Open Access Article
Open Access ArticleA palladium-catalyst stabilized in the chiral environment of a monoclonal antibody in water†
Yuichiro
Kobayashi
 a,
Keisuke
Murata
a,
Akira
Harada
a,
Keisuke
Murata
a,
Akira
Harada
 *b and
Hiroyasu
Yamaguchi
*b and
Hiroyasu
Yamaguchi
 *a
*a
aDepartment of Macromolecular Science, Graduate School of Science, Osaka University, 1-1 Machikaneyama-cho, Toyonaka, Osaka 560-0043, Japan. E-mail: hiroyasu@chem.sci.osaka-u.ac.jp
bThe Institute of Scientific and Industrial Research, Osaka University, 8-1 Mihogaoka, Ibaraki 567-0047, Japan. E-mail: harada@chem.sci.osaka-u.ac.jp
First published on 6th January 2020
Abstract
We report the first preparation of a monoclonal antibody (mAb) that can immobilize a palladium (Pd)-complex. The allylic amination reaction using a supramolecular catalyst consisting of the Pd-complex and mAb selectively gives the (R)-enantiomer product with an enantiomeric excess (ee) of 98 ± 2%. This is in sharp contrast to the reaction catalyzed by a conventional Pd-catalyst (ee < 2%).
Palladium (Pd)-complexes have attracted much attention because they promote various cross-coupling reactions such as Negishi coupling,1–3 Suzuki–Miyaura cross-coupling,4–6 and the Mizoroki–Heck reaction.7–10 Recent efforts have focused on controlling the synthesis products and increasing the catalytic activity due to the importance of chirality for living systems. For structural control of a product, both the first coordination sphere, which is formed by metal ions and ligands, and the second coordination sphere must be appropriately constructed. Synthesizing a ligand with the desired structure can realize the desired first coordination sphere. However, the second coordination sphere is composed by multiple non-covalent interactions such as hydrogen bonding, hydrophobic effects, and electro-static interactions. Hence, it is difficult to produce the desired second coordination sphere with a low molecular weight system.
To this end, hybrid catalysts with various combinations of Pd-complexes and biomolecules,11 such as ferritin,12 lipase,13 and biotin-streptavidin,14,15 have been developed to obtain structure-controlled products. A Pd-complex anchored in a biomolecule provides asymmetric environmental fields in the second coordination sphere. In this case, the Pd-complex is incorporated into biomacromolecules by employing a non-direct method. One example is the utilization of avidin–biotin interactions during the complex formation of avidin with a biotinylated Pd-complex.14,15 Because the space originally possessed by the biomacromolecules is used as the second coordination sphere of the Pd-complex, it is not suitable for the Pd-complex. If biomacromolecules can directly recognize Pd-complexes, complexation of the Pd-complex and biomacromolecules can construct an appropriate second coordination sphere, realizing a novel platform for a hybrid catalyst of the Pd-complex and biomacromolecules.
Our work has focused on monoclonal antibodies (mAbs) because the binding site of mAbs can be tailored to the antigen specifications.16–29 This intriguing feature allows asymmetric environmental fields to be constructed as the second coordination sphere for transition metal complexes, enabling a stereospecific reaction.30,31 However, to the best of our knowledge, a hybrid catalyst consisting of a Pd-complex and a mAb is yet to be reported because typical Pd-complexes are unstable in water and degrade during immunization. Currently, there is only one report on the complexation of a Pd-complex of a porphyrin.32 This may be due to the difficulty in preparing mAbs capable of recognizing Pd-complexes.
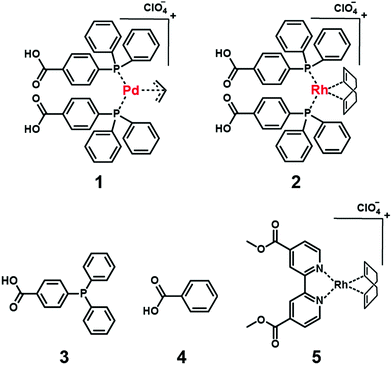
In this study, we describe our efforts to create a supramolecular catalyst using a mAb to immobilize an unstable Pd-complex (1) (Fig. 1 and Fig. S1–S3, Scheme S1, ESI†) for the allylic amination reaction. Because 1 degrades during immunization, a mAb capable of recognizing 1 is prepared by the cross-reactivity of mAb binding to the rhodium (Rh)-complex (2) (Fig. 1 and Fig. S4, S5, Scheme S2, ESI†) with the same ligand as 1. A supramolecular catalyst consisting of 1 and mAb catalyzes the allylic amination reaction, which selectively affords an (R)-enantiomer product.
