synthesized by Roy and coworkers66 (A and B), Renaudet and coworkers68 (C) and Nierengarten and coworkers69 (D). | ||
LecA and LecB are particularly interesting targets from the biomedical point of view since both are involved in the recognition, adhesion and internalization of P. aeruginosa by airway epithelial cells.70,71 During the initial phase of infection, multiple types of glycans on the cell surfaces bind diverse types of lectins on the bacteria. This heterovalent network of carbohydrate–protein interactions is also critical for biofilm development and stabilization.72 Recently, Zhang and coworkers developed heteromultivalent nanotherapeutics inspired by this natural strengthening mechanism for the carbohydrate–lectin interactions between the bacteria and the cells. Gold nanorods (AuNRs) decorated with lactose (Lac) and Fuc homoglycopolymers specifically blocked LecA and LecB, promoted bacterial aggregation and inhibited biofilm formation.73 The near-infrared (NIR)-light-induced photothermal effect of the AuNRs additionally endowed the system with bacterial killing properties (Fig. 4A). The authors further extended this biomimetic approach to the co-assembly of polymers combining galactosylated and fucosylated blocks together with an acid-sensitive block that switched from hydrophobic to hydrophilic upon protonation.74 The resulting micelles with a heteroglycomultivalent shell specifically recognized P. aeruginosa, inhibited biofilm formation, protected native cells from bacterial infection and selectively released phototherapeutic agents included in their hydrophobic core in an acidic microenvironment at the infection site (Fig. 4B). Notably, the AuNRs and the co-assembled micelles coated with only the Lac, Gal or Fuc homopolymers showed much lower biofilm inhibition and bacterial killing properties, indicating the necessity of heteromultivalency to achieve a synergistic effect.
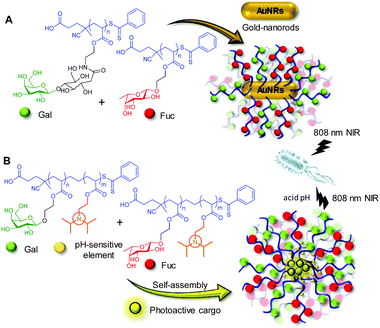 | ||
| Fig. 4 Schematic representation of the heteromultivalent nanotherapeutics based on gold nanorods decorated with Gal and Fuc glycopolymers developed by Zhang and coworkers (A).73 Inhibition of bacterial biofilm formation by using the acid-sensitive micelles that encapsulate a phototherapeutic reported by the same group (B).74 | ||
In some cases, it was found that targeting different lectins simultaneously with heteromultivalent glycoconjugates could be achieved with no explicit phase separation of the distinct glycotopes.75 In a very instructive study, Jiang and coworkers prepared glycopolymeric micelles from α-Man- and β-Gal-modified aliphatic polyesters, individually as well as in admixtures, or from a mixed α-Man/β-Gal-modified heteroglycopolymer analog.76 The purpose of the work was to investigate their interactions with the macrophage mannose receptor (MMR; CD206) and the macrophage galactose-binding lectin (MGL; CD301), both of which mediate clathrin-dependent endocytosis. Unexpectedly, the results showed that the nanoparticles built from the α-Man/β-Gal heteroglycopolymer were internalized by RAW 264.7 macrophages much more efficiently than not only the nanoparticles containing a single kind of monosaccharide, but also the nanoparticles combining the α-Man- and β-Gal glycotopes in separate polymers, even though the relative α-Man/β-Gal ratios were identical (Fig. 5). The same trend was observed for the affinity towards the plant lectins ConA (α-Man specific) and peanut agglutinin (PNA, β-Gal specific) by isothermal titration microcalorimetry (ITC). The authors speculated that the unlike polymer chains tend to aggregate separately, forming individual domains at the surfaces of the nanoparticles that make it less favorable for the establishment of concurrent interactions with MMR and MGL receptors at the cell membrane.
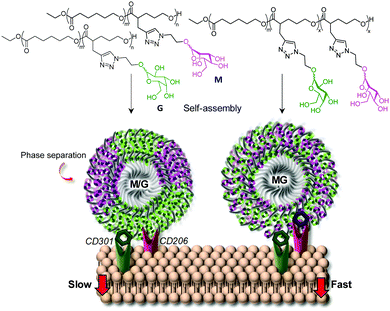 | ||
| Fig. 5 Schematic representation of the interactions of the macrophage mannose receptor (MMR; CD206) and the macrophage galactose-binding lectin (MGL; CD301) with nanoparticles self-assembled from α-Man (M) and β-Gal (G) glycopolymers or from heteroglycopolymers combining α-Man and β-Gal glycotopes and the consequences in the cell uptake rate as reported by Jiang and coworkers.76 | ||
The carbohydrate module method and the multidomain approaches claim that the observed lectin binding capability enhancements in heteromultivalent systems arise from the synchronized action of two or more discrete supramolecular events that can either be confined in a reduced binding site or expand on a larger 3D contacting area. In 2005, García Fernández, Ortiz Mellet and coworkers reported that high-density heteroclusters with alternating α-Man and β-Glc residues, built on a β-cyclodextrin (βCD) platform, exhibited ConA binding affinity enhancements that could not be rationalized on such grounds (Fig. 6).77 In their initial work, horseradish peroxidase-labelled ConA (HRP-ConA) was used for enzyme-linked lectin assay (ELLA)78–81 and ITC determinations. Although at the neutral pH of the experiments ConA is a homotetramer, the presence of the high molecular weight HRP label prevents cross-linking phenomena and essentially reduces the possibilities to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding mode. The α-Man motif is a weak HRP-ConA ligand in monovalent form but multivalent presentations can show very high avidities. On the other hand, monovalent β-Glc is not a ligand for ConA and only in a few cases measurable affinities for multivalent β-Glc arrays have been reported.82–86 In heteromultivalent α-Man/β-Glc conjugates, however, the presence of the β-Glc motif reinforced binding quite significantly. Thermodynamic data revealed that the free energy of binding decrease, as compared to homovalent α-Man clusters, had an entropic origin. This led the authors to propose that the presence of a very low affinity ligand facilitates ligand exchange processes, resulting in a higher contribution of the “memory-like” rebinding and sliding mechanisms to the multivalent effect (Fig. 6). The overall result can be interpreted as the enhancement of the affinity of a lectin towards a primary glycoligand by the presence of a low affinity secondary ligand or, alternatively, as the activation of the recognition of a secondary ligand enabled by the presence of a low proportion of a primary ligand. The term “heteromultivalent or heterocluster effect” was coined to refer to this new facet of multivalency.77,87
1 binding mode. The α-Man motif is a weak HRP-ConA ligand in monovalent form but multivalent presentations can show very high avidities. On the other hand, monovalent β-Glc is not a ligand for ConA and only in a few cases measurable affinities for multivalent β-Glc arrays have been reported.82–86 In heteromultivalent α-Man/β-Glc conjugates, however, the presence of the β-Glc motif reinforced binding quite significantly. Thermodynamic data revealed that the free energy of binding decrease, as compared to homovalent α-Man clusters, had an entropic origin. This led the authors to propose that the presence of a very low affinity ligand facilitates ligand exchange processes, resulting in a higher contribution of the “memory-like” rebinding and sliding mechanisms to the multivalent effect (Fig. 6). The overall result can be interpreted as the enhancement of the affinity of a lectin towards a primary glycoligand by the presence of a low affinity secondary ligand or, alternatively, as the activation of the recognition of a secondary ligand enabled by the presence of a low proportion of a primary ligand. The term “heteromultivalent or heterocluster effect” was coined to refer to this new facet of multivalency.77,87
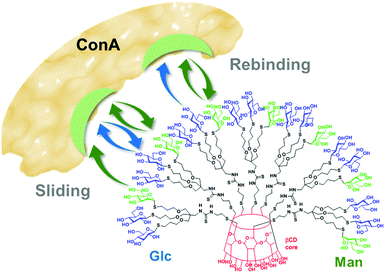 | ||
| Fig. 6 Schematic representation of the rebinding and sliding mechanisms in the interaction of heteromultivalent α-Man/β-Glc βCD-centered heteroglycoclusters with the α-Man-specific lectin ConA as discussed by García Fernández and coworkers.77,88,89 Upon initial binding of the putative Man glycotope, the secondary Glc residues can transiently bind to the carbohydrate recognition domains in the lectin and entropically favor both processes, enhancing the lifetime of the bound state. | ||
The heteromultivalent effect as expressed above implies synergistic phenomena influencing not only glycoligand–lectin binding affinity but also selectivity, and shows in heteroglycoclusters merging cognate and non-cognate glycotopes without phase separation. Its occurrence has been further corroborated in several laboratories with model “shuffled” heterovalent systems built on a variety of scaffolds. Thus, García Fernández and Ortiz Mellet made full use of the opportunities offered by βCD for selective functionalization to generate heteroglycocluster diversity;88,89 Hartmann,90–92 Kikkeri93 and Chen94 synthesized heteroglycosylated sequence-controlled oligopeptides, oligoamides and polymers, respectively, and Liu and Deng95 engineered heteroglyco-gold nanoparticles. Importantly, the heteromultivalent effect was found to be strongly dependent on the total ligand density. As a general rule, it is not apparent in the case of low valency heteroglycoconjugates,87,96–101 but manifests in heavily glycosylated architectures.
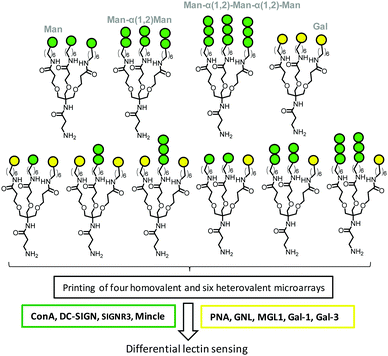
Interestingly, lectins that exhibit similar affinities for the same putative carbohydrate motif can significantly differ in their response to heteromultivalency when this motif is exposed conjointly with additional weakly binding or nonbinding glycans. As a corollary, heterogeneous glycan patterns have the potential to enhance lectin discrimination capabilities by virtue of the heteromultivalent effect in a more finely tuned manner than homogeneously glycosylated coatings do. This concept has been implemented by Gibson and coworkers102,103 and Kikkeri and coworkers104 for the development of analytical devices based on microarrays or gold nanoparticle glycotechnologies. For instance, the Man-binding lectins ConA, human Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin (CD-SIGN) and mouse Specific Intracellular adhesion molecule-3 Grabbing Non-integrin homolog-Related 3 C-type lectins (SIGNR3) and Macrophage inducible Ca2+-dependent lectin receptor (Mincle) and the Gal-binding lectins PNA, Galanthus nivalis lectin (GNL), mouse Macrophage Galactose-type Lectin-1 (MGL1) and human galectins 1 (Gal-1) and 3 (Gal-3) showed fully different recognition patterns when profiled against a battery of microarrays printed from homo- and heterotrivalent dendrons combining Man, α(1–2)-mannobiose, α(1–2)-mannotriose and Gal, as depicted in Fig. 7.104
Tanaka and coworkers further demonstrated that glycoheterogeneity affects the fate of glycocoated nanoconstructs in vivo by promoting differential interactions with protein receptor partners expressed in specific cells, tissues or organs.105–110 Thus, the arrangement of glycans relative to one another in a diverse series of structurally well-defined heteroglycoalbumins critically influenced pathway-selective excretion (urinary or gall bladder) or cell-specific targeting, thereby impacting biodistribution or tumor adhesion (Fig. 8). Their results open new avenues for the development of glycan-based imaging or therapeutic tracers.
