Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A versatile cholera toxin conjugate for neuronal targeting and tracing†
Jessica L.
Haigh
 ab,
Daniel J.
Williamson
a,
Emma
Poole
ab,
Daniel J.
Williamson
a,
Emma
Poole
 a,
Yuan
Guo
a,
Yuan
Guo
 c,
Dejian
Zhou
c,
Dejian
Zhou
 a,
Michael E.
Webb
a,
Michael E.
Webb
 a,
Susan A.
Deuchars
a,
Susan A.
Deuchars
 b,
Jim
Deuchars
b,
Jim
Deuchars
 *b and
W. Bruce
Turnbull
*b and
W. Bruce
Turnbull
 *a
*a
aSchool of Chemistry and Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds LS2 9JT, UK. E-mail: w.b.turnbull@leeds.ac.uk
bSchool of Biomedical Sciences, Faculty of Biological Sciences, University of Leeds, Leeds LS2 9JT, UK
cSchool of Food Science and Nutrition, University of Leeds, Leeds LS2 9JT, UK
First published on 28th April 2020
Abstract
Tracing of neurons plays an essential role in elucidating neural networks in the brain and spinal cord. Cholera toxin B subunit (CTB) is already widely used as a tracer although its use is limited by the need for immunohistochemical detection. A new construct incorporating non-canonical azido amino acids (azido-CTB) offers a novel way to expand the range and flexibility of this neuronal tracer. Azido-CTB can be detected rapidly in vivo following intramuscular tongue injection by ‘click’ chemistry, eliminating the need for antibodies. Cadmium selenide/zinc sulfide (CdSe/ZnS) core/shell nanoparticles were attached to azido-CTB by strain-promoted alkyne–azide cycloaddition to make a nano-conjugate. Following tongue injections the complex was detected in vivo in the brainstem by light microscopy and electron microscopy via silver enhancement. This method does not require membrane permeabilization and so ultrastructure is maintained. Azido-CTB offers new possibilities to enhance the utility of CTB as a neuronal tracer and delivery vehicle by modification using ‘click’ chemistry.
Cholera toxin produced by Vibrio cholerae is usually an infectious agent of the gut, but the toxin also has the potential to be developed for more versatile tracing and targeting of neurons. It is formed of two main parts: the catalytically active A subunit and the non-toxic pentamer of B subunits (CTB) that facilitate cell entry via binding ganglioside GM1.1,2 GM1 is ubiquitously expressed in vertebrates but is found in high relative abundance in the nervous system, including in the periphery of motor neurons at the neuro-muscular junction; for this reason CTB has been used extensively as a neuronal tracer.3–6 It is usually detected via antibody-mediated detection using fluorescence or 3′-Diaminobenzidine (DAB) immunohistochemistry,7–9 but direct fluorophore conjugates are also available10,11 as well as CTB–horseradish peroxidase (HRP) conjugates for detection via peroxidase chemistry.4,12 The ability to detect the tracer directly without the need of antibodies is advantageous. It does not require an amplification step and so the labelling process is quicker, and cheaper as no antibodies are required. Detecting proteins without immunohistochemistry also provides more scope for labelling multiple proteins without the need to consider cross-reactivity of primary and secondary antibodies as well as removing issues with antibody batch variability and limited shelf-life. Existing fluorescein–CTB conjugates are prone to bleaching, CTB–HRP conjugates are not compatible with immunofluorescence methods and neither enables easy detection by electron microscopy as membranes must be permeabilised to enable detection.
One method of detecting proteins without using antibodies is to use a bioorthogonal reaction such as copper-catalysed azide–alkyne cycloaddition (CuAAC) commonly referred to as ‘click chemistry’.13–15 As the azide group is small it can be incorporated into probe molecules without interfering with their behaviour in live organisms, while providing the opportunity for subsequent conjugation to fluorescent probes. Alternatively, strain-promoted azide–alkyne cycloaddition (SPAAC) allows preparation of conjugates under copper-free conditions that are suitable for use in live cells and animals.16–18 Azide-functionalised amino acids can be incorporated into proteins using the methionine surrogate azidohomoalanine (Aha).19,20 In methionine-depleted cultures, Aha is activated by methionyl-tRNA synthetase of methionine auxotroph Escherichia coli and replaces methionine in translated proteins. In this communication we report a novel azido-CTB toxoid and demonstrate its utility for neuronal tracing in vivo. Azido-CTB can be made in any lab with protein production facilities and a variety of molecules may be readily conjugated in a site-specific manner to suit individual needs.