We chose a η3-allyl palladium(II) complex as a late transition metal complex because it promotes various coupling reactions such as allylic amination reactions.33 To obtain mAbs capable of recognizing 1, we prepared antigens in which 1 was modified to keyhole limpet hemocyanin (KLH) (1-KLH, Scheme S3 and Table S1, ESI†) for immunization and immunized 1-KLH in saline emulsified 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in Freund's complete adjuvant for Balb/c mice four times at two-week intervals. To investigate antibody production, 1 was introduced into bovine serum albumin (BSA) to obtain antigens for assays (1-BSA), and then enzyme-linked immunosorbent assay (ELISA) measurements of the blood from immunized and non-immunized mice using a 1-BSA coated plate were performed. However, there were no differences in the absorbance ascribable to the enzymatic reaction product in ELISA for the immunized and non-immunized mice, showing that no antibodies for 1 have been produced (Fig. 2a). This is most probably due to the low stability of the Pd-complex in water. In fact, the proton nuclear magnetic resonance (1H NMR) spectrum of 1 was changed by immersing it in N,N-dimethylformamide-d7/deuterium oxide for 7 days, showing decomposition of 1. (Fig. S6, ESI†). The 1 was decomposed during immunization, which could not allow for creation of antibodies.
1 in Freund's complete adjuvant for Balb/c mice four times at two-week intervals. To investigate antibody production, 1 was introduced into bovine serum albumin (BSA) to obtain antigens for assays (1-BSA), and then enzyme-linked immunosorbent assay (ELISA) measurements of the blood from immunized and non-immunized mice using a 1-BSA coated plate were performed. However, there were no differences in the absorbance ascribable to the enzymatic reaction product in ELISA for the immunized and non-immunized mice, showing that no antibodies for 1 have been produced (Fig. 2a). This is most probably due to the low stability of the Pd-complex in water. In fact, the proton nuclear magnetic resonance (1H NMR) spectrum of 1 was changed by immersing it in N,N-dimethylformamide-d7/deuterium oxide for 7 days, showing decomposition of 1. (Fig. S6, ESI†). The 1 was decomposed during immunization, which could not allow for creation of antibodies.
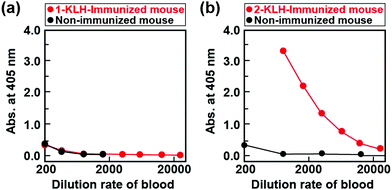 | ||
| Fig. 2 Results of ELISA for the blood obtained from the mouse immunized with transition-metal complex modified KLH (red) and the non-immunized mouse (black) [(a) 1-KLH and (b) 2-KLH]. | ||
To overcome this problem, we employed cross-reactivity of antibodies. Because Rh-complexes are stable in water, we immunized 2 with the same ligand as 1. The 2 was prepared from the chloro(1,5-cyclooctadiene)rhodium(I) dimer. The 2 was introduced to the proteins, such as KLH and BSA, in 0.1 M PBS buffer (pH 7.0) to obtain the antigens for immunization and assays, respectively, (2-KLH and 2-BSA) (Scheme S3, ESI†). The modification ratio was estimated by using the 2,4,6-trinitrobenzenesulfonic acid (TNBS) method,34–36 and over six hundred or eight of 2 were introduced into one KLH or BSA, respectively (Table S1, ESI†). Balb/c mice were immunized with 2-KLH in saline emulsified 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in Freund's complete adjuvant four times at two-week intervals. To confirm antibody production, we performed ELISA measurement of the blood from immunized and non-immunized mice using a 2-BSA coated plate. In contrast to 1, the absorbance ascribable to the enzymatic reaction product in ELISA for an immunized mouse was higher than that of an non-immunized mouse (Fig. 2b). Moreover, in the case of an immunized mouse, the absorbance of 2-BSA was higher than that of BSA (Fig. S7, ESI†). These results clearly show the preparation of antibodies for 2. The hybridomas secreting antibodies specific to 2 were cloned twice by the limiting dilution method and then the obtained mAbs were purified from the ascites fluid by affinity chromatography using a HiTrap IgM Purification HP column (GE healthcare). The class of mAbs was determined using the Iso Strip Mouse Monoclonal Antibody Isotyping Kit. As a result, the obtained mAb was immunoglobulin M (IgM), and the light chain was the kappa chain.
