Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR†
Abstract
G-quadruplexes (G4s) are frequently formed in the promoter regions of oncogenes, considered as promising drug targets for anticancer therapy. Due to high structure similarity of G4s, discovering ligands selectively interacting with only one G4 is extremely difficult. Here, mainly by NMR, we report that colchicine selectively binds to oncogene RET G4-DNA.
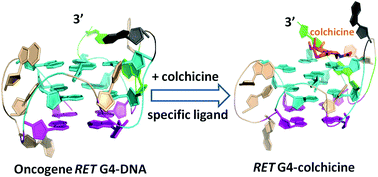


 Please wait while we load your content...
Please wait while we load your content...