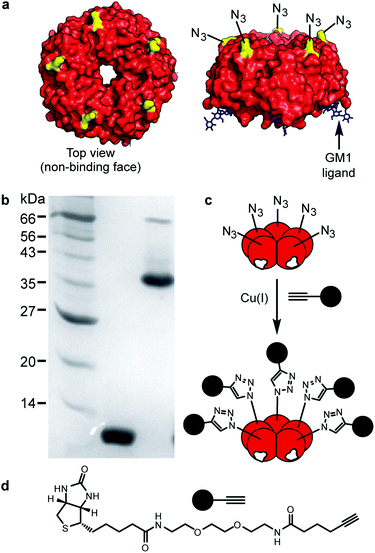
The isosteric replacement of methionine with Aha at the surface of CTB required the creation of a new mutant toxoid. Three sequential rounds of site-directed mutagenesis were used to replace all the native methionine residues with leucines (M37L, M68L, M101L) and a surface exposed lysine residue with methionine (K43M) (Fig. S1, ESI†). As the K43M mutation is on the non-binding face of the protein (Fig. 1a), future chemical modifications at this site were predicted not to interfere with CTB–GM1 binding. Methionine auxotroph E. coli B834 Gold (DE3) cells were transformed with the new construct, pSAB2.3 (Azido), and azido-CTB was overexpressed using a defined growth medium supplemented with Aha.19–21 The azido-CTB was purified using Ni-affinity chromatography and size exclusion chromatography before analysis by SDS-PAGE and mass spectrometry (Fig. S2, ESI†). For the wild type CTB protein, the tetra-mutant CTB is sufficiently stable to migrate as a pentamer during SDS-PAGE unless the sample has been boiled prior to loading (Fig. 1b). While the native CTB normally gives rise to a protein band around 45 kDa on a gel, azido-CTB is more negatively charged than native CTB which likely explains why it ran faster (apparent mass <40 kDa).
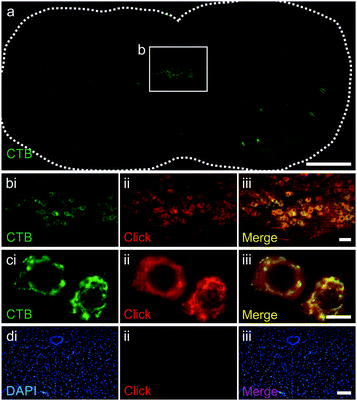
The ability of the purified azido-CTB protein to enter motor neurons, transport in a retrograde manner to the brainstem and to be detected in vivo via CuAAC, was tested using tongue injections in mice. Azido-CTB successfully traced the motor neurons of the hypoglossal nucleus that innervate the tongue; it was detected using CuAAC with a biotinylated alkyne (Kerafast) followed by Alexa-555 labelled streptavidin (streptavidin-555) in paraformaldehyde-fixed brain tissue (Fig. 2b–cii). Streptavidin labelling (shown in red) overlapped with immunohistochemical detection for CTB using an anti-CTB primary antibody and secondary antibody labelled with Alexa-488), it was detected in the same cells and to the same extent (Fig. 2a–ci). Control brainstem slices with no azido-CTB injection undergoing the click reaction and detection with streptavidin-555 gave no positive cells, verifying that the signal in injected tissue is from azido-CTB (Fig. 2d). Azido-CTB can therefore be used as a neuronal tracer. If used in conjunction with immunohistochemistry to identify other proteins such as ion channels and receptor subunits within the tissue, it will reduce limitations to the primary antibody species that can be used since CTB is detected without antibodies. Furthermore, azido-CTB has the potential to be used to make a fluorescently tagged tracer for delivery with any fluorescent molecule containing an alkyne group.