1 in Freund's complete adjuvant four times at two-week intervals. To confirm antibody production, we performed ELISA measurement of the blood from immunized and non-immunized mice using a 2-BSA coated plate. In contrast to 1, the absorbance ascribable to the enzymatic reaction product in ELISA for an immunized mouse was higher than that of an non-immunized mouse (Fig. 2b). Moreover, in the case of an immunized mouse, the absorbance of 2-BSA was higher than that of BSA (Fig. S7, ESI†). These results clearly show the preparation of antibodies for 2. The hybridomas secreting antibodies specific to 2 were cloned twice by the limiting dilution method and then the obtained mAbs were purified from the ascites fluid by affinity chromatography using a HiTrap IgM Purification HP column (GE healthcare). The class of mAbs was determined using the Iso Strip Mouse Monoclonal Antibody Isotyping Kit. As a result, the obtained mAb was immunoglobulin M (IgM), and the light chain was the kappa chain.
To quantitatively investigate the affinity of the mAb to 1, we determined the dissociation constant (Kd) of the complex between the mAb and 1 or 2. The Kd values of the complexes between the mAb and 1 or 2 were found to be 2.0 or 7.6 × 10−5 M, respectively (Fig. S8a and b, ESI†), showing that the affinities of mAb to 1 and that of mAb to 2 were almost the same. To understand this reason, the binding form of mAb to 2 was estimated using Kd of the complex between the mAb and the ligand of 2 (3), benzoic acid (4), or the Rh-complex (5) whose structure is different from 2 (Fig. 1). The mAb showed no affinity to 4 and 5 (>1.5 × 10−2 and >1.0 × 10−3 M, respectively), whereas the mAb showed affinity to 3 (3.4 × 10−4 M) (Fig. S8c and S9, ESI†). This result indicates that the mAb recognizes the molecular elements of 3 in complex 2. The ligand structure of 2 and that of 1 were the same, and thus the mAb can form complexes with 1. We prepared for the first time the mAb for a Pd-complex using the cross-reactivity of the mAb.
A reaction in a nanoconfined geometry often gives products whose symmetries differ from those obtained in a solution reaction. We carried out an allylic amination reaction using 1 in the absence and presence of the mAb (Table 1). Firstly, the reaction concentrations of 1 and mAb were optimized. 3-Buten-2-yl acetate (6) (10 mM) and benzyl amine (7) (20 mM) were reacted with 1 (1 μM) and the mAb (0.1 μM, binding site: 1 μM) in a mixed solution of 0.1 M phosphate borate buffer (PBB; pH 9.0) and DMSO (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at r.t. for 24 h (Fig. S10 and Scheme S4, ESI†). Under these conditions, the binding site of the mAb and 1 is the same. The yield and ee of the product (8) were determined by comparing the area ratio of 8 and allylbenzene as an internal standard in HPLC analysis (Fig. S11, ESI†) and calculated from the peak area ratio of (R)- and (S)-enantiomers of 8 of HPLC spectra (Fig. S12, ESI†). The yield and ee of 8 decreased with increasing concentration of 1 and mAb. This result suggests that the aggregation of IgM causes 1 to be adsorbed at locations other than where the product structure is controlled. The optimum reaction concentration of 1 and mAb was found to be 1 and 0.1 μM, respectively. The yield of 8 in the presence of the mAb (6%) was lower than that in the absence of the mAbs (38%) (Table 1, entries 1 and 2). Owing to the complex formation between 1 and mAb, it is difficult for the substrates such as 6 and 7 to access 1. The decline in the yield of 8 was also observed in the presence of BSA (11%) (Table 1, entry 3), suggesting that the presence of protein reduces the catalytic activity of 1. In the case of the absence of mAb, 1 gave racemic 8 (ee <
1) at r.t. for 24 h (Fig. S10 and Scheme S4, ESI†). Under these conditions, the binding site of the mAb and 1 is the same. The yield and ee of the product (8) were determined by comparing the area ratio of 8 and allylbenzene as an internal standard in HPLC analysis (Fig. S11, ESI†) and calculated from the peak area ratio of (R)- and (S)-enantiomers of 8 of HPLC spectra (Fig. S12, ESI†). The yield and ee of 8 decreased with increasing concentration of 1 and mAb. This result suggests that the aggregation of IgM causes 1 to be adsorbed at locations other than where the product structure is controlled. The optimum reaction concentration of 1 and mAb was found to be 1 and 0.1 μM, respectively. The yield of 8 in the presence of the mAb (6%) was lower than that in the absence of the mAbs (38%) (Table 1, entries 1 and 2). Owing to the complex formation between 1 and mAb, it is difficult for the substrates such as 6 and 7 to access 1. The decline in the yield of 8 was also observed in the presence of BSA (11%) (Table 1, entry 3), suggesting that the presence of protein reduces the catalytic activity of 1. In the case of the absence of mAb, 1 gave racemic 8 (ee <![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2%) (Table 1, entry 1 and Fig. S12a, ESI†). In contrast, interestingly, the reaction catalysed by 1 incorporated into the binding site of the mAb was found to proceed with excellent (R)-enantioselectivity with a 98 ± 2% ee (Table 1, entry 2 and Fig. S12b, ESI†). These results indicate that no catalytic reaction occurred outside the binding sites. Since the catalytic activity of free 1 is reduced by the presence of protein, the catalytic activity of free 1 would be negligible in the mAb system. This asymmetric allylic amination was not observed in the presence of BSA (ee < 2%) (Table 1, entry 3 and Fig. S12c, ESI†). These results indicate that the binding site of mAb functions as a reaction field for the asymmetric reaction.