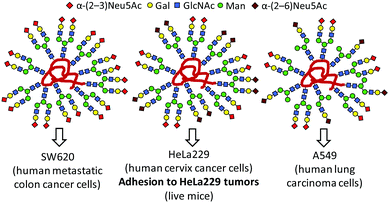 | ||
| Fig. 8 Differential cancer cell binding properties and in vivo tumor adhesion capabilities of heterogeneous-clustered glycoalbumins as reported by Tanaka and coworkers.109 | ||
The relevance of the heterocluster effect in cell biology was presumed since its formulation fifteen years ago on the basis of model glycocluster–lectin recognition studies.77,87 However, direct experimental evidence and theoretical support have been provided only very recently. The contribution by Wu and coworkers has been fundamental in this line. In their seminal work,111,112 the authors spotlighted unexplained data by Yanasigawa and coworkers,113 signalling that binding of cholera toxin B subunit (CTB) homopentamers to mouse embryonic neural precursor cells does not correlate with GM1 ganglioside (Gal-β-(1–3)-GalNAc-β-(1–4)[Neu5Ac-α-(2–3)]Gal-β-(1–4)-Glc-Ceramide) expression, even though GM1 is well known to be the primary ligand of the toxin. They also noted reports by Klassen and coworkers114–116 postulating cooperative effects in the binding of CTB to GM1. Most strikingly, the authors demonstrated that the GM2 ganglioside (GalNAc-β-(1–4)[Neu5Ac-α-(2–3)]Gal-β-(1–4)-Glc-Ceramide), which is a very weak ligand of the toxin, can contribute to CTB binding if present in a glycolipid mixture with a high affinity binder such as GM1 (Fig. 9).112 The experimental observations cannot be explained on the basis of allosteric regulation, but they are undoubtedly dynamic in nature and associated with the simultaneous presence of the high and low affinity ligands over a certain expression level in a relatively large peripheral area. The fact that going from the bulk to coating is a necessary requisite led the authors hypothesize that the heterocluster effect is mediated by a simple mechanism: reduction of dimensionality (RD). The basis of RD is that once the lectin has attached to a strong receptor in a fluid membrane, subsequent binding events are confined to a 2D surface. Surface binding rates can then be increased up to 1014 as compared with bulk binding rates, so that even a weak ligand, such as GM2 for CTB, can now participate in next binding events.
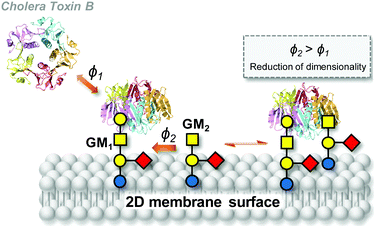 | ||
| Fig. 9 Schematic representation of the reduction of dimensionality (RD) concept in the binding of cholera toxin B subunit (CTB) homopentamers to GM1 (high-affinity ligand) and GM2 (low-affinity ligand) in the cell membrane as reported by Wu and coworkers.111,112ϕ1 and ϕ2 are the binding rates of GM1 and GM2 to CTB in the bulk and on the surface, respectively. After a lectin has attached to a high-affinity ligand, a low-affinity ligand encounters the bound lectin, completing the heteromultivalent binding (ϕ2 > ϕ1). In a next step, the lectin can be stabilized by two low-affinity ligands (not shown) and the free high-affinity ligand can then accept another lectin from the solution phase. | ||
Through RD, heteromultivalency can significantly alter lectin binding properties, including avidities, selectivities and kinetics, offering a new strategy to design high-affinity carriers for targeted drug delivery.117–120 As a proof of concept, Wu and coworkers demonstrated that the retention of liposomes by the pathogenic bacteria Pseudomonas aeruginosa was significantly enhanced when the strong LecA binder globotriaosylceramide (Gb3; Gal-α-(1–4)-Gal-β-(1–4)-Glc-Ceramide) was combined with lactosyl-ceramide or Gal-β-ceramide, which are much weaker LecA ligands, as compared with liposomes with a homogeneous array of Gb3.120 Heteromultivalent cooperative phenomena under RD also provide a rationale for the fact that human intestinal epithelial cells or murine lung cells are efficiently infected by the bacteria in spite of expressing Gb3 at very low levels.121,122
Kinetic Monte Carlo (kMC) simulations supported the underpinning role of RD in the heterocluster effect.123 The main conclusions of the kMC computational study are in full agreement with the experimental observations, namely: (a) the density of the low affinity ligand is a critical parameter for the activation of the heterocluster effect and a threshold density is needed to enable its contribution to lectin binding, (b) a tiny amount of high affinity ligand is sufficient to accelerate lectin binding to low affinity ligands and (c) heteromultivalent binding phenomena can modulate lectin binding kinetics and lectin bound states via ligand exchange processes on the surface. Note that ligand exchange under RD is equivalent to the entropy-driven mechanism hypothesized for high-density nanosized heteroglycoclusters.77,87 In conjunction with results reported by other laboratories on biologically relevant lectins binding to heteromultivalent systems,124–126 the ensemble of evidence establishes that heteromultivalency plays a key role in lectin–glycan recognition and that cells probably utilize this mechanism to regulate the downstream of lectin functions.
Multivalent glycosidase inhibition
Enzymatic substrate processing is generally considered to be preceded by a typical lock-and-key supramolecular recognition event that warrants enzyme specificity. In the case of a glycosidase (glycosyl hydrolase) the substrate is a glycoside that must reach the catalytic receptacle and fit the configurational and conformational requirements imposed by the architecture of the active site. In most cases, a rather strict complementarity between the glycone substitution pattern and the amino acids at the glycone hosting spot (the so-called −1 site) is achieved through an intricate network of hydrogen bonding, stacking and hydrophobic contacts. This matching relationship is mandatory for the glycoside to adopt the right conformation in the corresponding enzyme:substrate complex and the subsequent activation of the catalytic machinery. Contributions to binding from the aglycone moiety at the aglycone (+1) binding site are commonly less vital, although the aglycone nature can significantly affect the catalytic reaction rate or even impede enzymatic hydrolysis by steric and/or conformational biases, which can also be exploited in the design of enzyme regulators.127–129The ambition of reproducing the biunivocal enzyme–substrate relationship has monopolized the search for glycosidase inhibitors for biological studies or biomedical applications. Not surprisingly, the iconic specimens are monomeric glycomimetics with hydroxylation profiles of structural complementarity with the glycone moiety of the natural substrate, among which the nitrogen-in-the-ring monosaccharide analogues of the iminosugar category are by far the most popular.130 Appropriately selected iminosugars, either from natural origin or chemically synthesized, enable altering the cellular glycosylation profile, interfering in the metabolism of glycoconjugates and carbohydrates, modifying the carbohydrate-dependent properties of glycoproteins or hindering the carbohydrate-mediated interaction of host cells with infective agents, among others.131 Defying the prevalent philosophy, Gouin and coworkers advanced in 2009 the visionary idea that glycosidases might be responsive to multivalent arrangements of inhibitory motifs (inhitopes) in the same manner as lectins having a single CRD available are sensitive to multivalent arrays of carbohydrate ligands, provided that the kinetic and thermodynamic parameters of the individual binding phenomena are apt.132 The merit of their work is double: first, a potential analogy between carbohydrate/lectin and glycomimetic/glycosidase supramolecular binding mechanisms has been invoked for the first time; secondly, by testing a trivalent conjugate of the iminosugar 1-deoxynojirimycin (DNJ), a piperidine analogue of D-glucopyranose, towards a panel of glycosyl hydrolases, they determined a 2.6-fold enhancement in the inhibitory potency towards Jack bean α-mannosidase (JbMan) on a DNJ molar basis (Fig. 10A).
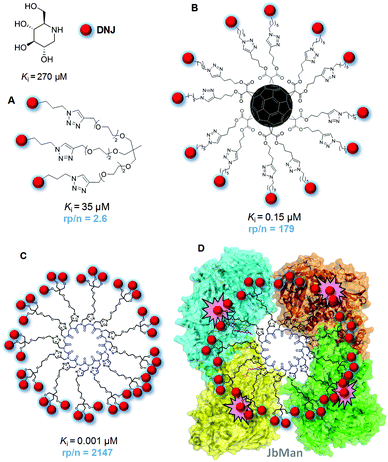 | ||
| Fig. 10 Structures, Jack bean α-mannosidase (JbMan) inhibition constant (Ki) values and normalized relative inhibition potencies (rp/n; DNJ molar basis) of the trivalent (A), dodecavalent (B) and 36-valent (C) DNJ conjugates reported by Gouin and coworkers;132 Compain, Ortiz Mellet, Nierengarten and coworkers (B)133 and Compain and coworkers.139 A schematic representation of the complex of the latter with JbMan, based on the reported crystal structure,163 showing two heterodimeric enzyme units with the four catalytic sites (highlighted in pink) occupied by DNJ inhitopes, is also shown (D). | ||
Although significant, the above multivalent enzyme inhibition (MEI) effect is moderate and Gouin's work would have had probably passed overlooked if one year later Compain, Ortiz Mellet, and Nierengarten had not published that the inhibitory potency of DNJ towards JbMan was boosted up to 179-fold on a DNJ molar basis when the inhitope was exposed in 12 copies on a C60 fullerene scaffold (Fig. 10B).133 This was a counterintuitive aftermath that advanced multivalency as a promising tool for the design of potent glycosidase inhibitors and encouraged to visualize MEI as a variant of the glycoside cluster effect. MEI soon became a trending topic in the glycosciences that has been discussed in several reviews.43,134–139 A consistent body of work from the four mentioned laboratories as well as by the groups of Cardona, Moreno-Vargas, Blériot and Li, often in multicollaborative networks, confirmed the reactiveness of JbMan and other glycosidases, such as α- and β-glucosidase, amyloglucosidase or hexosaminidases, to multivalent arrays of iminosugars.140–162 In addition to polyhydroxylated pyrimidine-type iminosugars like DNJ, iminosugars with other azaheterocyclic cores, e.g. pyrrolizidine,145,147 pyrrolidine157,158,160 or azepane156 derivatives, also proved competent to elicit MEI when conjugated to multi-armed platforms. A strong dependency of the inhibition potency enhancement elicited by multivalency from the total valency and architectural parameters was documented. Yet, only in the case of JbMan a parallelism with the behavior generally observed in carbohydrate–protein interactions could be supported throughout the different studies.
JbMan and other α-mannosidases of the glycosyl hydrolase (GH) family GH38 exhibit a rather open and accessible active site and often are multimeric in their functional form, a scenario that is reminiscent of that encountered in lectins. Thus, JbMan itself is a dimer of heterodimers, with each of the heterodimers possessing a catalytic receptacle. It seemed therefore logical to assume that it can share with lectins the same mechanisms leading to high avidities when faced to clustered ligand partners, that is, the sliding and rebinding processes associated with high local epitope density and the crosslinking and chelation phenomena characteristic of multipoint interactions.43 Indeed, experimental evidence for co-aggregation events involving multivalent iminosugar constructs and JbMan has been obtained by atomic force microscopy (AFM), transmission electron microscopy (TEM), mass spectrometry and analytical ultracentrifugation measurements.143,152,157 Recently, Compain and coworkers further obtained the first high-resolution crystal structures of apo JbMan and of its complex with a 36-valent DNJ-cyclopeptoid conjugate that displayed the largest inhibition enhancement observed for this enzyme so far (Fig. 10C and D).163 The data clearly demonstrated the interplay of bridging and chelation processes in the formation of a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 JbMan:multivalent inhibitor sandwich-type complex, with all four glycone sites occupied by DNJ subunits.