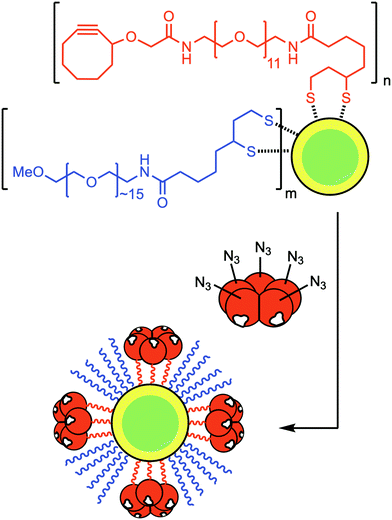
To further extend the flexibility of azido-CTB, we used its azide moiety for conjugation with nanoparticles prior to neuronal tracing. Correlative light and electron microscopy provides a powerful and complementary alternative to immunohistochemistry for studying sub-cellular localisation of probes. In this context, targeted nanoparticles can be used as nucleation sites for deposition of silver to facilitate their detection without the need to permeabilise the tissue. We therefore investigated the conjugation of CdSe/ZnS core/shell nanoparticles to azido-CTB and their ability to target neurons in vivo. The nanoparticles were first functionalised with a ligand mixture of dihydrolipoic acid (DHLA)-PEG11-cyclooctyne and a DHLA-PEG∼15-OMe spacer via cap-exchange at a total ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nanoparticle molar ratio of 3000
nanoparticle molar ratio of 3000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, with 10% or 25% of the ligands being DHLA-PEG11-cyclooctyne ligand (Fig. 3).22–24 Azido-CTB was then directly conjugated to the nanoparticles by SPAAC as confirmed by gel electrophoresis (ESI,† Fig. S3). Site-specific biorthogonal labelling of CTB with the nanoparticles means that the binding face of CTB remains unobstructed and thus endocytosis of the complex should be unaltered, unlike other studies using random derivations.25
1, with 10% or 25% of the ligands being DHLA-PEG11-cyclooctyne ligand (Fig. 3).22–24 Azido-CTB was then directly conjugated to the nanoparticles by SPAAC as confirmed by gel electrophoresis (ESI,† Fig. S3). Site-specific biorthogonal labelling of CTB with the nanoparticles means that the binding face of CTB remains unobstructed and thus endocytosis of the complex should be unaltered, unlike other studies using random derivations.25
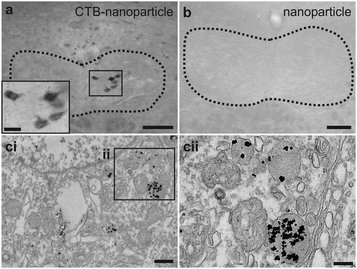
The CTB-functionalised nanoparticles (2 μL of 2.4 μM) were injected via the tongue in mice to assess whether the nano-conjugates could be endocytosed and transported in a retrograde direction in vivo. Silver enhanced brainstem slices revealed that the nano-conjugates were in neurons of the hypoglossal nucleus after injection (Fig. 4a). Staining was found optimal when using azido-CTB conjugated to nanoparticles prepared using 25% of the active DHLA-PEG11-cyclooctyne ligand as shown in Fig. 3. Given the reduced amount of CTB–nanoparticles delivered versus azido-CTB, there are fewer labelled cells. These nanoparticles were not detected in mice injected with just nanoparticles (Fig. 4b), indicating that they cannot transport in a retrograde manner without CTB. The silver enhanced slices were then prepared for EM where puncta corresponding to the metal deposits were found to be mainly localised to lysosomes (Fig. 4c). While CTB is known to travel in a retrograde manner to the Golgi,26,27 the appearance of CTB–nanoparticle conjugates in the lysosome is also in agreement with other studies,28,29 and may be the final fate of CTB after longer periods in vivo. Nonetheless, the easy application of azido-CTB as a nanoparticle-conjugate could further improve its use as a neuronal tracer for EM analysis. It does not require cellular permeabilisation, and thus avoids damage to cellular membranes, which occurs when using agents such as Triton X-100 or ethanol to allow penetrance of antibodies. The easy detection of CTB–nanoparticle conjugates via correlated light microscopy and EM, thus provides a much wider scope for possible experiments with tissue. These experiments highlight that alkynated substances can be readily and efficiently “clicked” onto azido-CTB prior to injection, increasing the possibility for rapid testing of CTB as a powerful delivery vehicle to carry different cargoes into neurons.