2%) (Table 1, entry 1 and Fig. S12a, ESI†). In contrast, interestingly, the reaction catalysed by 1 incorporated into the binding site of the mAb was found to proceed with excellent (R)-enantioselectivity with a 98 ± 2% ee (Table 1, entry 2 and Fig. S12b, ESI†). These results indicate that no catalytic reaction occurred outside the binding sites. Since the catalytic activity of free 1 is reduced by the presence of protein, the catalytic activity of free 1 would be negligible in the mAb system. This asymmetric allylic amination was not observed in the presence of BSA (ee < 2%) (Table 1, entry 3 and Fig. S12c, ESI†). These results indicate that the binding site of mAb functions as a reaction field for the asymmetric reaction.
| Entry | Catalyst | Conc. of Pd (μM)/mAb (μM) | Yield of 8a (%) | eeb (%) |
|---|---|---|---|---|
| a Calculated from the peak area ratio of HPLC spectra using allylbenzene as an internal standard. b Calculated from the peak area ratio of (R)- and (S)-enantiomers of 8 of HPLC spectra. c Since the mAb is IgM with 10 binding sites, the concentrations of 1 and the binding site of mAb are equivalent. | ||||
| 1 | 1 | 1/0 | 38 | <2 |
| 2 | 1 + mAb | 1/0.1c | 6 | 98 ± 2 |
| 3 | 1 + BSA | 1/0.1 | 11 | <2 |
In summary, we have demonstrated that Pd-complex catalysed allyl amination in the presence of mAb is a useful synthetic strategy to control the resultant product asymmetry. The substrate is converted to a racemic product by a Pd-catalyst without a mAb. In contrast, the (R)-enantiomer (98 ± 2% ee) is obtained in the presence of the complex between the mAb and the Pd-catalyst.
This work was supported by the JSPS KAKENHI grant number JP15H05807 in Precisely Designed Catalysts with Customized Scaffolding.
Conflicts of interest
There are no conflicts to declare.Notes and references
- E. Negishi, Q. Hu, Z. H. Huang, M. X. Qian and G. W. Wang, Aldrichimica Acta, 2005, 38, 71 CAS.
- E. Negishi, G. W. Wang, H. H. Rao and Z. Q. Xu, J. Org. Chem., 2010, 75, 3151 CrossRef CAS PubMed.
- D. Haas, J. M. Hammann, R. Greiner and P. Knochel, ACS Catal., 2016, 6, 1540 CrossRef CAS.
- N. Miyaura and A. Suzuki, Chem. Commun., 1979, 866 RSC.
- N. Miyaura, K. Yamada and A. Suzuki, Tetrahedron Lett., 1979, 20, 3437 CrossRef.
- N. Miyaura and A. Suzuki, Chem. Rev., 1995, 95, 2457 CrossRef CAS.
- T. Mizoroki, K. Mori and A. Ozaki, Bull. Chem. Soc. Jpn., 1971, 44, 581 CrossRef CAS.
- R. F. Heck and J. P. Nolley, J. Org. Chem., 1972, 37, 2320 CrossRef CAS.
- I. P. Beletskaya and A. V. Cheprakov, Chem. Rev., 2000, 100, 3009 CrossRef CAS PubMed.
- A. B. Dounay and L. E. Overman, Chem. Rev., 2003, 103, 2945 CrossRef CAS PubMed.
- F. Schwizer, Y. Okamoto, T. Heinisch, Y. F. Gu, M. M. Pellizzoni, V. Lebrun, R. Reuter, V. Kohler, J. C. Lewis and T. R. Ward, Chem. Rev., 2018, 118, 142 CrossRef CAS PubMed.