1 JbMan:multivalent inhibitor sandwich-type complex, with all four glycone sites occupied by DNJ subunits.
Differently from JbMan, other glycosidases reported to experience MEI are monomeric and/or have buried catalytic sites, which are hardly compatible with the carbohydrate–lectin affinity enhancement-like mechanisms sustained for the former. Notably, in some cases multivalency switched on, rather than enhanced, the inhibitory capacity of a glycomimetic towards a glycosidase while simultaneously up- or down-regulating the inhibitory activity against others. To reconcile the experimental observations, García Fernández, Nierengarten, Ortiz Mellet and coworkers launched the hypothesis that multimerization of a glycomimetic onto a nanometric scaffold elicits binding modes to the glycosidases that can be radically different from the binding mode of the monomer.164 The authors capitalized on the unique stereoelectronic and chemical properties of the so-called sp2-iminosugar glycomimetics,165 in which the underlining amine group of iminosugars is replaced into a trigonal planar pseudoamide-type nitrogen (N-carbonyl, N-thiocarbonyl, N-imino group), with a substantial sp2-hybridization nature (e.g., as in carbamate,166 thiocarbamate,167–169 urea,170–172 thiourea,173–180 isourea,180–187 isothiourea,187–195 guanidine,196,197 sulfamide198 or thiohydantoin199 functionalities), to access stable O-, S-, N- and C-pseudoglycosides.200–211 Through click-type multiconjugation strategies, they then succeeded in preparing homogeneous multivalent glycoside analogues on a fullerene C60 scaffold that exhibited dual recognition capabilities towards lectins and glycosidases.164 A competitive lectin–glycosidase binding assay was next implemented that enabled mapping the enzyme regions interacting with the high-valency sp2-iminosugar conjugates. Briefly, it consists of determining the effects of reference inhibitors of the enzyme known to bind at glycone, glycone/aglycone or surface binding sites in the steady-state partition of the multivalent inhibitor between the lectin, labelled with horseradish peroxidase (HRP; reporter enzyme), and the glycosidase. If the reference inhibitor and the multivalent ligand under study compete for the same site in the glycosidase, the capability of the enzyme to sequester the ligand will lessen in the presence of the former (Fig. 11). By using this technique, JbMan/multivalent enzyme inhibitor complex formation was found to involve substantial implications for the glycone site, which is in agreement with the reported crystal structure.163 In sharp contrast, in the case of the α-glucosidase from Saccharomyces cerevisiae (yeast maltase; GH13) the multivalent derivatives bind at low affinity non-glycone binding sites of the enzyme, leading to inhibition by a “recognition and blockage” mechanism. This operational model likely applies also to the isomaltase (Saccharomyces cerevisiae), β-glucosidase (bovine liver), and α-galactosidase (green coffee beans) enzymes, all of them belonging to glycosyl hydrolase families known to possess relatively deep catalytic sites (GH13, GH1, and GH27, respectively).164
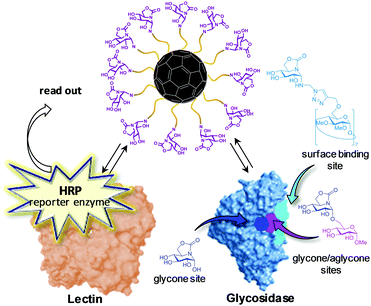 | ||
| Fig. 11 Schematic representation of the lectin–glycosidase competitive binding assay developed by Ortiz Mellet, García Fernández and coworkers to map the binding mode involved in multivalent enzyme inhibition (MEI). The assay construction further implies a non-labelled lectin fixed to the surfaces of microplate wells, thereby enabling quantification of the effect of the reference glycosidase inhibitors in the lectin crosslinking capabilities of the probed compound. The data can be directly correlated with the displacement of the equilibrium between the lectin–multivalent inhibitor complex and the glycosidase–multivalent inhibitor complex. Note that the choice of reference inhibitor depends on the glycosidase under study. The structures depicted here correspond to examples of α-glucosidase inhibitors.164 | ||
The monovalent/multivalent shift in preferred binding mode supported by the above data provides an immediate explanation for the multivalency-dependent “on–off” switching effect observed for some inhitope-glycosidase pairs. It may happen, for instance, that a monovalent inhitope does not match the glycone site but is able to bind at aglycone and/or surface sites, thus benefitting from multivalency when exposed in a multicopy manner and eventually impairing the access of the substrate to the catalytic locus. Alternatively, it is conceivable that upon binding at secondary surface binding sites the multivalent ligand could stabilize enzyme conformations facilitating, instead of hampering, substrate processing, in the same manner as some polysaccharides act as enhancers of amylases.212 This reasoning provides a reliable explanation for the apparently contradictory observation by Gouin and coworkers on dextran-based DNJ and 1-deoxymannojirimycin (DMJ) polymers, with valencies ranging from 20 to 900, behaving as activators of several glycosidases (two galactosidases, a fucosidase and a bacterial mannoside phosphorylase).213
As already demonstrated for classical monovalent inhibitors, MEI offers the possibility of developing glycosidase ligands that stabilize the proper folding of disease-causative misfolded mutant forms of the enzyme, a therapeutic paradigm termed pharmacological chaperone therapy.214,215 This concept has already been demonstrated by Compain and coworkers for Gaucher disease, an autosomal recessive lysosomal storage disorder (LSD) due to β-glucocerebrosidase (GCase) dysfunction,216–218 and by Higaki, García Fernández, Ortiz Mellet and coworkers for α-mannosidosis, another LSD involving malfunctioning of lysosomal α-mannosidase (LAMAN).219 Compain and coworkers also reported a beneficial effect of multivalent DNJ derivatives in cystic fibrosis by rescuing the misfolded mutant cystic fibrosis transmembrane conductance regulator (CFTR) protein from endoplasmic reticulum-associated degradation (ERAD) in patient cells.220 The results are compatible with a calnexin-dependent mechanism of action, but not with the inhibition of ER α-glucosidases I and II as is the case for monovalent DNJ derivatives.
Nguyen and coworkers221,222 used computational studies to implement a non-iminosugar-based strategy for MEI-mediated selective inhibition of heparanase, a β-endoglucuronidase whose main function is cleaving the internal β-(1–4) glycosidic bond between glucuronic acid (GlcA) and N-sulfated glucosamine (GlcNS) along heparan sulfate (HS) chains in proteoglycans in the extracellular matrix. The authors first identified the disulfated (at the N′- and 6′-O-positions) disaccharide GlcNS(6S)α-(1–4)GlcA as the preferred unit for supramolecular binding at the −2 and −1 glycone binding sites. They next designed glycopolymers exposing this component for maximal heparanase inhibition and minimal anticoagulant activity, from which a 12-valent representative was determined to be the most potent heparanase inhibitor with a picomolar inhibitory concentration (meaning an over 1000-fold enhancement as compared with the monovalent control) and tight binding characteristics. This is notable because heparanase is a monomeric enzyme with a hindered active site. While the molecular basis for MEI is not discussed, it is interesting to speculate that the dissociation rate of the disaccharide motif, which is indeed the same present in the reaction product of enzymatic hydrolysis of HS, must be fast enough to allow rebinding processes to operate, as far as the spacers and ligand densities are adjusted to prevent steric clashes. The main motivation for this research is that HS degradation by heparanase in the extracellular matrix has been correlated with tumor angiogenesis and metastasis. Rewardingly, the authors found that the optimal glycopolymers showed potent antimetastatic effects against 4T1 mammary carcinoma cells and inhibited experimental metastasis into the lungs in vivo (Fig. 12).223 MEI was also invoked by Nishimura and coworkers to account for the efficiency of nanosomes exposing β-linked N-acetylglucosamine (GlcNAc) suicide substrates at inhibiting the lysosomal β-hexosaminidases and triggering apoptosis in cancer cells as well as in a mouse model, although the underlying molecular mechanism remains unclear.224
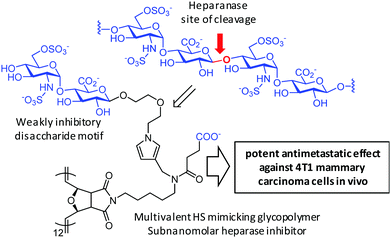 | ||
| Fig. 12 Structures of the heparansulfate (HS) region where heparanase acts and of the multivalent glycopolymer prepared by Tanaka and coworkers225 exposing the GlcNS(6S)α-(1–4)GlcA disaccharide, devoid of the scissile bond, to achieve potent heparanase inhibition. | ||
Multivalency-promoted lectin/glycosidase cross-talk behaviours
The conclusion that multivalent glycomimetics can alter glycosidase functioning upon binding in lectin-like regions beyond the catalytic area led to the conjecture that glycotope-related versions might behave similarly. In other words, multivalency might turn a putative glycosidase substrate, a glycoside, into an inhibitor by eliciting “aberrant” supramolecular binding modes.164 Confirmation of this hypothesis was provided in 2015 by Siriwardena, García Fernández, Ortiz Mellet, Szunerits and coworkers after evaluating glycocoated nanodiamonds (NDs) against a panel of commercial glycosidases (Fig. 13).225 The authors found that α-O-glucosides and also α-O-mannosides, when conjugated on ND particles, were not only resistant towards the hydrolytic action of the corresponding matching glycosidases, but acquired the ability to inhibit them. Moreover, the glycocoated NDs further became inhibitors of mismatching enzymes for which they do not serve as substrates even when in their monovalent form. A step ahead, it was established that glycosidase inhibition was sensitive to heterogeneous arrangements of the Glc and Man motifs in the same manner as the binding affinity to ConA lectin was. For instance, homoglucosylated-NDs (Glc-NDs) and homomannosylated-NDs (Man-NDs) inhibited S. cerevisiae α-glucosidase with inhibition constant (Ki) values of 22 and 9.4 μM, respectively, whereas Glc/Man-NDs (Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Man 1
Man 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with identical total saccharide loadings were about 5-fold more potent inhibitors of this enzyme (Ki 1.9 μM). Conversely, inhibition of E. coli β-galactosidase by Glc-NDs (Ki 1.9 μM) and Man-NDs (Ki 13.2 μM) was thwarted when both glycotopes were exposed together in Glc/Man-NDs (Ki 268 μM). In other words, the supramolecular events underlining multivalent enzyme inhibition for a given inhibitory motif/enzyme pair can be significantly altered by the presence of a second partner, leading to positive or negative synergies, what represents an extension of the heterocluster effect.
1) with identical total saccharide loadings were about 5-fold more potent inhibitors of this enzyme (Ki 1.9 μM). Conversely, inhibition of E. coli β-galactosidase by Glc-NDs (Ki 1.9 μM) and Man-NDs (Ki 13.2 μM) was thwarted when both glycotopes were exposed together in Glc/Man-NDs (Ki 268 μM). In other words, the supramolecular events underlining multivalent enzyme inhibition for a given inhibitory motif/enzyme pair can be significantly altered by the presence of a second partner, leading to positive or negative synergies, what represents an extension of the heterocluster effect.
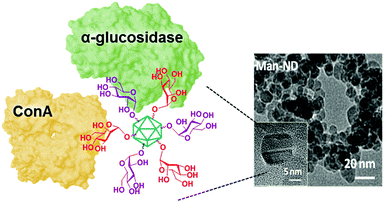 | ||
| Fig. 13 Schematic representation of the Glc/Man heterovalent glyconanodiamonds exhibiting simultaneous lectin binding and glycosidase inhibition capabilities reported by Sirwardena, García Fernández, Ortiz Mellet, Szunerits and coworkers.225 | ||
Additional evidence for enzyme inhibition by multivalent glycosystems has been provided using glycofullerenes226 and glycocyclodextrin conjugates.227,228 Integrated mechanistic studies exploiting ELLA and lectin/glycosidase competitive assays were conducted for homogeneous constructs as well as for mixed glycoside/glycomimetic (sp2-iminosugar) heterovalent arrays.226,227 Collectively, the ensemble of results substantiates the vision that multivalent presentations of a glycotope or an inhitope can promote binding modes to glycosidases that show significant analogies to those governing carbohydrate–lectin supramolecular interactions. Multivalent arrays can thus simultaneously act on lectins and glycosidases in a multimodal manner. Moreover, a given glycotope or inhitope moiety may elicit different responses depending on the presence or absence of a second glyco(mimetic) motif, even if a priori irrelevant towards the lectin/glycosidase target, supporting a unified framework for the glycoside cluster effect, the heteromultivalent effect and multivalent enzyme inhibition: the generalized multivalent effect.43 By changing the total and partial valencies and adjusting the overall topology of the (hetero)multivalent construct, “on” or “off” statuses for a range of lectins and glycosidases can be triggered, markedly altering the selectivity profile encountered for monovalent derivatives.