In summary, azido-CTB presents a useful tool for diversifying and enhancing its use as both a tracer and a delivery vehicle compared to current commercially available CTB conjugates due to ease of directed conjugation via CuAAC and SPAAC. Azido-CTB may be further developed to create interchangeable conjugated proteins via click chemistry; it could deliver multiple cargoes at once, or cargo along with a reporter dye or even a nanoparticle. This has been proven with CTB–nanoparticle conjugates produced from azido-CTB; where the nanoparticles were detectable after injection in vivo. Azido-CTB presents a diverse tool for tracing that is subsequently detected both with fluorescence and EM, for use in live and fixed tissue, and for use in extended time-lapse experiments. Moreover, other theranostic functions can be readily incorporated via conjugating to other functional nanoparticles (e.g. magnetic nanoparticles for magnetic resonance imaging;30 gold nanorods for photothermal treatment and/or photoacoustic imaging).31,32 With the increasing array of molecules and nanoparticles able to be ‘clicked’ to azides, there is a wide scope of opportunity for azido-CTB to be utilised for efficient and rapid attachment of novel tracers, cargoes and/or incorporating with multiple theranostic functions.
The authors thank the BBSRC for funding through Doctoral Training Partnership BB/J014443/1, and research grant BB/M005666/1, and EPSRC for funding through Doctoral Training Partnership EP/M50807X/1, with equipment support from the Wellcome Trust grants WT094232MA and 094232/Z/10/Z.
Conflicts of interest
There are no conflicts to declare.References
- T. Beddoe, A. W. Paton, J. Le Nours, J. Rossjohn and J. C. Paton, Trends Biochem. Sci., 2010, 35, 411–418 CrossRef CAS PubMed.
- E. A. Merritt, S. Sarfaty, F. van den Akker, C. L'Hoir, J. A. Martial and W. G. Hol, Protein Sci., 1994, 3, 166–175 CrossRef CAS PubMed.
- B. Lindh, H. Aldskogius and T. Hökfelt, Histochemistry, 1989, 92, 367–376 CAS.
- P. J. Dederen, A. A. Gribnau and M. H. Curfs, Histochem. J., 1994, 26, 856–862 CrossRef CAS.
- A. Gramsbergen, J. Ijkema-Paassen and M. F. Meek, Exp. Neurol., 2000, 161, 183–193 CrossRef CAS.
- P.-H. Luppi, K. Sakai, D. Salvert, P. Fort and M. Jouvet, Brain Res., 1987, 402, 339–345 CrossRef CAS.
- R. Flink and J. Westman, J. Comp. Neurol., 1986, 250, 265–281 CrossRef CAS.
- F. Datiche, P. H. Luppi and M. Cattarelli, Brain Res., 1995, 671, 27–37 CrossRef CAS.
- N. Luquin, S. Sierra, A. J. Rico, V. Gomez-Bautista, E. Roda, L. Conte-Perales, R. Franco, P. McCormick, J. L. Labandeira-Garcia and J. L. Lanciego, Neurobiol. Dis., 2012, 47, 347–357 CrossRef CAS PubMed.
- K. E. Fasanella, J. A. Christianson, R. S. Chanthaphavong and B. M. Davis, J. Comp. Neurol., 2008, 509, 42–52 CrossRef PubMed.