- S. Abe, J. Niemeyer, M. Abe, Y. Takezawa, T. Ueno, T. Hikage, G. Erker and Y. Watanabe, J. Am. Chem. Soc., 2008, 130, 10512 CrossRef CAS PubMed.
- M. Filice, O. Romero, A. Aires, J. M. Guisan, A. Rumbero and J. M. Palomo, Adv. Synth. Catal., 2015, 357, 2687 CrossRef CAS.
- J. Pierron, C. Malan, M. Creus, J. Gradinaru, I. Hafner, A. Ivanova, A. Sardo and T. R. Ward, Angew. Chem., Int. Ed., 2008, 47, 701 CrossRef CAS PubMed.
- A. Chatterjee, H. Mallin, J. Klehr, J. Vallapurackal, A. D. Finke, L. Vera, M. Marsh and T. R. Ward, Chem. Sci., 2016, 7, 673 RSC.
- B. L. Iverson and R. A. Lerner, Science, 1989, 243, 1184 CrossRef CAS PubMed.
- A. Harada, K. Okamoto, M. Kamachi, T. Honda and T. Miwatani, Chem. Lett., 1990, 917 CrossRef CAS.
- A. G. Cochran and P. G. Schultz, Science, 1990, 249, 781 CrossRef CAS PubMed.
- V. A. Roberts, B. L. Iverson, S. A. Iverson, S. J. Benkovic, R. A. Lerner, E. D. Getzoff and J. A. Tainer, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 6654 CrossRef CAS PubMed.
- R. A. Lerner, S. J. Benkovic and P. G. Schultz, Science, 1991, 252, 659 CrossRef CAS PubMed.
- P. Ghosh, D. Shabat, S. Kumar, S. C. Sinha, F. Grynszpan, J. Li, L. Noodleman and E. Keinan, Nature, 1996, 382, 339 CrossRef CAS PubMed.
- D. A. Blake, P. Chakrabarti, M. Khosraviani, F. M. Hatcher, C. M. Westhoff, P. Goebel, D. E. Wylie and R. C. Blake, J. Biol. Chem., 1996, 271, 27677 CrossRef CAS PubMed.
- A. Harada, H. Fukushima, K. Shiotsuki, H. Yamaguchi, F. Oka and M. Kamachi, Inorg. Chem., 1997, 36, 6099 CrossRef CAS PubMed.
- M. Khosraviani and R. C. Blake, Bioconjugate Chem., 2000, 11, 267 CrossRef CAS PubMed.
- K. M. Nicholas, P. Wentworth, C. W. Harwig, A. D. Wentworth, A. Shafton and K. D. Janda, PNAS, 2002, 99, 2648 CrossRef CAS PubMed.
- R. Ricoux, H. Sauriat-Dorizon, E. Girgenti, D. Blanchard and J. P. Mahy, J. Immunol. Methods, 2002, 269, 39 CrossRef CAS PubMed.
- H. Yamaguchi, K. Tsubouchi, K. Kawaguchi, E. Horita and A. Harada, Chem. – Eur. J., 2004, 10, 6179 CrossRef CAS PubMed.
- R. C. Blake, A. R. Pavlov, M. Khosraviani, H. E. Ensley, G. E. Kiefer, H. Yu, X. Li and D. A. Blake, Bioconjugate Chem., 2004, 15, 1125 CrossRef CAS PubMed.
- J. P. Mahy, J. D. Marechal and R. Ricoux, Chem. Commun., 2015, 51, 2476 RSC.
- H. Yamaguchi, T. Hirano, H. Kiminami, D. Taura and A. Harada, Org. Biomol. Chem., 2006, 4, 3571 RSC.
- T. Adachi, A. Harada and H. Yamaguchi, Sci. Rep., 2019, 9, 13551 CrossRef PubMed.
- A. P. Savitsky, M. V. Demcheva, E. Y. Mantrova and G. V. Ponomarev, FEBS Lett., 1994, 355, 314 CrossRef CAS PubMed.
- M. Johannsen and K. A. Jorgensen, Chem. Rev., 1998, 98, 1689 CrossRef CAS PubMed.
- K. Satake, T. Okuyama, M. Ohashi and T. Shinoda, J. Biochem., 1960, 47, 654 CrossRef CAS.
- L. C. Mokrasch, Anal. Biochem., 1967, 18, 64 CrossRef CAS.
- S. L. Snyder and P. Z. Sobocinski, Anal. Biochem., 1975, 64, 284 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, 1H and 13C NMR and affinities between the mAb and the catalyst. See DOI: 10.1039/c9cc08756g |
| This journal is © The Royal Society of Chemistry 2020 |