The shift in the perception of multivalency, from a natural and safe strategy to achieve useful responses in carbohydrate-mediated supramolecular processes to a multichannel switcher with the potential to act on a range of receptor/enzyme recognition events, depicts a much more complex scenario than classically assumed. On the one hand, the generalized multivalent effect calls for a careful evaluation of the potential risks derived from multivalency-associated “biological messiness”,229 expanding from carbohydrate receptors to glycoprocessing enzymes. On the other hand, the new evidence also informs the possibility of taking advantage of multivalency to finely shape the supramolecular properties of carbohydrates in an intrinsically multifactorial biological context. It is conceivable, for instance, that purposely tailoring glycoligands to specifically interact with biomedically relevant lectin/glycosidase subsets opens new channels in multitargeted drug design. García Fernández, Renaudet and Ortiz Mellet have recently implemented this notion in the development of mannosyl-coated glycoclusters with the ability to simultaneously and distinctly target the macrophage mannose receptor and lysosomal storage disorder-associated lysosomal glycosidases.230 After screening a series of structurally diverse candidates, the authors encountered that the MMR avidity, as determined by a modified ELLA protocol,231,232 was essentially dependent on the total Man valency. In contrast, glycosidase selectivity was strongly reliant on the overall glycoarchitecture topology: a heptavalent β-cyclodextrin conjugate (Fig. 14, upper pathway) behaved as a selective inhibitor of β-glucocerebrosidase (IC50 0.1 μM), whereas a hexadecavalent RAFT cyclopeptide derivative (Fig. 14, lower pathway) turned to be a highly selective inhibitor of lysosomal α-mannosidase. Since binding to the MMR inherently elicits macrophage uptake, these compounds epitomize the first examples of intrinsically site-specific, self-deliverable glycosidase regulators.
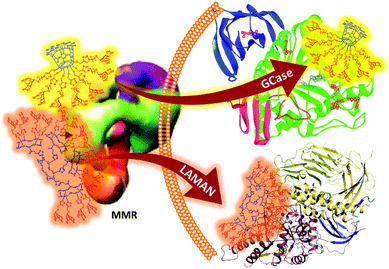 | ||
| Fig. 14 Structures of the mannosyl-coated heptavalent and decahexavalent glycoclusters built on β-cyclodextrin (highlighted in yellow) and RAFT cyclopeptide (highlighted in orange), reported by García Fernández, Renaudet and Ortiz Mellet, with the ability to simultaneously and distinctly target the MMR and the LSD-associated glycosidases β-glucocerebrosidase (GCase) and lysosomal α-mannosidase (LAMAN), respectively.230 | ||
Conclusions and future outlook
Pioneering work in 1999 by Kahne and coworkers already described that lectin specificity could be altered in heterovalent as compared with homovalent glycosurfaces.233 Ten years later, Penadés and coworkers reported that lactoside substituents became resistant to the action of β-galactosidase when multivalently exposed at the periphery of micelles or gold nanoparticles while retaining the ability to bind the galactose-specific agglutinin from Viscum album.234 The authors speculated that β-galactosidase recognition probably occurred, but that binding of a first enzyme molecule to the multivalent conjugate reduces the accessibility of other lactose residues to new enzyme molecules. These early manifestations of the heteromultivalent effect and multivalent enzyme inhibition have been largely supported by an increasing number of publications highlighting the relevance of those phenomena, not only in model systems but also in a biological context. The last few years have witnessed crucial advances in our understanding of multivalency-induced carbohydrate recognition promiscuity under the new generalized multivalent effect paradigm. Yet, much work is still needed to unveil the precise mechanisms at play; this is critical both to prevent potential risks and to program specific activities.The increasing awareness of the multilateral character of (hetero)multivalency, with additional reports expanding the range of MEI responsive glycosidases235,236 and extending the generalized multivalent effect to the glycosyltransferase enzyme category237–239 and anti-carbohydrate antigen antibodies,240 will likely fuel research in carbohydrate supramolecular chemistry with a new perspective. The current embodiments exploiting the heteromultivalent effect to reach optimal cell targeting in vivo76 and multivalent inhibitors to regulate the activity of disease-associated enzymes, e.g. in the context of LSDs,216–219 cystic fibrosis220 or cancer,225 pave the way for future developments. The possibility of combining enhanced glycoreceptor capabilities and specific glycosidase inhibitory properties in designing multitargeted glycodrug prototypes is particularly appealing in this sense. Glycoclusters targeting the MMR and LSD-causative glycosidase pairs are representative examples,230 but many other therapies could benefit from this concept. Thus, GlcNAc-coated nanoparticles having a hard metal core such as quantum dots or gold nanoparticles and a soft shell, namely nanosomes (NSs), were recently found to be efficiently internalized by human hepatocarcinoma HepG2 cells in vitro, probably through lectin-mediated endocytosis.224 NSs bearing a multivalent presentation of a GlcNAc-based suicide inhibitor additionally inhibited very competently lysosomal β-hexosaminidase, which is a promising target that induces a lysosomal membrane permeabilization (LMP)-mediated cell death pathway. This can be seen as an example of self-deliverable anticancer therapeutic nanomedicine benefiting from multivalent enzyme inhibition, a notion that deserves to be explored.
Advancing the fundamental knowledge of the generalized multivalent effect will also shed light on other supramolecular events essential for cell life, e.g. the role of glycoheterogeneity in the packing processes involving polysaccharides at the glycocalyx and the extracellular matrix.241 It is pertinent to note that many of the expressions of the generalized multivalent effect have a dimensional character; that is, they imply nanometric entities and relay on surface contacts, a biomimetic mechanism. It is then conceivable that other non-carbohydrate or glycomimetic motifs, or even the own nanoparticle shape and surface nature,242 could actively influence the supramolecular chemistry of (hetero)multivalent glycoligands towards both lectins and enzymes. Multifunctional molecular nanoparticles designed to simultaneously bind nucleic acids and lectins represent a particularly interesting case to study in this sense.243–247 Further outlook includes the conception of hybrid (hetero)glycomaterials to interrogate the compositional and functional complexities of the cell surface, which in turn has the potential to lead to unanticipated advances in precision glyconanotechnology.52,248
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge financial support through contract numbers SAF2016-76083-R (MINECO, FEDER, UE) and RTI2018-097609-B-C21 (MICIU, AEI, FEDER, UE). M. G.-C. is a FPI fellow (co-financed by FSE, EU). We also acknowledge the CSIC (URICI) for supporting open access publication and the CITIUS (Univ. Seville) for technical support.Notes and references
- C. Fasting, C. A. Schalley, M. Weber, O. Seitz, S. Hecht, B. Koksch, J. Dernedde, C. Graf, E.-W. Knapp and R. Haag, Angew. Chem., Int. Ed., 2012, 51, 10472 CrossRef CAS PubMed.
- C. S. Mahon and D. A. Fulton, Nat. Chem., 2014, 6, 665 CrossRef CAS PubMed.
- E. Mahon and M. Barboiu, Org. Biomol. Chem., 2015, 13, 10590 RSC.
- K. C. Tjandra and P. Thordarson, Bioconjugate Chem., 2019, 30, 503 CrossRef CAS PubMed.
- M. Mammen, S.-K. Choi and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 2754 CrossRef PubMed.
- C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357 CrossRef CAS PubMed.
- J. J. Lundquist and E. J. Toone, Chem. Rev., 2002, 102, 555 CrossRef CAS PubMed.
- L. L. Kiessling, J. E. Gestwicki and L. E. Strong, Angew. Chem., Int. Ed., 2006, 45, 2348 CrossRef CAS PubMed.
- Y. C. Lee and R. T. Lee, Acc. Chem. Res., 1995, 28, 321 CrossRef CAS.
- N. Jayaraman, Chem. Soc. Rev., 2009, 38, 3463 RSC.
- A. Bernardi, J. Jiménez-Barbero, A. Casnati, C. De Castro, T. Darbre, F. Fieschi, J. Finne, H. Funken, K.-E. Jaeger, M. Lahmann, T. K. Lindhorst, M. Marradi, P. Messner, A. Molinaro, P. V. Murphy, C. Nativi, S. Oscarson, S. Penadés, F. Peri, R. J. Pieters, O. Renaudet, J.-L. Reymond, B. Richichi, J. Rojo, F. Sansone, C. Schäffer, W. B. Turnbull, T. Velasco-Torrijos, S. Vidal, S. Vincent, T. Wennekes, H. Zuilhof and A. Imberty, Chem. Soc. Rev., 2013, 42, 4709 RSC.
- S. Cecioni, A. Imberty and S. Vidal, Chem. Rev., 2015, 115, 525 CrossRef CAS PubMed.
- C. Müller, G. Despras and T. K. Lindhorst, Chem. Soc. Rev., 2016, 45, 3275 RSC.
- Y. Chen, A. Star and S. Vidal, Chem. Soc. Rev., 2013, 42, 4532 RSC.
- F. Peri, Chem. Soc. Rev., 2013, 42, 4543 RSC.
- N. Spinelli, E. Defrancq and F. Morvan, Chem. Soc. Rev., 2013, 42, 4557 RSC.
- K. Hatano, K. Matsuoka and D. Terunuma, Chem. Soc. Rev., 2013, 42, 4574 RSC.
- M. C. Galan, P. Dumy and O. Renaudet, Chem. Soc. Rev., 2013, 42, 4599 RSC.
- T. R. Branson and W. B. Turnbull, Chem. Soc. Rev., 2013, 42, 4613 RSC.
- F. Sansone and A. Casnati, Chem. Soc. Rev., 2013, 42, 4623 RSC.
- N. Jayaraman, K. Maiti and K. Naresh, Chem. Soc. Rev., 2013, 42, 4640 RSC.
- Y. M. Chabre and R. Roy, Chem. Soc. Rev., 2013, 42, 4657 RSC.
- M. Marradi, F. Chiodo, I. García and S. Penadés, Chem. Soc. Rev., 2013, 42, 4728 RSC.
- A. Martínez, C. Ortiz Mellet and J. M. García Fernández, Chem. Soc. Rev., 2013, 42, 4746 RSC.
- P. Bojarová, R. R. Rosencrantz, L. Elling and V. Kren, Chem. Soc. Rev., 2013, 42, 4774 RSC.
- J. Arnaud, A. Audfray and A. Imberty, Chem. Soc. Rev., 2013, 42, 4798 RSC.
- L. Reymond, M. Bergmann and T. Darbre, Chem. Soc. Rev., 2013, 42, 4814 RSC.
- M. Gingras, Y. M. Chabre, M. Roya and R. Roy, Chem. Soc. Rev., 2013, 42, 4823 RSC.
- V. Wittmann and R. J. Pieters, Chem. Soc. Rev., 2013, 42, 4492 RSC.
- X. Zhang, G. Huang and H. Huang, Drug Delivery, 2018, 25, 18–40 CAS.
- Z. Ma and X. X. Zhu, J. Mater. Chem. B, 2019, 7, 1361 RSC.
- S. Gim, Y. Zhu, P. H. Seeberger and M. Delbianco, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2019, 11, e1558 Search PubMed.
- L. L. Kiessling, Bioorg. Med. Chem., 2018, 26, 5229 CrossRef CAS PubMed.
- C. W. Cairo, J. E. Gestwicki, M. Kanai and L. L. Kiessling, J. Am. Chem. Soc., 2002, 124, 1615 CrossRef CAS PubMed.
