Open Access Article
Open Access ArticleStabilizing synthetic DNA for long-term data storage with earth alkaline salts†
A. Xavier
Kohll
a,
Philipp L.
Antkowiak
 a,
Weida D.
Chen
a,
Bichlien H.
Nguyen
b,
Wendelin J.
Stark
a,
Weida D.
Chen
a,
Bichlien H.
Nguyen
b,
Wendelin J.
Stark
 a,
Luis
Ceze
a,
Luis
Ceze
 c,
Karin
Strauss
c,
Karin
Strauss
 b and
Robert N.
Grass
b and
Robert N.
Grass
 *a
*a
aInstitute for Chemical and Bioengineering, ETH Zurich, Vladimir-Prelog-Weg 1, 8093 Zurich, Switzerland. E-mail: rograss@ethz.ch
bMicrosoft, One Microsoft Way, Redmond, WA 98052, USA
cPaul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, WA, USA
First published on 24th February 2020
Abstract
Rapid aging tests (70 °C, 50% RH) of solid state DNA dried in the presence of various salt formulations, showed the strong stabilizing effect of calcium phosphate, calcium chloride and magnesium chloride, even at high DNA loadings (>20 wt%). A DNA-based digital information storage system utilizing the stabilizing effect of MgCl2 was tested by storing a DNA file, encoding 115 kB of digital data, and the successful readout of the file by sequencing after accelerated aging.
The unprecedented growth of digital data calls forward new technological solutions to bridge the growing gap between data generation and storage limitations.1 In archival storage, one is more concerned about information density, durability, and energy cost than access speed to the data.2 In this storage domain, DNA data storage has become an attractive idea to replace traditional media such as magnetic tape.3,4 The natural biopolymer offers some unique advantages: high data storage density, ease of replication, and extended storage lifetimes.2 DNA can potentially store up to 455 exabytes of information per gram,5 is easily replicated via polymerase chain reaction (PCR), and can be preserved for centuries to millennia.5,6
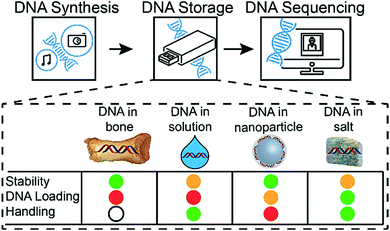
In addition to the encoding of digital information into DNA sequences and the subsequent decoding, DNA-based information storage systems are composed of three major parts: DNA synthesis, DNA storage, and DNA sequencing. Increasing throughput and cost reduction for DNA synthesis and sequencing makes archival DNA data storage economically attractive.7 Also, the total amount of digital data stored in DNA recently increased to over 200 MB.4 However, fewer efforts have been made to investigate an adequate physical DNA storage system. An effective DNA data storage system needs to satisfy the following requirements: High DNA loading, increased DNA stability, and simple sample handling (physical storage and accessibility). No current DNA storage media is optimal in all three aspects.
Without any protection, DNA is a relatively fragile biomolecule, prone to degradation, for example, by hydrolysis or oxidation.8,9 The prevention of DNA degradation is possible, for instance, by storing DNA in a dehydrated and anaerobic environment or at very low temperatures. As a result, existing DNA storage solutions include DNA storage at low temperatures (e.g., freezer at −80 °C) or at room temperature under anoxic and anhydrous conditions (e.g., DNAshells®).10 However, all of these storage systems exhibit low DNA loading (DNA mass/total mass of the storage system). Most recently, significant development efforts have gone into maximising the logical encoding efficiency (bit per nucleotide as well as bit per g of DNA).11–13 Hence, with the goal of having a system with optimal storage density, storage lifetime and energy requirements, the above mentioned storage solutions are less attractive for a potential DNA-based archival information storage system.
The vulnerability of DNA to degradation described above is in contradiction to the stability of DNA in select ancient fossils, such as biomineralized DNA in the collagen/calcium phosphate matrix of bones.15 In this matrix, DNA can be kept stable enough to be sequenced after thousands of years of storage.14 Evidently, the encapsulation of DNA into the inorganic matrix significantly increases its stability. Using this concept of encapsulation, Paunescu et al.15 have developed a DNA storage procedure in which the DNA is encapsulated with glass16 particles, achieving DNA stability comparable to natural fossils.6 However, even under optimized conditions, this storage technique can only load 3.4 wt% of DNA,17 requires several handling steps, potentially impeding automation, and the encapsulation reaction is relatively slow (4 days for completion).