- W. L. Conte, H. Kamishina and R. L. Reep, Nat. Protoc., 2009, 4, 1157–1166 CrossRef CAS PubMed.
- H. Liu, I. J. Llewellyn-Smith and A. I. Basbaum, J. Histochem. Cytochem., 1995, 43, 489–495 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS.
- C. W. Tornoe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057–3064 CrossRef CAS.
- L. Li and Z. Zhang, Molecules, 2016, 21, 1393 CrossRef.
- J. Dommerholt, F. P. J. T. Rutjes and F. L. van Delft, Top. Curr. Chem., 2016, 374, 16 CrossRef PubMed.
- E. M. Sletten and C. R. Bertozzi, Acc. Chem. Res., 2011, 44, 666–676 CrossRef CAS PubMed.
- J. C. Jewett and C. R. Bertozzi, Chem. Soc. Rev., 2010, 39, 1272–1279 RSC.
- K. L. Kiick, E. Saxon, D. A. Tirrell and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 19–24 CrossRef CAS PubMed.
- F. Tobola, E. Sylvander, C. Gafko and B. Wiltschi, Interface Focus, 2019, 9, 20180072 CrossRef PubMed.
- B. Wiltschi, in Synthetic Gene Networks: Methods and Protocols, ed. W. Weber and M. Fussenegger, Humana Press, Totowa, NJ, 2012, pp. 211–225 Search PubMed.
- Y. Guo, C. Sakonsinsiri, I. Nehlmeier, M. A. Fascione, H. Zhang, W. Wang, S. Pöhlmann, W. B. Turnbull and D. Zhou, Angew. Chem., Int. Ed., 2016, 55, 4738–4742 CrossRef CAS PubMed.
- Y. Guo, I. Nehlmeier, E. Poole, C. Sakonsinsiri, N. Hondow, A. Brown, Q. Li, S. Li, J. Whitworth, Z. Li, A. Yu, R. Brydson, W. B. Turnbull, S. Pöhlmann and D. Zhou, J. Am. Chem. Soc., 2017, 139, 11833–11844 CrossRef CAS PubMed.
- B. C. Mei, K. Susumu, I. L. Medintz, J. B. Delehanty, T. J. Mountziaris and H. Mattoussi, J. Mater. Chem., 2008, 18, 4949–4958 RSC.
- S. K. Chakraborty, J. A. J. Fitzpatrick, J. A. Phillippi, S. Andreko, A. S. Waggoner, M. P. Bruchez and B. Ballou, Nano Lett., 2007, 7, 2618–2626 CrossRef CAS PubMed.
- D. C. Machin, D. J. Williamson, P. Fisher, V. J. Miller, G. C. Wildsmith, J. F. Ross, C. Wasson, A. MacDonald, B. I. Andrews, D. Ungar, W. B. Turnbull and M. E. Webb, ChemRxiv, 2019 DOI:10.26434/chemrxiv.9125201.v1.
- W. I. Lencer, T. R. Hirst and R. K. Holmes, Biochim. Biophys. Acta, 1999, 1450, 177–190 CrossRef CAS.
- W. A. Walker, M. Tarannum and J. L. Vivero-Escoto, J. Mater. Chem. B, 2016, 4, 1254–1262 RSC.
- K. C. Joseph, S. U. Kim, A. Stieber and N. K. Gonatas, Proc. Natl. Acad. Sci. U. S. A., 1978, 75, 2815 CrossRef CAS PubMed.
- T.-H. Shin, Y. Choi, S. Kim and J. Cheon, Chem. Soc. Rev., 2015, 44, 4501–4516 RSC.
- Y.-S. Chen, Y. Zhao, S. J. Yoon, S. S. Gambhir and S. Emelianov, Nat. Nanotechnol., 2019, 14, 465–472 CrossRef CAS PubMed.
- W. Zhang, F. Wang, Y. Wang, J. Wang, Y. Yu, S. Guo, R. Chen and D. Zhou, J. Controlled Release, 2016, 232, 9–19 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc01085e |
| This journal is © The Royal Society of Chemistry 2020 |