- J. E. Gestwicki, C. W. Cairo, L. E. Strong, K. A. Oetje and L. L. Kiessling, J. Am. Chem. Soc., 2002, 124, 14922 CrossRef CAS PubMed.
- H.-J. Gabius, Folia Biologica, 2017, 63, 121 Search PubMed.
- H.-J. Gabius, Trends Biochem. Sci., 2015, 40, 341 CrossRef CAS PubMed.
- J. W. Dennis, Trends Biochem. Sci., 2015, 40, 673 CrossRef CAS PubMed.
- J. W. Dennis, Curr. Opin. Chem. Biol., 2017, 41, 1 CrossRef CAS PubMed.
- A. Varki and S. Kornfeld, in Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, C. Bertozzi, G. W. Har and M. E. Etzler, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 3rd edn, 2017, ch. 1 Search PubMed.
- J. M. Tarbell and L. M. Cancel, J. Intern. Med., 2016, 280, 97 CrossRef CAS PubMed.
- A. Varki, Glycobiology, 2017, 27, 3 CrossRef CAS PubMed.
- C. Ortiz Mellet, J.-F. Nierengarten and J. M. García Fernández, J. Mater. Chem. B, 2017, 5, 6428 RSC.
- M. Delbianco, P. Bharate, S. Varela-Aramburu and P. H. Seeberger, Chem. Rev., 2016, 116, 1693 CrossRef CAS PubMed.
- J. L. Jiménez Blanco, J. M. Benito, C. Ortiz Mellet and J. M. García Fernández, J. Drug Delivery Sci. Technol., 2017, 42, 18 CrossRef.
- S. S. Park, H.-W. Hsieh and J. Gervay-Hague, Molecules, 2018, 23, 1742 CrossRef PubMed.
- O. Francesconi and S. Roelens, ChemBioChem, 2019, 20, 1329 CrossRef CAS PubMed.
- T. Neva, C. Ortiz Mellet, J. M. García Fernández and J. M. Benito, J. Carbohydr. Chem., 2019, 38, 470 CrossRef CAS.
- S. Bheren and U. Westerlind, Molecules, 2019, 24, 1004 CrossRef PubMed.
- Multivalency: concepts, research and applications, ed. J. Huskens, L. J. Prins, R. Haag and B. J. Ravoo, Wiley, Oxford UK, 2018 Search PubMed.
- Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, C. Bertozzi, G. W. Har and M. E. Etzler, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 3rd edn, 2017 Search PubMed.
- M. L. Huang and K. Godula, Glycobiology, 2016, 26, 797 CrossRef CAS PubMed.
- Y. Nishida, H. Uzawa, T. Toba, K. Sasaki, H. Kondo and K. Kobayashi, Biomacromolecules, 2000, 1, 68 CrossRef CAS PubMed.
- K. Sasaki, Y. Nishida, T. Tsurumi, H. Uzawa, H. Kondo and K. Kobayashi, Angew. Chem., Int. Ed., 2002, 41, 4463 CrossRef CAS PubMed.
- Y. Nishida, H. Dohi and K. Kobayashi, Trends Glycosci. Glycotechnol., 2005, 17, 59 CrossRef CAS.
- C. A. Hunter and H. L. Anderson, Angew. Chem., Int. Ed., 2009, 48, 7488 CrossRef CAS PubMed.
- A. Miyachi, H. Dohi, P. Neri, H. Mori, H. Uzawa, Y. Seto and Y. Nishida, Biomacromolecules, 2009, 10, 1846 CrossRef CAS PubMed.
- H. Tran, P. I. Kitov, E. Paszkiewicz, J. M. Sadowska and D. R. Bundle, Org. Biomol. Chem., 2011, 9, 3658 RSC.
- Y. Terada, Y. Hoshino and Y. Miura, Chem. – Asian J., 2019, 14, 1021 CrossRef CAS PubMed.
- K. E. Moog, M. Barz, M. Bartneck, F. Beceren-Braun, N. Mohr, Z. Wu, L. Braun, J. Dernedde, E. A. Liehn, F. Tacke, T. Lammers, H. Kunz and R. Zentel, Angew. Chem., Int. Ed., 2017, 56, 1416 CrossRef CAS PubMed.
- M. Bartneck, C. T. Schlöβer, M. Barz, R. Zentel, C. Trautwein, T. Lammers and F. Tacke, ACS Nano, 2017, 11, 9689 CrossRef CAS PubMed.
- R. P. McEver, Cardiovasc. Res., 2015, 107, 331 CrossRef CAS PubMed.
- K. Ley, Trends Mol. Med., 2003, 9, 263 CrossRef CAS PubMed.
- T. A. Springer, Cell, 1994, 76, 301 CrossRef CAS PubMed.
- I. Deguise, D. Lagnoux and R. Roy, New J. Chem., 2007, 31, 1321 RSC.
- R. S. Bagul, M. Hosseini, T. C. Shiao, N. K. Saadeh and R. Roy, Polym. Chem., 2017, 8, 5354 RSC.
- D. Goyard, B. Thomas, E. Gillon, A. Imberty and O. Renaudet, Front. Chem., 2019, 7, 666 CrossRef PubMed.
- J. P. Ribeiro, S. Villringer, D. Goyard, L. Coche-Guerente, M. Höferlin, O. Renaudet, W. Römer and A. Imberty, Chem. Sci., 2018, 9, 7634 RSC.
- S. P. Vincent, K. Buffet, I. Nierengarten, A. Imberty and J.-F. Nierengarten, Chem. – Eur. J., 2016, 22, 88 CrossRef CAS PubMed.
- A. Imberty, M. Wimmerova, E. P. Mitchell and N. Gilboa-Garber, Microbes Infect., 2004, 6, 221 CrossRef CAS PubMed.
- C. Chemani, A. Imberty, S. de Bentzmann, M. Pierre, M. Wimmerova, B. P. Guery and K. Faure, Infect. Immun., 2009, 77, 2065 CrossRef CAS PubMed.
- H. C. Flemming and J. Wingender, Nat. Rev. Microbiol., 2010, 8, 623 CrossRef CAS.
- Y. Zhao, Q. Guo, X. Dai, X. Wei, Y. Yu, X. Chen, C. Li, Z. Cao and X. Zhang, Adv. Mater., 2019, 31, 1806024 CrossRef PubMed.
- Y. Zhao, C. Yu, Y. Yu, X. Wei, X. Duan, X. Dai and X. Zhang, ACS Appl. Mater. Interfaces, 2019, 11, 39648 CrossRef CAS PubMed.
- G. C. Daskhan, H.-T. T. Tran, P. J. Meloncelli, T. L. Lowary, L. J. West and C. W. Cairo, Bioconjugate Chem., 2018, 29, 343 CrossRef CAS PubMed.
- L. Wu, Y. Zhang, Z. Li, G. Yang, Z. Kochovski, G. Chen and M. Jiang, J. Am. Chem. Soc., 2017, 139, 14684 CrossRef CAS PubMed.
- M. Gómez-García, J. M. Benito, D. Rodríguez-Lucena, J. Yu, K. Chmurski, C. Ortiz Mellet, R. Gutiérrez Gallego, A. Maestre, J. Defaye and J. M. García Fernández, J. Am. Chem. Soc., 2005, 127, 7970 CrossRef.
- M. François-Heude, A. Méndez-Ardoy, V. Cendret, P. Lafite, R. Daniellou, C. Ortiz Mellet, J. M. García Fernández, V. Moreau and F. Djedaïni-Pilard, Chem. – Eur. J., 2015, 21, 1978 CrossRef.
- L. Gallego-Yerga, M. Lomazzi, F. Sansone, C. Ortiz Mellet, A. Casnati and J. M. García Fernández, Chem. Commun., 2014, 50, 7440 RSC.
- N. Smiljanic, V. Moreau, D. Yockot, J. M. Benito, J. M. García Fernández and F. Djedaïni-Pilard, Angew. Chem., Int. Ed., 2006, 45, 5465 CrossRef CAS PubMed.
- I. Baussanne, J. M. Benito, C. Ortiz Mellet, J. M. García Fernández and J. Defaye, ChemBioChem, 2001, 2, 777 CrossRef CAS.
- Y. Oda, K.-I. Kasai and S.-I. Ishii, J. Biochem., 1981, 89, 285 CrossRef CAS PubMed.
- I. Otsuka, T. Hongo, H. Nakade, A. Narumi, R. Sakai, T. Satoh, H. Kaga and T. Kakuchi, Macromolecules, 2007, 40, 8930 CrossRef CAS.
- B.-B. Ke, L.-S. Wan and Z.-K. Xu, Langmuir, 2010, 26, 8946 CrossRef CAS PubMed.
- M. Kori, M. Sato, F. Abo, T. Masakatsu, K. Tatsuo and T. Nakahira, Eur. Polym. J., 2011, 47, 2351 CrossRef.
- L. Valtola, A. Rahikkala, J. Raula, E. I. Kaupinnen, H. Tenhu and S. Hietala, Eur. Polym. J., 2014, 59, 282 CrossRef CAS.
- J. L. Jiménez Blanco, C. Ortiz Mellet and J. M. García Fernández, Chem. Soc. Rev., 2013, 42, 4518 RSC.
- M. Gómez-García, J. M. Benito, R. Gutierrez-Gallego, A. Maestre, C. Ortiz Mellet, J. M. García Fernández and J. L. Jiménez Blanco, Org. Biomol. Chem., 2010, 8, 1849 RSC.
- M. Gómez-García, J. M. Benito, A. P. Butera, C. Ortiz Mellet, J. M. García Fernández and J. L. Jiménez Blanco, J. Org. Chem., 2012, 77, 1273 CrossRef PubMed.
- D. Ponader, P. Maffre, J. Aretz, D. Pussak, N. M. Ninnemann, S. Schmidt, P. H. Seeberger, C. Rademacher, G. U. Nienhaus and L. Hartmann, J. Am. Chem. Soc., 2014, 136, 2008 CrossRef CAS.
- S. A. Hill, C. Gerke and L. Hartman, Chem. – Asian J., 2018, 13, 3611 CrossRef CAS PubMed.
- K. S. Bücher, P. B. Konietzny, N. L. Snyder and L. Hartmann, Chem. – Eur. J., 2019, 25, 3301 Search PubMed.
- S. Toraskar, M. Gade, S. Sangabathuni, H. V. Thulasiram and R. Kikkeri, ChemMedChem, 2017, 12, 1116 CrossRef CAS PubMed.
- L. Xue, X. Xiong, K. Chen, Y. Luan, G. Chen and H. Chen, Polym. Chem., 2016, 7, 4263 RSC.
- Z. Liu, Y. Zhu, W. Ye, T. Wu, D. Miao, W. Deng and M. Liu, Polym. Chem., 2019, 10, 4006 RSC.
- M. Ortega-Muñoz, F. Pérez-Balderas, J. Morales-Sanfrutos, F. Hernández-Mateo, J. Isac-García and F. Santoyo-González, Eur. J. Org. Chem., 2009, 2454 CrossRef.
- M. Smadhi, S. de Bentzmann, A. Imberty, M. Gingras, R. Abderrahim and P. G. Goekjian, Beilstein J. Org. Chem., 2014, 10, 1981 CrossRef PubMed.
- S. Jiang, S. Niu, Z. H. Zhao, Z. J. Li and Q. Li, Carbohydr. Res., 2015, 414, 39 CrossRef CAS PubMed.
- G. C. Daskhan, C. Pifferi and O. Renaudet, ChemistryOpen, 2016, 5, 477 CrossRef CAS PubMed.
- T. Freichel, S. Eierhoff, N. L. Snyder and L. Hartmann, J. Org. Chem., 2017, 82, 9400 CrossRef CAS PubMed.
- M. Taouai, K. Chakroun, A. Vallin-Butruille, C. Cézard, M. Romdhani, D. Mathiron, D. Lesur, R. Abidi and M. Benazza, ARKIVOC, 2018, 186 Search PubMed.