While the storage system in glass particles employs the biomimetic concept of encapsulation, it uses a different inorganic material (SiO2) as that found in nature (calcium phosphates).
We herein investigate the influence of calcium phosphate (CaP), calcium chloride (CaCl2), magnesium chloride (MgCl2), and several other salts on DNA stability for DNA data storage purposes, primarily focusing on high DNA loadings (>10 wt%) as well as simple DNA storage and retrieval procedures.
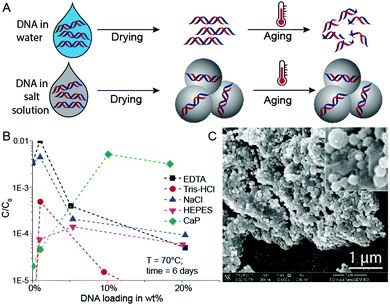
In initial experiments, we selected a model DNA system and exposed it in different matrices to accelerated aging conditions. The model DNA is a synthetic 148mer double-stranded DNA sequence with specific priming regions for qPCR quantification (see ESI†). Accelerated aging-induced degradation of DNA was performed by exposing samples of varying wt% loadings to 60 or 70 °C at 50% relative humidity.6 Water molecules, which can be structurally essential for DNA to maintain its double helix conformation, are neglected in these DNA wt% calculations.9 qPCR was used to determine the amount of intact and amplifiable DNA after exposure to accelerated aging for up to 33 days. Fig. 1A shows a simplified schematic of the used sample drying and accelerated aging process. For detailed protocols, see the ESI.†
Since the mineral phase of bones is primarily composed of CaP, we tried to protect DNA by coprecipitating the biomolecule with CaP.18 This idea of trapping DNA in CaP has been used for some time for DNA transfection.19 Additionally, Lindahl and others previously mentioned the stabilizing effects of DNA adsorption to hydroxyapatite, and it has already been reported that CaP shields DNA from degradation by DNase I.9,20,21 Therefore, we compared the stability of DNA dried in the presence of calcium and phosphate ions with DNA dried in the presence of standard buffer components (Tris–HCl, EDTA, NaCl, HEPES, Fig. 1B). During these initial experiments, we searched for an optimum between DNA loading in CaP and DNA stability at accelerated aging conditions.
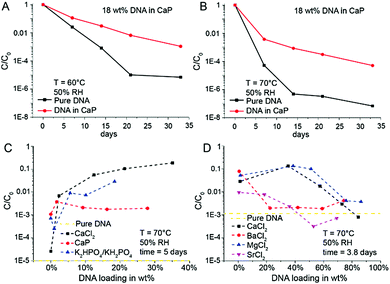
Intuitively, the more protecting material surrounds the DNA, the higher the DNA protection should be. This is true for low DNA loadings (≤1 wt%) in EDTA and NaCl (Fig. 1B), but at higher DNA loadings the protective nature of the chemicals rapidly disappears. However, the CaP system behaves quite differently: A steep stability increase from very low DNA loadings in CaP (<1 wt%) to loadings beyond 10 wt% can be observed (see Fig. 1B). Nearly two orders of magnitude more DNA is preserved in the 18 wt% DNA in CaP samples in comparison to 1 wt% samples. Fig. 1C and Fig. S1 (ESI†) show SEM pictures of the DNA with CaP precipitate at 18 wt% DNA loading. We further investigated high DNA loadings in CaP over extended periods of time: 18 wt% DNA in CaP was aged over 33 days at 60 and 70 °C at 50% RH compared to unprotected DNA (Fig. 2A and B). These results also highlight that in the presence of CaP, DNA indeed exhibits greater stability against thermal degradation for extended periods of time. This effect can be explained by the chemical interaction of the calcium ions with the phosphate backbone of the DNA during precipitation. The co-precipitation of DNA and calcium phosphate can be evidenced when coprecipitating genomic DNA, resulting in needle-like structures (see ESI†).