- L. Otten and M. I. Gibson, RSC Adv., 2015, 5, 53911 RSC.
- L. Otten, D. Vlachou, S.-J. Richards and M. I. Gibson, Analyst, 2016, 141, 4305 RSC.
- M. Gade, C. Alex, S. L. Ben-Arye, J. T. Monteiro, S. Yehuda, B. Lepenies, V. Padler-Karavani and R. Kikkeri, ChemBioChem, 2018, 19, 1170 CrossRef CAS PubMed.
- A. Ogura, T. Tahara, S. Nozaki, K. Morimoto, Y. Kizuka, S. Kitazume, M. Hara, S. Kojima, H. Onoe, A. Kurbangalieva, N. Taniguchi, Y. Watanabe and K. Tanaka, Sci. Rep., 2016, 6, 21797 CrossRef CAS PubMed.
- T. Tanaka, Org. Biomol. Chem., 2016, 14, 7597 RSC.
- R. Sibgatullina, K. Fujiki, T. Murase, T. Yamamoto, T. Shimoda, A. Kurbangalieva and K. Tanaka, Tetrahedron Lett., 2017, 58, 1929 CrossRef CAS.
- L. Latypova, R. Sibgatullina, A. Ogura, K. Fujiki, A. Khabibrakhmanova, T. Tahara, S. Nozaki, S. Urano, K. Tsubokura, H. Onoe, Y. Watanabe, A. Kurbangalieva and K. Tanaka, Adv. Sci., 2017, 4, 1600394 CrossRef.
- A. Ogura, S. Urano, T. Tahara, S. Nozaki, R. Sibgatullina, K. Vong, T. Suzuki, N. Dohmae, A. Kurbangalieva, Y. Watanabe and K. Tanaka, Chem. Commun., 2018, 54, 8693 RSC.
- K. Nakamura, K. Tsubokura, A. Kurbangalieva, Y. Nakao, T. Murase, T. Shimoda and K. Tanaka, J. Carbohydr. Chem., 2019, 38, 127 CrossRef CAS.
- N. C. Worstell, P. Krishnan, J. D. Weatherston and H. J. Wu, PLoS One, 2016, 11, e0153265 CrossRef.
- P. Krishnan, A. Singla, C.-A. Lee, J. D. Weatherston, N. C. Worstell and H.-J. Wu, Colloids Surf., B, 2017, 160, 281 CrossRef CAS PubMed.
- M. Yanagisawa, T. Ariga and R. K. Yu, Glycobiology, 2006, 16, 19G CrossRef CAS PubMed.
- H. Lin, E. N. Kitova and J. S. Klassen, J. Am. Soc. Mass Spectrom, 2014, 25, 104 CrossRef CAS PubMed.
- J. Li, X. Fan, E. N. Kitova, C. Zou, C. W. Cairo, L. Eugenio, K. K. Ng, Z. J. Xiong, G. G. Prive and J. S. Klassen, Anal. Chem., 2016, 88, 4742 CrossRef CAS.
- L. Han, E. N. Kitova and J. S. Klassen, J. Am. Soc. Mass Spectrom, 2016, 27, 1878 CrossRef CAS.
- K. Momoeda, K. Hirota, T. Utsuki, Y. Tsuchida, K. Hanaoka and M. Iwamori, J. Biochem., 1996, 119, 1189 CrossRef CAS PubMed.
- M. E. Breimer, G. C. Hansson, K.-A. Karlsson, G. Larson and H. Leffler, Glycobiology, 2012, 22, 1721 CrossRef CAS.
- N. C. Worstell, A. Singla, P. Saenkham, T. Galbadage, P. Sule, D. Lee, A. Mohr, J. S.-I. Kwon, J. D. Cirillo and H.-J. Wu, Sci. Rep., 2018, 8, 8419 CrossRef.
- N. C. Worstell, A. Singla and H.-J. Wu, Colloids Surf., B, 2019, 175, 84 CrossRef CAS.
- A. Singla, N. C. Worstell, P. Saenkham, T. Galbadage, P. Sule, K. Sha, C. L. Cannon, J. D. Cirillo and H.-J. Wu, Glycobiology, 2018, 28, 1061 Search PubMed.
- S. B. Simbassa, A. Singla, K. N. Shah, Q. Chen, B. Chirra, N. Dangol, R. Raina, H.-J. Wu and C. Cannon, Pediatr. Pulmunol., 2019, 54, S266 Search PubMed.
- H.-K. Choi, D. Lee, A. Singla, J. S.-I. Kwon and H.-J. Wu, Glycobiology, 2019, 29, 397 CrossRef CAS PubMed.
- L. E. Wilkins, N. Badi, F. Du Prez and M. I. Gibson, ACS Macro Lett., 2018, 7, 1498 CrossRef CAS PubMed.
- B. Martyn, C. I. Biggs and M. I. Gibson, J. Polym. Sci., Part A: Polym. Chem., 2019, 57, 40 CrossRef CAS.
- S. Zhang, R.-O. Moussodia, S. Vértesy, S. André, M. L. Klein, H.-J. Gabius and V. Percec, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 5585 CrossRef CAS.
- C. D. Navo, F. Corzana, E. M. Sánchez-Fernández, J. H. Busto, A. Avenoza, M. M. Zurbano, E. Nanba, K. Higaki, C. Ortiz Mellet, J. M. García Fernández and J. M. Peregrina, Org. Biomol. Chem., 2016, 14, 1473 RSC.
- J. Castilla, R. Rísquez, K. Higaki, E. Nanba, K. Ohno, Y. Suzuki, Y. Díaz, C. Ortiz Mellet, J. M. García Fernández and S. Castillón, Eur. J. Med. Chem., 2015, 90, 258 CrossRef CAS PubMed.
- J. Castilla, R. Rísquez, D. Cruz, K. Higaki, E. Nanba, K. Ohno, Y. Suzuki, Y. Díaz, C. Ortiz Mellet, J. M. García Fernández and S. Castillón, J. Med. Chem., 2012, 55, 6857 CrossRef CAS PubMed.
- G. Horne, F. X. Wilson, J. Tinsley, D. H. Williams and R. Storer, Drug Discovery Today, 2011, 16, 107 CrossRef CAS PubMed.
- Iminosugars: From Synthesis to Therapeutic Applications, ed. P. Compain and O. R. Martin, Wiley-VCH, Weinheim, Germany, 2007 Search PubMed.
- J. Diot, M. I. Garcia-Moreno, S. G. Gouin, C. Ortiz Mellet, K. Haupt and J. Kovensky, Org. Biomol. Chem., 2009, 7, 357 RSC.
- P. Compain, C. Decroocq, J. Iehl, M. Holler, D. Hazelard, T. Mena Barragan, C. Ortiz Mellet and J.-F. Nierengarten, Angew. Chem., Int. Ed., 2010, 49, 5753 CrossRef CAS PubMed.
- P. Compain and A. Bodlenner, ChemBioChem, 2014, 15, 1239 CrossRef CAS PubMed.
- S. Gouin, Chem. – Eur. J., 2014, 20, 11616 CrossRef CAS PubMed.
- R. Zelli, J.-F. Longevial, P. Dumy and A. Marra, New J. Chem., 2015, 39, 5050 RSC.
- N. Kanfar, E. Bartolami, R. Zelli, A. Marra, J.-Y. Winum and P. Dumy, Org. Biomol. Chem., 2015, 13, 9894 RSC.
- C. Matassini, C. Parmeggiani, F. Cardona and A. Goti, Tetrahedron Lett., 2016, 57, 5407 CrossRef CAS.
- P. Compain, Chem. Rec., 2020, 20, 10 CrossRef CAS PubMed.
- C. Decroocq, D. Rodríguez-Lucena, V. Russo, T. Mena Barragán, C. Ortiz Mellet and P. Compain, Chem. – Eur. J., 2011, 17, 13825 CrossRef CAS PubMed.
- F. Cardona, G. Isoldi, F. Sansone, A. Casnati and A. Goti, J. Org. Chem., 2012, 77, 6980 CrossRef CAS PubMed.
- C. Decroocq, A. Joosten, R. Sergent, T. Mena Barragán, C. Ortiz Mellet and P. Compain, ChemBioChem, 2013, 14, 2038 CrossRef CAS PubMed.
- Y. Brissonnet, C. Ortiz Mellet, S. Morandat, M. I. Garcia Moreno, D. Deniaud, S. E. Matthews, S. Vidal, S. Šesták, K. El Kirat and S. G. Gouin, J. Am. Chem. Soc., 2013, 135, 18427 CrossRef CAS PubMed.
- A. Joosten, J. P. Schneider, M. L. Lepage, C. Tarnus, A. Bodlenner and P. Compain, Eur. J. Org. Chem., 2014, 1866 CrossRef CAS.
- G. D’Adamio, C. Parmeggiani, A. Goti, A. J. Moreno-Vargas, E. Moreno-Clavijo, I. Robina and F. Cardona, Org. Biomol. Chem., 2014, 12, 6250 RSC.
- C. Bonduelle, J. Huang, T. Mena-Barragán, C. Ortiz Mellet, C. Decroocq, E. Etamé, A. Heise, P. Compain and S. Lecommandoux, Chem. Commun., 2014, 50, 3350 RSC.
- C. Matassini, M. Marradi, F. Cardona, C. Parmeggiani, I. Robina, A. J. Moreno-Vargas, S. Penadés and A. Goti, RSC Adv., 2015, 5, 95817 RSC.
- C. Matassini, S. Mirabella, A. Goti, I. Robina, A. J. Moreno-Vargas and F. Cardona, Beilstein J. Org. Chem., 2015, 11, 2631 CrossRef CAS PubMed.
- R. Zelli, E. Bartolami, J.-F. Longevial, Y. Bessin, P. Dumy, A. Marra and S. Ulrich, RSC Adv., 2016, 6, 2210 RSC.
- R. Zelli, S. Tommasone, P. Dumy, A. Marra and A. Dondoni, Eur. J. Org. Chem., 2016, 5102 CrossRef CAS.
- F. Stauffert, A. Bodlenner, T. M. N. Trinh, M. I. García-Moreno, C. Ortiz Mellet, J.-F. Nierengarten and P. Compain, New J. Chem., 2016, 40, 7421 RSC.
- M. L. Lepage, J. P. Schneider, A. Bodlenner, A. Meli, F. De Riccardis, M. Schmitt, C. Tarnus, N.-T. Nguyen-Huynh, Y.-N. François, E. Leize-Wagner, C. Birck, A. Cousido-Siah, A. Podjarny, I. Izzo and P. Compain, Chem. – Eur. J., 2016, 22, 5151 CrossRef CAS PubMed.
- T. Hurtaux, G. Sfihi-Loualia, Y. Brissonnet, J. Bouckaert, J.-M. Mallet, B. Sendid, F. Delplace, E. Fabre, S. G. Gouin and Y. Guerardel, Carbohydr. Res., 2016, 429, 123 CrossRef CAS PubMed.
- D. Alvarez-Dorta, Y. Brissonnet, A. Saumonneau, D. Deniaud, J. Bernard, X. Yan, C. Tellier, F. Daligault and S. G. Gouin, ChemistrySelect, 2017, 2, 9552 CrossRef CAS.
- T. M. N. Trinh, M. Holler, J. P. Schneider, M. I. García-Moreno, J. M. García Fernández, A. Boldlenner, P. Compain, C. Ortiz Mellet and J.-F. Nierengarten, J. Mater. Chem. B, 2017, 5, 6546 RSC.
- D. Alvarez-Dorta, D. T. King, T. Legigan, D. Ide, I. Adachi, D. Deniaud, J. Désiré, A. Kato, D. Vocadlo, S. G. Gouin and Y. Blériot, Chem. – Eur. J., 2017, 23, 9022 CrossRef CAS PubMed.
- S. Mirabella, G. D’Adamio, C. Matassini, A. Goti, S. Delgado, A. Gimeno, I. Robina, A. J. Moreno-Vargas, S. Šesták, J. Jimenez-Barbero and F. Cardona, Chem. – Eur. J., 2017, 23, 14585 CrossRef CAS PubMed.
- C. Matassini, C. Vanni, A. Goti, A. Morrone, M. Marradi and F. Cardona, Org. Biomol. Chem., 2018, 16, 8604 RSC.
- J. F. Nierengarten, J. P. Schneider, T. M. Nguyet Trinh, A. Joosten, M. Holler, M. L. Lepage, A. Bodlenner, M. I. García-Moreno, C. Ortiz Mellet and P. Compain, Chem. – Eur. J., 2018, 24, 2483 CrossRef CAS PubMed.
- M. Martínez-Bailén, E. Jiménez-Ortega, A. T. Carmona, I. Robina, J. Sanz-Aparicio, D. Talens-Perales, J. Polaina, C. Matassini, F. Cardona and A. J. Moreno-Vargas, Bioorg. Chem., 2019, 89, 103026 CrossRef.
- M. Li, K.-R. Wang, J.-X. Yang, Y.-T. Peng, Y.-X. Liu, H.-X. Zhang and X.-L. Li, J. Mater. Chem. B, 2019, 7, 1379 RSC.
- M. M. Pichon, F. Stauffert, A. Bodlenner and P. Compain, Org. Biomol. Chem., 2019, 17, 5801 RSC.
- E. Howard, A. Cousido-Siah, M. L. Lepage, J. P. Schneider, A. Bodlenner, A. Mitschler, A. Meli, I. Izzo, A. Alvarez, A. Podjarny and P. Compain, Angew. Chem., Int. Ed., 2018, 57, 8002 CrossRef CAS PubMed.
- R. Rísquez-Cuadro, J. M. García Fernández, J.-F. Nierengarten and C. Ortiz Mellet, Chem. – Eur. J., 2013, 19, 16791 CrossRef PubMed.
- J. L. Jiménez Blanco, V. M. Díaz Pérez, C. Ortiz Mellet, J. Fuentes, J. M. García Fernández, J. C. Díaz Arribas and F. J. Cañada, Chem. Commun., 1997, 1969 RSC.
- V. M. Díaz Pérez, M. I. García-Moreno, C. Ortiz Mellet, J. Fuentes, J. M. García Fernández, J. C. Díaz Arribas and F. J. Cañada, J. Org. Chem., 2000, 65, 136 CrossRef PubMed.
- J. M. García Fernández, C. Ortiz Mellet, J. M. Benito and J. Fuentes, Synlett, 1998, 316 CrossRef.
- V. M. Díaz Pérez, M. I. García-Moreno, C. Ortiz Mellet and J. M. García Fernández, Synlett, 2003, 341 Search PubMed.
- T. Mena-Barragán, M. I. García-Moreno, A. Sevšek, T. Okazaki, E. Nanba, K. Higaki, N. I. Martin, R. J. Pieters, J. M. García Fernández and C. Ortiz Mellet, Molecules, 2018, 23, 927 CrossRef.
- M. I. García-Moreno, J. M. Benito, C. Ortiz Mellet and J. M. García Fernández, J. Org. Chem., 2001, 66, 7604 CrossRef.
- M. I. García-Moreno, C. Ortiz Mellet and J. M. García Fernández, Eur. J. Org. Chem., 2004, 1803 CrossRef.
- A. de la Fuente, T. Mena-Barragán, R. A. Farrar-Tobar, X. Verdaguer, J. M. García Fernández, C. Ortiz Mellet and A. Riera, Org. Biomol. Chem., 2015, 13, 6500 RSC.
- V. M. Díaz Pérez, M. I. García-Moreno, C. Ortiz Mellet, J. M. García Fernández, C. Ortiz Mellet, J. M. Benito and J. Fuentes, Synlett, 2003, 341 Search PubMed.
- M. I. García-Moreno, C. Ortiz Mellet and J. M. García Fernández, Tetrahedron, 2007, 63, 7879 CrossRef.
- M. Aguilar, T. M. Gloster, M. I. García-Moreno, C. Ortiz Mellet, G. J. Davies, A. Llebaria, J. Casas, M. Egido-Gabás and J. M. García Fernández, ChemBioChem, 2008, 9, 2612 CrossRef CAS.
- P. Alfonso, V. Andreu, A. Pino-Ángeles, A. A. Moya-García, M. I. García-Moreno, J. C. Rodríguez-Rey, F. Sánchez-Jiménez, M. Pocoví, C. Ortiz Mellet, J. M. García Fernández and P. Giraldo, ChemBioChem, 2013, 14, 943 CrossRef CAS.
- M. De la Mata, D. Cotán, M. Oropesa-Ávila, J. Garrido-Maraver, M. D. Cordero, M. Villanueva Paz, A. Delgado Pavón, E. Alcocer-Gómez, I. de Lavera, P. Ybot-González, A. P. Zaderenko, C. Ortiz Mellet, J. M. García Fernández and J. A. Sánchez Alcázar, Sci. Rep., 2015, 5, 10903 CrossRef CAS.
- Y. Yu, T. Mena-Barragán, K. Higaki, J. Johnson, J. Drury, R. Lieberman, N. Nakasone, H. Ninomiya, T. Tsukimura, H. Sakuraba, Y. Suzuki, E. Nanba, C. Ortiz Mellet, J. M. García Fernández and K. Ohno, ACS Chem. Biol., 2014, 9, 1460 CrossRef CAS PubMed.
- T. Mena-Barragán, A. Narita, D. Matias, G. Tiscornia, E. Nanba, K. Ohno, Y. Suzuki, K. Higaki, J. M. Garcia Fernández and C. Ortiz Mellet, Angew. Chem., Int. Ed., 2015, 54, 11696 CrossRef.
- M. I. García-Moreno, M. de la Mata, E. M. Sánchez-Fernández, J. M. Benito, A. Díaz-Quintana, S. Fustero, E. Nanba, K. Higaki, J. A. Sánchez-Alcázar, J. M. García Fernández and C. Ortiz Mellet, J. Med. Chem., 2017, 60, 1829 CrossRef PubMed.
- M. I. García-Moreno, P. Díaz-Pérez, C. Ortiz Mellet and J. M. García Fernández, Chem. Commun., 2002, 848 RSC.
- M. I. García-Moreno, P. Díaz-Pérez, C. Ortiz Mellet and J. M. García Fernández, J. Org. Chem., 2003, 68, 8890 CrossRef.
- Z. Luan, K. Higaki, M. Aguilar-Moncayo, H. Ninomiya, K. Ohno, M. I. García-Moreno, C. Ortiz Mellet, J. M. García Fernández and Y. Suzuki, ChemBioChem, 2009, 10, 2780 CrossRef CAS.
- Z. Luan, K. Higaki, M. Aguilar-Moncayo, L. Li, H. Ninomiya, E. Nanba, K. Ohno, M. I. García-Moreno, C. Ortiz Mellet, J. M. García Fernández and Y. Suzuki, ChemBioChem, 2010, 11, 2453 CrossRef CAS PubMed.
- G. Tiscornia, E. Lorenzo Vivas, L. Matalonga, I. Berniakovich, M. Barragán Monasterio, C. E. Argaiz, L. Gort, F. González, C. Ortiz Mellet, J. M. García Fernández, A. Ribes, A. Veiga and J. C. Izpisua Belmonte, Hum. Mol. Genet., 2013, 22, 633 CrossRef CAS PubMed.
- J. Rodríguez-Lavado, M. de la Mata Fernández, J. L. Jiménez Blanco, M. I. García-Moreno, J. M. Benito, A. Díaz-Quintana, J. A. Sánchez Alcázar, K. Higaki, E. Nanba, K. Ohno, Y. Suzuki, C. Ortiz Mellet and J. M. García Fernández, Org. Biomol. Chem., 2014, 12, 2289 RSC.
- E. M. Sánchez-Fernández, E. Álvarez, C. Ortiz Mellet and J. M. García Fernández, J. Org. Chem., 2014, 79, 11722 CrossRef PubMed.
- M. Aguilar-Moncayo, T. M. Gloster, J. P. Turkenburg, M. I. García-Moreno, C. Ortiz Mellet, G. J. Davies and J. M. García Fernández, Org. Biomol. Chem., 2009, 7, 2738 RSC.
- M. Aguilar-Moncayo, M. I. García-Moreno, A. Trapero, M. Egido-Gabás, A. Llebaria, J. M. García Fernández and C. Ortiz Mellet, Org. Biomol. Chem., 2011, 9, 3698 RSC.
- M. Aguilar-Moncayo, T. Takai, K. Higaki, T. Mena-Barragán, Y. Hirano, K. Yura, L. Li, Y. Yu, H. Ninomiya, M. I. García-Moreno, S. Ishii, Y. Sakakibara, K. Ohno, E. Nanba, C. Ortiz Mellet, J. M. García Fernández and Y. Suzuki, Chem. Commun., 2012, 48, 6514 RSC.
- T. Takai, K. Higaki, M. Aguilar-Moncayo, T. Mena-Barragán, Y. Hirano, K. I. Yura, L. Yu, H. Ninomiya, M. I. García-Moreno, Y. Sakakibara, K. Ohno, E. Nanba, C. Ortiz Mellet, J. M. García Fernández and Y. Suzuki, Mol. Ther., 2013, 21, 526 CrossRef CAS PubMed.
- S. Silva, E. M. Sánchez-Fernández, C. Ortiz Mellet, A. Tatibouët, A. P. Rauter and P. Rollin, Eur. J. Org. Chem., 2013, 7941 CrossRef CAS.
- H. Suzuki, U. Ohto, K. Higaki, T. Mena-Barragan, M. Aguilar-Moncayo, C. Ortiz Mellet, E. Nanba, J. M. Garcia Fernandez, Y. Suzuki and T. Shimizu, J. Biol. Chem., 2014, 289, 14560 CrossRef CAS PubMed.
- T. Mena-Barragán, M. I. García-Moreno, E. Nanba, K. Higaki, A. L. Concia, P. Clapés, J. M. García Fernández and C. Ortiz Mellet, Eur. J. Med. Chem., 2016, 121, 880 CrossRef PubMed.
- A. de la Fuente, R. Rísquez-Cuadro, X. Verdaguer, J. M. García Fernández, E. Nanba, K. Higaki, C. Ortiz Mellet and A. Riera, Eur. J. Med. Chem., 2016, 121, 926 CrossRef CAS PubMed.
- M. Aguilar, V. M. Díaz-Pérez, M. I. García-Moreno, C. Ortiz Mellet and J. M. García Fernández, J. Org. Chem., 2008, 73, 1995 CrossRef CAS PubMed.
- R. Kooij, H. M. Branderhorst, S. Bonte, S. Wieclawska, N. I. Martin and R. J. Pieters, Med. Chem. Commun., 2013, 4, 387 RSC.
- M. Benltifa, M. I. García-Moreno, C. Ortiz Mellet, J. M. García Fernández and A. Wadouachi, Bioorg. Med. Chem. Lett., 2008, 18, 2805 CrossRef CAS PubMed.
- M. Aguilar-Moncayo, C. Ortiz Mellet, J. M. García Fernández and M. I. García-Moreno, J. Org. Chem., 2009, 74, 3595 CrossRef CAS PubMed.
- E. M. Sánchez-Fernández, R. Rísquez-Cuadro, M. Aguilar-Moncayo, M. I. García-Moreno, C. Ortiz Mellet and J. M. García Fernández, Org. Lett., 2009, 11, 3306 CrossRef PubMed.
- E. M. Sánchez-Fernández, R. Rísquez-Cuadro, M. Chasseraud, A. Ahidouch, C. Ortiz Mellet, H. Ouadid-Ahidouch and J. M. García Fernández, Chem. Commun., 2010, 46, 5328 RSC.