These results suggest that DNA precipitated in the presence of CaP would be a straightforward and effective way for stable storage, and further suggest that the chemical nature of bone is responsible for the long-term preservation of DNA in ancient fossils. However, our results also show that CaP is not always a robust storage system, since DNA stability results varied between different experiments and drying conditions. One reason for the varying stability results is that the precipitation process of DNA with CaP is difficult to control. In extreme cases, DNA stability results varied significantly when changing the sample drying process to precipitate the DNA (e.g., changing from vacuum centrifuge drying to freeze-drying, see Fig. S2, ESI†). Such effects are reported in the DNA transfection literature.22–25 A set of control experiments was performed to gain further insights into the stabilizing effects of the individual chemical species used to produce the CaP system. Model DNA was stored separately in the presence of calcium ions without phosphate (CaCl2) and in the presence of phosphate ions without calcium (KH2PO4/K2HPO4). The results show that CaCl2 stabilizes DNA to a higher degree than CaP. CaCl2 significantly increased DNA stability especially at loadings beyond 20 wt% (Fig. 2C).
Since CaCl2 shows a significant DNA stabilizing effect, we were interested in comparing various earth alkaline chlorides. As shown in Fig. 2D, barium chloride and strontium chloride do not exhibit exceptional DNA stabilization effects, magnesium chloride effectively protected DNA from thermal degradation at high loadings (>10 wt%). Even though most prior work was performed on DNA in aqueous solution, the stabilization effect of Mg2+ is not unexpected since Lindahl et al. and Marguet et al. both highlighted the stabilizing impact of MgCl2 on DNA in solution.26,27 Our results on DNA preservation in solids indicate that both the calcium and magnesium cation result in chemical interactions with the DNA backbone (phosphate group), to limit its thermal degradation at very high solid loadings. This also applies to the storage of genomic DNA (see ESI†). A detailed FTIR study by Serec et al. has previously shown that the presence of magnesium ions in solid state DNA result in a red shift and intensity decrease of the asymmetric PO2 stretching band, indicating an increase of the magnesium–phosphate binding to the DNA backbone.28
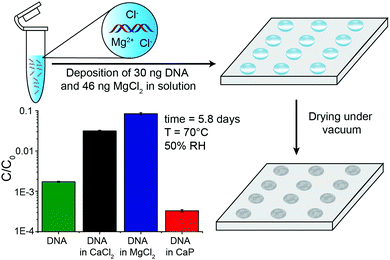
To provide more detail on the usefulness of earth alkaline salts in long-term storage of information in DNA, we performed an additional series of experiments, where the model DNA used in previous experiments was replaced with a DNA sample encoding actual digital information. Additionally, we went from drying samples in individual Eppendorf tubes to a potentially scalable and automatable 2D set-up. The utilized DNA sequences represent a 115 kB image encoded in 7′323 distinct DNA sequences according to the methods described by Organick et al.4 To assess the reproducibility of the salt-based storage system with a different DNA source, the DNA file was first stored in Eppendorf tubes, with a selection of stabilizing salts (MgCl2, CaCl2, and CaP). Similar stability trends as for the model DNA were measured, and the results are presented in the ESI† (Fig. S3). Second, a potentially scalable and automation-friendly storage system (a 2D substrate) was selected and tested with the DNA file stored in the inorganic matrix. For this we chose a recently published cartridge-based system.29 This system allows the automated manipulation, deposition, and retrieval of physically separated DNA spots with the help of a electrowetting-based digital microfluidic (DMF) device. These spots on a Teflon-coated glass slide can be dried and rehydrated separately and, therefore, allow to selectively address a specific DNA pool without rehydration of multiple DNA pools. As a simple proof of concept, we limited ourselves to manually depositing and retrieving DNA salt spots from such a Teflon-coated glass slide used in the previously mentioned automated system. The measurement data of DNA stability in the presence of MgCl2, CaCl2, and CaP after accelerated aging on the cartridge again revealed that DNA stored with MgCl2 or CaCl2 is degraded the least (Fig. 3). Finally, to fully demonstrate the salt-based DNA storage system on a glass slide, a successful readout (sequencing) of the stored DNA file, after accelerated aging, is essential. Therefore, five spots each of the DNA file with or without protection by MgCl2 underwent accelerated aging for ca. 6 days at 70 °C and were subsequently sequenced. The digital file in all the salt-protected DNA spots was successfully decodable without a single error. However, none of the unprotected DNA samples could be read after accelerated aging. Fig. 4A summarizes the results of the storage and sequencing experiment, showing high sequence losses for the non-protected DNA samples, which is in line with the qPCR results of Fig. 3. The consistency in low sequence losses for the five individually sequenced MgCl2 samples additionally depicts the reliability of the storage medium. Fig. S4–S6 (ESI†) show detailed sequencing statistics on insertion, deletion and substitution probabilities.
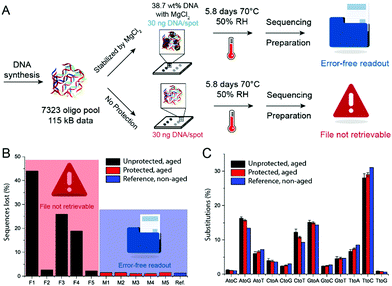 | ||
| Fig. 4 (A) Schematic representation of the DNA storage and sequencing experiment. (B) Total sequences lost after sequencing of protected (with MgCl2) and unprotected DNA samples. (C) % of substitutions of different nucleobases in case of substitution (probability ≈ 1%, see ESI†). Mean values and error bars (s.d.) from n = 5 independent experiments, five spots on the same glass slide. | ||
Compared to previously described DNA storage solutions,6,10,17 the earth alkaline salt storage system presented is simple, fast, can be easily automated, and results in a significant DNA stabilizing effect at very high DNA loadings (>30 wt%), even if stored at 50% relative humidity. Decreasing humidity would most probability lead to a further increase of DNA stability.7 A qualitative comparison of DNA storage systems is given in Fig. 5 (see also ESI†). As shown previously by Newman et al.29 the storage of DNA spots on physically addressable 2D substrates enables very high volumetric storage capacities. As shown here, the addition of appropriate salts can have a substantial effect on the stability of the deposited DNA. In accelerated aging tests, DNA in salt achieves stability (Fig. S9, ESI†) comparable to DNA hermetically encapsulated in glass particles and overcomes the long and rather complex material preparation routines required.6,17 Under the tested rapid aging conditions, it outperforms DNA storage in commercial storage buffers both in stability and in DNA loading. The implementation on 2D surfaces additionally allows the physical addressing of DNA pools by the use of digital microfluidic devices, as recently reported by Newman et al.29 Our results with calcium and magnesium chloride additionally suggest that the presence of the earth alkaline ions is responsible for the stability of DNA in ancient bone samples.14,30 It remains challenging and time consuming to predict the DNA longevity in the salt-based system at ambient temperature due the uncertainty over whether the extrapolation of stability data from high temperature to room temperature persists.31 We hope to overcome this uncertainty by ongoing long-term testing at lower storage temperatures and the construction of a kinetic degradation model.
Conflicts of interest
Bichlien H. Nguyen and Karin Strauss are Microsoft employees.Notes and references
- D. Reinsel, J. Gantz and J. Rydning, IDC Data Age 2025, 2017.
- L. Ceze, J. Nivala and K. Strauss, Nat. Rev. Genet., 2019, 20, 456–466 CrossRef CAS PubMed.
- D. Carmean, L. Ceze, G. Seelig, K. Stewart, K. Strauss and M. Willsey, Proc. IEEE, 2019, 107, 63–72 CAS.
- L. Organick, S. D. Ang, Y.-J. Chen, R. Lopez, S. Yekhanin, K. Makarychev, M. Z. Racz, G. Kamath, P. Gopalan, B. Nguyen, C. N. Takahashi, S. Newman, H.-Y. Parker, C. Rashtchian, K. Stewart, G. Gupta, R. Carlson, J. Mulligan, D. Carmean, G. Seelig, L. Ceze and K. Strauss, Nat. Biotechnol., 2018, 36, 242–249 CrossRef CAS.
- G. M. Church, Y. Gao and S. Kosuri, Science, 2012, 337, 1628 CrossRef CAS.
- R. N. Grass, R. Heckel, M. Puddu, D. Paunescu and W. J. Stark, Angew. Chem., Int. Ed., 2015, 54, 2552–2555 CrossRef CAS.
- N. Goldman, P. Bertone, S. Chen, C. Dessimoz, E. M. LeProust, B. Sipos and E. Birney, Nature, 2013, 494, 77–80 CrossRef CAS PubMed.
- J. Bonnet, M. Colotte, D. Coudy, V. Couallier, J. Portier, B. Morin and S. Tuffet, Nucleic Acids Res., 2009, 38, 1531–1546 CrossRef PubMed.
- T. Lindahl, Nature, 1993, 366, 529–531 CrossRef.
- D. Clermont, S. Santoni, S. Saker, M. Gomard, E. Gardais and C. Bizet, Biopreserv. Biobank., 2014, 12, 176–183 CrossRef CAS PubMed.
- Y. Erlich and D. Zielinski, Science, 2017, 355, 950–954 CrossRef CAS PubMed.
- Y. Choi, T. Ryu, A. C. Lee, H. Choi, H. Lee, J. Park, S. H. Song, S. Kim, H. Kim, W. Park and S. Kwon, Sci. Rep., 2019, 9, 6582 CrossRef PubMed.
- L. Anavy, I. Vaknin, O. Atar, R. Amit and Z. Yakhini, Nat. Biotechnol., 2019, 37, 1229–1236 CrossRef CAS PubMed.
- J. Dabney, M. Knapp, I. Glocke, M.-T. Gansauge, A. Weihmann, B. Nickel, C. Valdiosera, N. Garcia, S. Paabo, J.-L. Arsuaga and M. Meyer, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 15758–15763 CrossRef CAS PubMed.
- D. Paunescu, M. Puddu, J. O. B. Soellner, P. R. Stoessel and R. N. Grass, Nat. Protoc., 2013, 8, 2440–2448 CrossRef CAS.
- B. Liu, Y. Yao and S. Che, Angew. Chem., Int. Ed., 2013, 52, 14186–14190 CrossRef CAS.
- W. D. Chen, A. X. Kohll, B. H. Nguyen, J. Koch, R. Heckel, W. J. Stark, L. Ceze, K. Strauss and R. N. Grass, Adv. Funct. Mater., 2019, 1901672 CrossRef.
- K. M. Iyer and W. S. Khan, General Principles of Orthopedics and Trauma, Springer International Publishing, 2019, pp. 1–779 Search PubMed.
- F. L. Graham and A. J. Van der EB, Virology, 1973, 52, 456–467 CrossRef CAS.
- L. J. Del Valle, O. Bertran, G. Chaves, G. Revilla-López, M. Rivas, M. T. Casas, J. Casanovas, P. Turon, J. Puiggalí and C. Alemán, J. Mater. Chem. B, 2014, 2, 6953–6966 RSC.
- Y. Yang, G. Wang, G. Zhu, X. Xu, H. Pan and R. Tang, Chem. Commun., 2015, 51, 8705–8707 RSC.
- E. T. Schenborn and V. Goiffon, in Transcription Factor Protocols, ed. M. J. Tymms, Humana Press, Totowa, NJ, 2000, pp. 135–145 Search PubMed.
- W. Li, D. Liu, Q. Wang, H. Hu and D. Chen, J. Mater. Chem. B, 2018, 6, 3466–3474 RSC.
- C. E. Pedraza, D. C. Bassett, M. D. McKee, V. Nelea, U. Gbureck and J. E. Barralet, Biomaterials, 2008, 29, 3384–3392 CrossRef CAS.
- H. Kuroda, R. H. Kutner, N. G. Bazan and J. Reiser, J. Virol. Methods, 2009, 157, 113–121 CrossRef CAS.
- E. Marguet and P. Forterre, Extremophiles, 1998, 2, 115–122 CrossRef CAS PubMed.
- T. Lindahl and B. Nyberg, Biochemistry, 1972, 11, 3610–3618 CrossRef CAS PubMed.
- K. Serec, S. D. Babić, R. Podgornik and S. Tomić, Nucleic Acids Res., 2016, 44, 8456–8464 CrossRef CAS PubMed.
- S. Newman, A. P. Stephenson, M. Willsey, B. H. Nguyen, C. N. Takahashi, K. Strauss and L. Ceze, Nat. Commun., 2019, 10, 1706 CrossRef CAS PubMed.
- P. Turon, J. Puiggalí, O. Bertrán and C. Alemán, Chem. – Eur. J., 2015, 21, 18893–18898 CrossRef PubMed.
- M. Celina, K. T. Gillen and R. A. Assink, Polym. Degrad. Stab., 2005, 90, 395–404 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, material characterization and details to Fig. 5. See DOI: 10.1039/d0cc00222d |
| This journal is © The Royal Society of Chemistry 2020 |