- E. M. Sánchez-Fernández, R. Rísquez-Cuadro, C. Ortiz Mellet, J. M. García Fernández, P. M. Nieto-Mesa and J. Angulo-Álvarez, Chem. – Eur. J., 2012, 18, 8527 CrossRef PubMed.
- G. Allan, H. Ouadid-Ahidouch, E. M. Sánchez-Fernández, R. Rísquez-Cuadro, J. M. García Fernández, C. Ortiz Mellet and A. Ahidouch, PLoS One, 2013, 8, e76411 CrossRef CAS PubMed.
- E. M. Sánchez-Fernández, V. Gómez-Perez, R. García-Hernández, J. M. García Fernández, G. B. Plata, J. M. Padrón, C. Ortiz Mellet, S. Castanys-Cuello and F. Gamarro-Conde, RSC Adv., 2015, 5, 21812 RSC.
- E. M. Sánchez-Fernández, R. Gonçalves-Pereira, R. Rísquez-Cuadro, G. B. Plata, J. M. Padrón, J. M. García Fernández and C. Ortiz Mellet, Carbohydr. Res., 2016, 429, 113 CrossRef PubMed.
- E. M. Sánchez Fernández, C. D. Navo, N. Martínez-Sáez, R. Gonçalves-Pereira, V. J. Somovilla, A. Avenoza, J. H. Busto, G. J. L. Bernardes, G. Jiménez-Osés, F. Corzana, J. M. García Fernández, C. Ortiz Mellet and J. M. Peregrina, Org. Lett., 2016, 18, 3890 CrossRef PubMed.
- A. I. Arroba, E. Alcalde-Estévez, M. García-Ramírez, D. Cazzoni, P. de la Villa, E. M. Sánchez-Fernández, C. Ortiz Mellet, J. M. García Fernández, C. Hernández, R. Simó and A. M. Valverde, Biochim. Biophys. Acta, Mol. Basis Dis., 2016, 1862, 1663 CrossRef CAS PubMed.
- N. Gueder, G. Allan, M.-S. Telliez, F. Hague, J. M. García Fernández, E. M. Sánchez-Fernández, C. Ortiz Mellet, A. Ahidouch and H. Ouadid-Ahidouch, J. Cell. Physiol., 2017, 232, 3631 CrossRef CAS PubMed.
- E. Alcalde-Estévez, A. I. Arroba, E. M. Sánchez-Fernández, C. Ortiz Mellet, J. M. García Fernández, L. Masgrau and A. M. Valverde, Food Chem. Toxicol., 2018, 111, 456 CrossRef PubMed.
- E. Schaeffer, E. M. Sánchez-Fernández, R. Gonçalves-Pereira, V. Flacher, D. Lamon, M. Duval, J.-D. Fauny, J. M. García Fernández, C. G. Mueller and C. Ortiz Mellet, Eur. J. Med. Chem., 2019, 169, 111 CrossRef CAS PubMed.
- E. M. Sánchez-Fernández, M. García-Moreno, A. I. Arroba, M. Aguilar-Diosdado, J. M. Padrón, R. García-Hernández, F. Gamarro, S. Fustero, J.-E. Sánchez-Aparicio, L. Masgrau, J. M. García Fernández and C. Ortiz Mellet, Eur. J. Med. Chem., 2019, 182, 111604 CrossRef PubMed.
- S. Cuyvers, E. Dornez, J. A. Delcour and C. M. Courtin, Crit. Rev. Biotechnol., 2012, 32, 93 CrossRef CAS PubMed.
- Y. Brissonnet, S. Ladevèze, D. Tezé, E. Fabre, D. Deniaud, F. Daligault, C. Tellier, S. Šesták, M. Remaud-Simeon, G. Potocki-Veronese and S. G. Gouin, Bioconjugate Chem., 2015, 26, 766 CrossRef CAS PubMed.
- E. M. Sánchez-Fernández, J. M. García Fernández and C. Ortiz Mellet, Chem. Commun., 2016, 52, 5497 RSC.
- J. M. García Fernández and C. Ortiz Mellet, ACS Med. Chem. Lett., 2019, 10, 1020 CrossRef PubMed.
- C. Decroocq, D. Rodrıíguez-Lucena, K. Ikeda, N. Asano and P. Compain, ChemBioChem, 2012, 13, 661 CrossRef CAS PubMed.
- A. Joosten, C. Decroocq, J. de Sousa, J. Schneider, E. Etamé, A. Bodlenner, T. D. Butters and P. Compain, ChemBioChem, 2014, 15, 309 CrossRef CAS PubMed.
- E. Laigre, D. Hazelard, J. Casas, J. Serra-Vinardell, H. Michelakakis, I. Mavridou, J. M. F. G. Aerts, A. Delgado and P. Compain, Carbohydr. Res., 2016, 429, 98 CrossRef CAS PubMed.
- R. Rísquez-Cuadro, R. Matsumoto, F. Ortega-Caballero, E. Nanba, K. Higaki, J. M. García Fernández and C. Ortiz Mellet, J. Med. Chem., 2019, 62, 5832 CrossRef PubMed.
- P. Compain, C. Decroocq, A. Joosten, J. de Sousa, D. Rodriguez-Lucena, T. D. Butters, J. Bertrand, R. Clément, C. Boinot, F. Becq and C. Norez, ChemBioChem, 2013, 14, 2050 CrossRef CAS PubMed.
- E. T. Sletten, R. S. Loka, F. Yu and H. M. Nguyen, Biomacromolecules, 2017, 18, 3387 CrossRef CAS PubMed.
- R. S. Loka, F. Yu, E. T. Sletten and H. M. Nguyen, Chem. Commun., 2017, 53, 9163 RSC.
- R. S. Loka, E. T. Sletten, U. Barash, I. Vlodavsky and H. M. Nguyen, ACS Appl. Mater. Interfaces, 2019, 11, 244 CrossRef CAS PubMed.
- R. Koide and S.-I. Nishimura, Angew. Chem., Int. Ed., 2019, 58, 1413 CrossRef PubMed.
- A. Siriwardena, M. Khanal, A. Barras, O. Bande, T. Mena-Barragan, C. Ortiz Mellet, J. M. García Fernández, R. Boukherroubb and S. Szunerits, RSC Adv., 2015, 5, 100568 RSC.
- M. Abellán Flos, M. I. García Moreno, C. Ortiz Mellet, J. M. García Fernández, J.-F. Nierengarten and S. P. Vincent, Chem. – Eur. J., 2016, 22, 11450 CrossRef PubMed.
- M. I. García-Moreno, F. Ortega-Caballero, R. Rísquez-Cuadro, C. Ortiz Mellet and J. M. García Fernández, Chem. – Eur. J., 2017, 23, 6295 CrossRef.
- U. Alali, A. Vallin, A. Bil, T. Khanchouche, D. Mathiron, C. Przybylski, R. Beaulieu, J. Kovensky, M. Benazza and V. Bonnet, Org. Biomol. Chem., 2019, 17, 7228 RSC.
- D. S. Tawfik, Nat. Chem. Biol., 2010, 6, 692 CrossRef PubMed.
- M. González-Cuesta, D. Goyard, E. Nanba, K. Higaki, J. M. García Fernández, O. Renaudet and C. Ortiz Mellet, Chem. Commun., 2019, 55, 12845 RSC.
- M. François-Heude, A. Méndez-Ardoy, V. Cendret, P. Lafite, R. Daniellou, C. Ortiz Mellet, J. M. García Fernández, V. Moreau and F. Djedaïni-Pilard, Chem. – Eur. J., 2015, 21, 1978 CrossRef PubMed.
- L. Gallego-Yerga, M. Lomazzi, F. Sansone, C. Ortiz Mellet, A. Casnati and J. M. García Fernández, Chem. Commun., 2014, 50, 7440 RSC.
- N. Horan, L. Yan, H. Isobe, G. M. Whitesides and D. Kahne, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11782 CrossRef CAS PubMed.
- A. G. Barrientos, J. M. de la Fuente, M. Jiménez, D. C. Solis, F. J. Cañada, M. Martin-Lomas and S. Penades, Carbohydr. Res., 2009, 344, 1474 CrossRef CAS PubMed.
- Y. Brissonnet, G. Compain, B. Renoux, E.-M. Krammer, F. Daligault, D. Deniaud, S. Papot and S. G. Gouin, RSC Adv., 2019, 9, 40263 RSC.
- Y. Brissonnet, C. Assailly, A. Saumonneau, J. Bouckaert, M. Maillasson, C. Petitot, B. Roubinet, B. Didak, L. Landemarre, C. Bridot, R. Blossey, D. Deniaud, X. Yan, J. Bernard, C. Tellier, C. Grandjean, F. Daligault and S. G. Gouin, Chem. – Eur. J., 2019, 25, 2358 CrossRef CAS PubMed.
- M. Durka, K. Buffet, J. Iehl, M. Holler, J. F. Nierengarten and S. P. Vincent, Chem. – Eur. J., 2012, 18, 641 CrossRef CAS PubMed.
- T. Hurtaux, G. Sfihi-Loualia, Y. Brissonnet, J. Bouckaert, J.-M. Mallet, B. Sendid, F. Delplace, E. Fabre, S. G. Gouin and Y. Guérardel, Carbohydr. Res., 2016, 429, 123 CrossRef CAS PubMed.
- A. Tikad, H. Fu, C. M. Sevrain, S. Laurent, J.-F. Nierengarten and S. P. Vincent, Chem. – Eur. J., 2016, 22, 13147 CrossRef CAS PubMed.
- C. Pifferi, B. Thomas, D. Goyard, N. Berthet and O. Renaudet, Chem. – Eur. J., 2017, 23, 16283 CrossRef CAS PubMed.
- S. I. S. Hendrikse, L. Su, T. P. Hogervorst, R. P. M. Lafleur, X. Lou, G. A. van der Marel, J. D. C. Codee and E. W. Meijer, J. Am. Chem. Soc., 2019, 141, 13877 CrossRef CAS PubMed.
- S.-H. Cha, J. Hong, M. McGuffie, B. Yeom, J. S. VanEpps and N. A. Kotov, ACS Nano, 2015, 9, 9097 CrossRef CAS PubMed.
- A. Díaz-Moscoso, N. Guilloteau, C. Bienvenu, A. Méndez-Ardoy, J. L. Jiménez Blanco, J. M. Benito, L. le Gourrierec, C. Di Giorgio, P. Vierling, J. Defaye, C. Ortiz Mellet and J. M. García Fernández, Biomaterials, 2011, 32, 7263 CrossRef PubMed.
- N. Symens, A. Méndez-Ardoy, A. Díaz-Moscoso, E. Sánchez-Fernández, K. Remaut, J. Demeester, J. M. García Fernández, S. C. de Smedt and J. Rejman, Bioconjugate Chem., 2012, 23, 1276 CrossRef CAS PubMed.
- E. M. Aguilar Moncayo, N. Guilloteau, C. Bienvenu, J. L. Jiménez Blanco, C. Di Giorgio, P. Vierling, J. M. Benito, C. Ortiz Mellet and J. M. García Fernández, New J. Chem., 2014, 38, 5215 RSC.
- A. Méndez-Ardoy, A. Díaz-Moscoso, C. Ortiz Mellet, C. Di Giorgio, P. Vierling, J. M. Benito and J. M. García Fernández, RSC Adv., 2015, 5, 76464 RSC.
- A. I. Carbajo-Gordillo, J. Rodríguez-Lavado, J. L. Jiménez Blanco, J. M. Benito, C. Di Giorgio, I. Vélaz, C. Tros de Ilarduya, C. Ortiz Mellet and J. M. García Fernández, Chem. Commun., 2019, 55, 8227 RSC.
- M. L. Huang, C. J. Fisher and K. Godula, Exp. Biol. Med., 2016, 241, 1042 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |