Biomaterial strategies to replicate gynecological tissue
Ines
Cadena
a,
Athena
Chen
 b,
Aaron
Arvidson
a and
Kaitlin C.
Fogg
b,
Aaron
Arvidson
a and
Kaitlin C.
Fogg
 *ac
*ac
aDepartment of Chemical, Biological, and Environmental Engineering, Oregon State University, Corvallis, Oregon, USA
bDepartment of Pathology, School of Medicine, Oregon Health & Science University, Portland, Oregon, USA
cDepartment of Biomedical Engineering, School of Medicine, Oregon Health & Science University, Portland, Oregon, USA
First published on 19th November 2020
Abstract
Women's health is an important and understudied area of research. The current standard of care for many gynecological diseases such as cancer or autoimmune-linked disorders such as endometriosis is surgery; however, the underlying mechanisms of action of many gynecological diseases are poorly understood. The field of tissue engineering has the potential to transform the field of women's health by developing in vitro models of healthy and diseased tissue that could be used to identify novel treatment strategies as well as gain a better understanding of complex signaling dynamics. Identification of the appropriate biomaterials, cell types, and stimuli (the tissue engineering triad) needed to build these in vitro models can be gleaned by interrogating the underlying extracellular matrix, cell organization, and soluble factors present in the tissue. In this review, we provide a general overview of the biology and components of the major tissues that make up the female reproductive system (ovaries, fallopian tubes, the uterus, and cervix) as well as a comprehensive survey of the different biomaterials that have been chosen to build in vitro models of these tissues. Furthermore, for each tissue, we recommend guiding principles in the design of in vitro models and discuss their potential to be used in drug screening and mechanistic studies.
1. Introduction
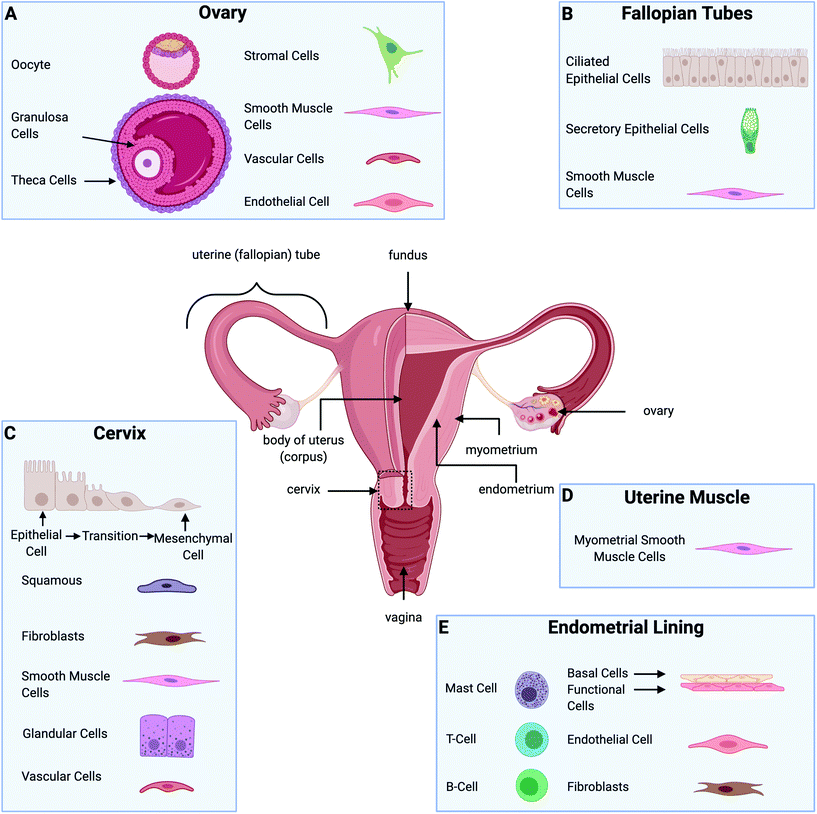
The female reproductive system's main organs include the ovaries, fallopian tubes, uterus (cervix and corpus), and vagina, illustrated in Fig. 1. Together, they are responsible for providing hormonal support, producing ova, and maintaining a pregnancy to term, with all of these functions depending on the dynamic interactive physiology of various gynecological tissues.1 Gynecological disorders are a source of significant suffering. In the U.S., between 2012 and 2016 approximately 94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 women were diagnosed with gynecologic cancer, and 1
000 women were diagnosed with gynecologic cancer, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400
400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 women visited the emergency department with gynecological complaints.2 Among the most common gynecological complaints were pelvic diseases, such as endometriosis and polycystic ovarian syndrome, and gynecological cancers such as ovarian cancer, uterine sarcomas, endometrial cancer, and cervical cancer.3 The current standard of care for the majority of gynecological diseases is surgery, chemotherapy, and radiation.4–7Table 1 describes the different treatment options available for the gynecological diseases covered in this review. However, the underlying mechanisms of action are often poorly understood, and there are limited personalized standards of care for patients with metastatic cancer due to its heterogeneous manifestations.8–10 While the removal of reproductive organs may address the symptoms resulting from a gynecological diseases, this carries substantial consequences to the endocrine system.8–10 Beyond fertility, the endocrine system has an essential role in the development and maintenance of tissue structure11 and regulates gene expression for numerous biological processes12 Consequentially, removing the tissue disrupts the endocrine system and can have detrimental effects on women's health. Furthermore, some diseases, such as uterine sarcomas, respond poorly to conventional chemotherapy and radiotherapy.13 Thus, there is a need for tissue-engineered models of reproductive tissue in order to gain a better understanding of disease progression, screen potential therapies,14,15 or restore damaged tissues.16–20
000 women visited the emergency department with gynecological complaints.2 Among the most common gynecological complaints were pelvic diseases, such as endometriosis and polycystic ovarian syndrome, and gynecological cancers such as ovarian cancer, uterine sarcomas, endometrial cancer, and cervical cancer.3 The current standard of care for the majority of gynecological diseases is surgery, chemotherapy, and radiation.4–7Table 1 describes the different treatment options available for the gynecological diseases covered in this review. However, the underlying mechanisms of action are often poorly understood, and there are limited personalized standards of care for patients with metastatic cancer due to its heterogeneous manifestations.8–10 While the removal of reproductive organs may address the symptoms resulting from a gynecological diseases, this carries substantial consequences to the endocrine system.8–10 Beyond fertility, the endocrine system has an essential role in the development and maintenance of tissue structure11 and regulates gene expression for numerous biological processes12 Consequentially, removing the tissue disrupts the endocrine system and can have detrimental effects on women's health. Furthermore, some diseases, such as uterine sarcomas, respond poorly to conventional chemotherapy and radiotherapy.13 Thus, there is a need for tissue-engineered models of reproductive tissue in order to gain a better understanding of disease progression, screen potential therapies,14,15 or restore damaged tissues.16–20
| Tissue | Pathology | Current treatments available | Ref. |
|---|---|---|---|
| Ovary | Polycystic ovarian syndrome | Oral Contraceptive treatments | 4–7 |
| Metformin for adolescents (weight loss, improves ovulation) | |||
| Anti-androgens | |||
| Estrogen-projection pills (improves hyperandrogenism) | |||
| Spironolactone (decrease excessive hair growth-androgen receptor blocker) | |||
| Ovarian cancer | Surgical resection of tumors (debulking) | 37–40 | |
| Chemotherapy (platinum-based-regimen) | |||
| Hysterectomy and bilateral salpingo-oophorectomy | |||
| Omental biopsy and/or omentectomy | |||
| PARP inhibitors | |||
| Lymphadenectomy | |||
| Endometrial lining | Endometriosis | Surgical excision of lesions | 41 and 42 |
| Ablation of lesions | |||
| Lysis of adhesions | |||
| Hormone therapy | |||
| Endometrial cancer | Bilateral salpingo-oophorectomy | 10, 43–46 | |
| Lymphadenectomy | |||
| Radiation | |||
| Chemotherapy | |||
| Hormone therapy (progestins) | |||
| Hysterectomy (simple or radical) | |||
| Uterine muscle | Uterine sarcomas | Surgical resection of tumors | 10, 47–50 |
| Hysterectomy (and bilateral salpingo-oophorectomy) | |||
| Hormonal therapy | |||
| Chemotherapy | |||
| Radiation | |||
| Cervix | Cervical cancer | Immunotherapy | 51–55 |
| Chemoradiation | |||
| Hysterectomy (simple or radical and bilateral salpingo-oophorectomy) |
In vitro preclinical models are being increasingly explored as alternatives to conventional animal models as they are faster and less expensive.21 Cells cultured in two dimensions (2D) on tissue culture plastic are widely used for in vitro studies. However, they fail to resemble the in vivo tissue.22 In contrast, three dimensional (3D) in vitro models provide a better approximation of the in vivo tissue by providing a third dimension of biophysical cues. 3D culture models range from cancer cell spheroids, cell-seeded 3D scaffolds,23–25 cells embedded in hydrogels,26–28 microfluidic chips,29 cell patterning,16,30 and organoids.20 The goals of 3D culture models are to mimic the microenvironment, interrogate the effect of the extracellular matrix (ECM), and provide an alternative to animal models of human disease.
The ECM plays a vital role in cell behavior.31 It is responsible for providing mechanical and structural support to cells and tissues, as well as controlling different cell functions such as cell cycle, morphogenesis, apoptosis, and migration.32 Cell–ECM and cell–cell interactions influence fundamental cell behaviors related to the function of the whole organs. Fig. 2 illustrates the most common components found in the ECM of each of the gynecological tissues discussed in this review. 3D culture systems that incorporate biomaterials are essential for studying the role of the ECM in healthy tissue homeostasis as well as disease progression.26,33–36 Furthermore, recreating in vitro tissue requires biological matrices with specific characteristics and tunable properties that can provide an appropriate condition for the attachment, growth, proliferation, and signaling of different types of cells. A guiding principle in the design of 3D culture environments is the accurate presentation of the various signals (growth factors, hormones, ECM, mechanics, and others) in tunable microenvironments that will allow multiple cell types to grow. Specifically, gynecological tissues are highly dynamic, plastic, with continuously changing ECM due to hormone-responsive processes such as menstruation, pregnancy, and menopause. Thus, it is difficult to model the native tissue in vitro. However, numerous biomaterials have been evaluated in different in vitro models of female reproductive tissues to interrogate the cell response to hormone stimuli and the role of the ECM.23,56 The selection of which biomaterial to use in an in vitro model depends on each tissue's characteristics, the disease, and the specific hypothesis being addressed.
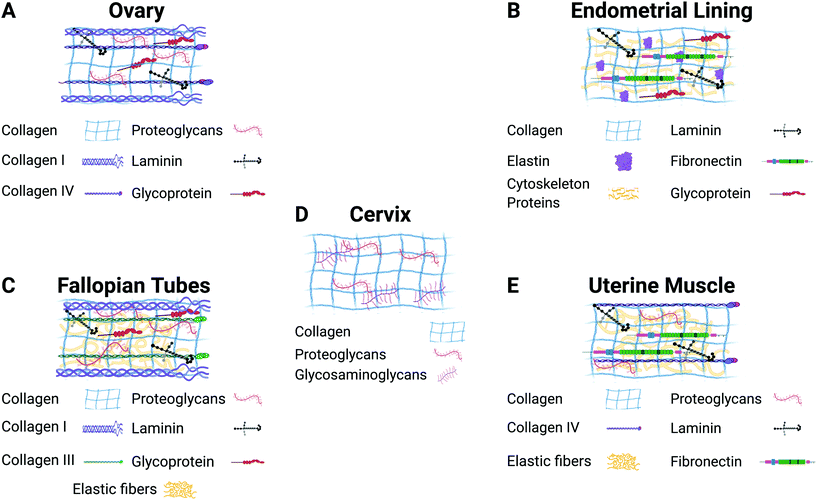 | ||
| Fig. 2 Description of the major components of the extracellular matrix in each gynecological tissue. Figure created with BioRender.com. | ||
In this review, we provide a general overview of the tissues that make up the female reproductive system and a description of different biomaterials used in 3D in vitro models to simulate healthy tissues and disorders affecting women's health. There is a specific emphasis on the guiding principles for designing 3D culture environments and their potential to be used as tools to look for alternative treatment strategies.
2. Building in vitro models with biomaterials
Biomaterials currently used in in vitro models are constructed from either natural polymers or synthetic polymers.56,57 Natural biomaterials can provide similar biological cues to those found in the body.34,58,59 However, they can be difficult to precisely control in terms of spatiotemporal cues or substrate stiffness.60,61 In contrast, synthetic polymers provide more experimental control but need to be modified to provide biological cues.26,32,62,63Among the most used natural biomaterials are collagen and Matrigel. Collagen is the most abundant ECM constituent; it corresponds to approximately 30% of the total mammalian protein mass.64 The collagen family consists of 28 collagen types (I–XXVIII), where collagen type I is the main structural protein in the interstitial ECM,65 and collagen type IV is a crucial component for observed differences in shape, structure, and function of cells.64,66 Due to its bioavailability, role in cell–ECM interactions, and association with disease progression, collagen I is a frequently used biomaterial in 3D in vitro models.32,67,68 Moreover, collagen I is hydrophilic and has a porous structure. These properties enable the diffusion of nutrients and oxygen, allowing cells to attach and grow.67 However, collagen I hydrogels have some limitations. Collagen hydrogels are limited in protein concentration by the biological sources available, and without chemical modification, it is difficult to decouple protein concentration and substrate stiffness.36 Additionally, collagen alone may not provide sufficient biochemical cues to induce cells to respond as they would in vivo.69
Matrigel is another commonly used natural matrix that promotes cell attachment and proliferation across a wide range of cell types.34 Matrigel is a tumor-derived product extracted from Engelbreth–Holm–Swarm mouse sarcomas comprised of basement membrane components.70 It is widely used for in vitro adhesion, invasion, and capillary formation assays as it provides cells with ECM and growth factor cues present in many tissues.71 Moreover, Matrigel constituent proteins stimulate cell–matrix interactions and induce differentiation.72 Matrigel hydrogels have biomimetic cues that provide a suitable environment to support cell adhesion and allow the diffusion of nutrients.34,73–75 However, Matrigel does not contain high concentrations of some ECM components such as collagen type I and hyaluronan, limiting its ability to mimic in vivo tissue.60 Matrigel, when combined with other biomaterials such as collagen type I, can improve the simulation of gynecological tissue and tumor models.34 For example, Park et al. found that the combination of collagen type I and Matrigel simulated the architecture and physiology of the native endometrial tissue.76 Nevertheless, Matrigel still has limitations due to its low batch-to-batch reproducibility, which creates uncertainty in cell-culture experiments.60
Other natural polymers include alginate,77 gelatin,78 chitosan,79 fibrin,80 hyaluronic acid,81 and decellularized matrices.16 These natural biomaterials are biocompatible and can be used to replicate specific types of ECM. However, they often lack mechanical integrity, and are frequently blended with other polymers. The limitations of natural biomaterials have driven the search for synthetic alternatives.60
Synthetic biomaterials are an alternative to natural biomaterials. Among the most common are polyethylene glycol (PEG) and PEG copolymers such as poly(L-lactide) (PLLA), poly(D,L-lactide-co-glycolide) (PLGA), and poly(-caprolactone) (PCL). However, synthetic polymers also have limitations due to their bio-inert nature and without modifications often fail to support desired cell behaviors and tissue formation.82 They can be chemically modified or combined with other natural biomaterials to design in vitro models and study cell–cell and cell–ECM interactions.83 These modified synthetic biomaterials are biocompatible, biodegradable, and reproducible.62 Furthermore, they can provide more experimental control over ECM properties in the microenvironment.63 For example, PEG is one of the most studied and widely used synthetic polymers due to its biocompatibility and versatility.84,85 This biomaterial also presents advantages in cell culture as it can be chemically modified and is hydrophilic, enabling cell encapsulation.60 The use of natural and synthetic biomaterials, or a combination of the two, can lead to the formation of advanced in vitro models that resemble in vivo tissue.
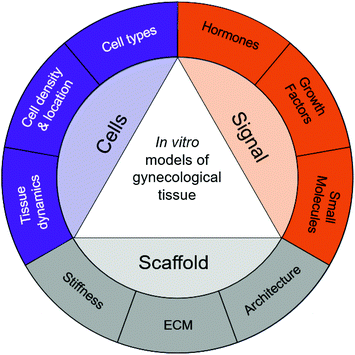
As with all tissue-engineered constructs, developing 3D models of gynecological tissue requires cells, scaffold, and signal (the tissue engineering triad). The choice of cell types, biomaterials, and growth factors must be tailored to the tissue of interest as well as the disease state.21 The size and the thickness of 3D models must take nutrient transport into account as well, as oxygen diffusion is limited to 200 μm.86 Furthermore, if the construct is going to exceed the limit of oxygen transport or the role of vasculature in disease is being evaluated, vascularization must be considered as an added parameter.87,88Table 3 provides an overview of the biomaterials and cell types used to design in vitro models for gynecological tissue, and Fig. 3 demonstrates the properties of gynecological tissue that can inspire biomimetic in vitro models. As researchers have a wide array of biomaterials to choose from, this paper reviews the native function, architecture, and ECM components to be emulated.
3. Biomaterials for in vitro models of gynecological tissues
3.1. Ovaries and fallopian tubes
| Tissue | Prominent extracellular matrix | Cell types present | Ref. |
|---|---|---|---|
| Ovary | • Collagen | • Germ cell | 77, 89–91 |
| ○ Type I and IV | ○ Oocyte | ||
| • Fibronectin | • Somatic cells | ||
| • Laminin | ○ Granulosa cells | ||
| • Glycoproteins | ○ Theca cells | ||
| • Proteoglycans | • Stromal cells | ||
| • Vascular cells | |||
| • Smooth muscle cells | |||
| • Endothelial cells | |||
| Fallopian tubes | • Fibers | • Ciliated epithelial cells | 92 and 93 |
| ○ Collagens (I and III) | • Secretory epithelial cells | ||
| ○ Elastic and reticular | • Smooth muscle cells | ||
| • Nonfibrillar molecules | |||
| ○ Glycoproteins | |||
| ○ Proteoglycans (decorin, biglycan, fibromodulin, and versican) | |||
| Endometrial lining | • Collagen | • Endometrial epithelial cells | 28, 94–103 |
| • Fibronectin | ○ Endometrial basalis | ||
| • Laminin | ○ Endometrial functionalis | ||
| • Elastin | • Endometrial Stromal cells | ||
| • Cytoskeletal proteins | ○ Fibroblasts | ||
| • Glycoproteins | • Immune cells | ||
| ○ Leukocytes (T and B cells, mast cells) | |||
| • Endothelial cells | |||
| Uterine muscle | • Structural proteins | • Myometrial smooth muscle cells | 104–106 |
| ○ Collagen | |||
| ○ Elastin | |||
| • Substrate adhesion molecules | |||
| ○ Fibronectin | |||
| ○ Laminin | |||
| ○ Collagen IV | |||
| • Proteoglycans | |||
| Cervix | • Structural proteins | • Epithelial cells | 107–110 |
| ○ Collagen fibers | ○ Squamous | ||
| • Glycosaminoglycans other proteins | ○ Glandular | ||
| • Proteoglycans | • Stromal cells | ||
| ○ Fibroblasts | |||
| • Smooth muscle cells | |||
| ○ Cervical smooth muscle | |||
| ○ Vascular smooth muscle | |||
| • Endothelial cells | |||
In vitro studies of artificial ovaries for fertility preservation have used fibrin as a supporting biomaterial due to its physical properties and crosslinking ability (Table 3).80 However, this biomaterial has low mechanical strength compared with other polymers, and it needs to be combined with other natural or synthetic polymers to mimic the cell microenvironment.80 When mouse follicles were encapsulated in a fibrin alginate mixture, the hydrogels provided excellent architecture for cell attachment, proliferation, differentiation, and cell–cell signaling.114In vitro models of the ovary have also used alginate58,90 or PEG113,115 as supporting materials. These biomaterials also promote the survival, maturation, and autonomous function of follicles. The majority of the 3D in vitro models focus on the study of fertility preservation (not covered in this review, but covered in others111,116,117) and diseases such as polycystic ovarian syndrome and ovarian cancer.
| Tissue | General description | Biomaterial(s) | Cell(s) | Ref. |
|---|---|---|---|---|
| Ovary | 3D culture model to assess cell matrix interactions of epithelial ovarian cancer cells, crucial in cancer progression and anti-cancer drug resistance | PEG-based hydrogel | Human epithelial ovarian cancer cell lines: OV-MZ-6, SKOV-3 | 35 |
| Thermal responsive hydrogel tri-block copolymer for 3D ovarian cancer culture | PLGA-PEG-PLGA | Ovarian cancer cell line (HO8910) | 62 | |
| Ovarian follicles for fertility preservation and follicle development | Alginate (alone or combined with fibrin) | Mouse ovarian follicles | 58,90 | |
| PEG-based hydrogel | Ovarian preantral follicles | 115 | ||
| Primary follicles (isolated from B6CBAF1 mice) co-cultured with human adipose-derived stem cells (human ADSCs (Zen-Bio)) | 113 | |||
| 3D culture of mouse follicles for Toxicity and High-Throughput (HTP) Analysis | Fibrin alginate hydrogel matrix | Two-layered secondary follicles from female F1 hybrids (C57BL/6JRccHsd inbred × CBA/J CrHsd) | 114 | |
| Human endometrial organoids constructed to study the effects of androgen levels in PCOS | Novel scaffold-free multicellular endometrial organoid | Primary endometrial epithelial and stromal cells from endometrial tissues | 119 | |
| Micro-culture device to examine the influence of macrophages on ovarian cancer spheroid spreading | Spheroid co-culture model with collagen type I hydrogel | HGSOC cell lines OVCAR3 and OV90 (ATCC), OVCA433 (NCI 60 panel (NIH, Bethesda, MD)), primary human macrophages | 122 | |
| 3D organotypic model of ovarian cancer to simulate the metastatic microenvironment | Collagen type I | Ovarian cancer cell line (OVCAR4) | 129 | |
| Primary fibroblasts (NOF) | ||||
| Mesothelial (HPMC) cells | ||||
| Biomaterial-based platform of ovarian cancer spheroid growth to analyze cell–ECM interactions | GelMA-based hydrogels | Ovarian cancer cell line OV-MZ-6 | 132 | |
| Collagen I hydrogels used as a platform to investigate the influence of collagen I concentration on ovarian cancer proliferation | Collagen type I | Primary human omental fibroblasts | 133 | |
| Ovarian cancer cell lines: OVCAR3, OV90 (ATCC), OVCA433 and OVCAR8 (NCI 60 panel) | ||||
| Fallopian tubes | 3D spheroid culture model of primary fallopian tube secretory epithelial cells to study the biology and etiology of fallopian tube tissues and compared with 2D cell cultures | 3D spheroids coated in poly-2 hydroxyethyl methacrylate (polyHEMA, sigma) | Fallopian tube secretory epithelial cells | 92 |
| Collagen I substrates with varying curvature to examine cell–cell and cell–substrate interactions on FTE invasion | Collagen types I and III | Mouse fallopian tube epithelial (FTE) cells | 182 | |
| Influence of ECM cues in fallopian tube epithelial cell invasion in a microfluidic lumen model | Collagen gel mixture (collagen type I and III) | Mouse fallopian tube epithelial cells expressing mutated p53 | 139 and 181 | |
| 3D organoid in vitro model to mimic the human fallopian tube epithelium | Matrigel | Fallopian tube epithelial cells | 141 | |
| 3D alginate culture system to study the role of the fallopian fimbriae in serous tumorigenesis | Alginate | Fallopian fimbriae | 143 | |
| Uterine muscle | Magnetic 3D bioprinting model to study uterine contractility over time | 3D bioprinted human myometrium cells into rings | Human uterine smooth muscle cells (HUtSMCs): C-12575 and C-12576, PromoCell Primary human uterine smooth muscle cells (SMCs) | 104 |
| 3D in vitro model for myometrial smooth muscle cells studies | Fibrinogen-PEG | hTERT cells | 105 | |
| PVA | PHM cells | |||
| human primary myometrium cells | ||||
| Uterine scaffolds to repair damaged uteruses in rabbits, resulting in live births | PGA-PGLA | Rabbit autologous myometrial and endometrial cells | 146 | |
| Two forms of collagen (monomeric and fibrillar) were analyzed to show the differences in cell morphology, proliferation, and interaction of uterine fibroids | PureCol collagen solution | Leiomyoma samples (LSMCs) | 152 | |
| Endometrial lining | Multicellular model to study angiogenesis and trophoblast invasion in the endometrium | GelMA | Cell lines: | 28 |
| HUVECs: C2517A | ||||
| HESCs: CRL-4003 | ||||
| EECs: FC-0078 | ||||
| HTR-8/SVneo: CRL-3271 | ||||
| 3D organoid models mimic the physiology of the endometrium and endometrial stromal and epithelial cells interactions | Matrigel | Endometrial stromal cells and epithelial cells | 71, 74 and 120 | |
| Collagen/Matrigel matrix to study endometrial cancer invasion | Collagen type I–Matrigel | Human endometrial adenocarcinoma cell line KLE | 76 | |
| Human endometrial tissues | ||||
| Collagen scaffold-based model of the endometrium containing both epithelial and stromal cells | Bovine collagen type I | Decidual stromal cells and epithelial cells | 97 | |
| Multicellular model to analyze the influence of hormones on cell–cell interactions | PEG-VS | Ishikawa human endometrial adenocarcinoma | 100 | |
| hTERT immortalized human endometrial stromal cells | ||||
| Primary isolated endometrial stromal and epithelial cells | ||||
| 3D model to analyze physiologic changes that occur in endometrial regeneration during the proliferative phase | Collagen type I-Matrigel | Human telomerase-immortalized endometrial stromal cells | 162 | |
| Ishikawa endometrial adenocarcinoma cells | ||||
| Developed a multicellular model of the endometrium to analyze the dissolution rate of ECM under the effect of SrtA | PEG-VS | Ishikawa human endometrial adenocarcinoma cells | 169 | |
| PEG-NB | hTERT-immortalized human endometrial stromal cells | |||
| 3D fibrin matrix of endometrial explants reflected the environment established in the peritoneal surface as a result of the retrograde menstruation | Fibrin matrix | Fragments of human endometrium | 168 | |
| Cervix | 3D engineered model of the cervical ECM to study the mechanical and biochemical effects of progesterone on engineered cervical tissue | Collagen scaffolds | Human cervical fibroblasts | 110 |
| 3D organotypic culture of epithelial cell cultures from human ectocervix, transformation zone, and endocervix | Collagen type I | Human cervical tissue from the CHTN (epithelial cells, stromal cells) | 109 | |
| 3D printed model with a cervical cancer cell line and gelatin/alginate/fibrinogen hydrogels simulated the tumor microenvironment | Gelatin-alginate-fibrinogen | HeLa cells | 178 | |
| Organotypic culture for analyzing the ability of human lymphocytes to infiltrate human papillomavirus (HPV)-associated (pre)neoplastic lesions of the uterine cervix | Organotypic raft cultures (collagen gel) | SiHa cell line (cervical carcinoma-derived keratinocyte cell line) | 180 | |
| Peripheral blood mononuclear cells (PBMC) |
There are currently multiple in vitro models of PCOS that have been developed to simulate the behavior of endometrial epithelial cells. Endometrial organoids of epithelial cells organized within Matrigel droplets enabled 3D passaging of normal and PCOS-derived human endometrial cells, demonstrating potential application for long-term culture.120 Similarly, a scaffold-free endometrial organoid replaced the biomaterial with stromal cells to provide a supportive layer for the epithelial cells, much like in the native tissue. These scaffold free organoids simulated the effects of excess androgen, leading to the understanding of new mechanisms associated with PCOS and endometrial neoplasia.119 Methods to expand endometrial epithelial cells in vitro are critical to enabling future development of PCOS models. Similar to cancer organoids, biomaterial strategies may help to support endometrial epithelial cells to survive and proliferate in vitro.121
In vitro models have been developed to simulate the various stages of ovarian cancer progression. Ovarian cancer attachment to the omentum or peritoneal cavity is a critical step in the metastatic cascade. This stage is influenced by ECM cues, as demonstrated by studies where the ECM ligand density was varied on a 3D hydrogel matrix.125 PEGDA/GelMA hydrogels enable changing the stiffness and ligand density independently, allowing exploration of hypotheses regarding mechanical and chemical cues in the tumor microenvironment.125 Organotypic models can be used as a platform to evaluate cell adhesion and invasion.126,127 A 3D co-culture model based on omentum with three layers of cell types and ECM (fibronectin and collagen type I) was used to analyze the effect of drug combinations on cell adhesion and proliferation.128,129
After ovarian cancer cells attach and clear through the mesothelial cell lining, they spread along the underlying ECM, remodel the ECM, and proliferate.130,131 The underlying mechanism of these steps has also been investigated using 3D in vitro models. Ovarian cancer spheroid spreading in response to macrophage-derived soluble cues has been evaluated on a high density collagen I hydrogels122 and modified gelatin hydrogels.132 After spreading along the ECM, ovarian cancer cells proliferate, which has also been demonstrated to depend on ECM cues. Collagen I hydrogels were used as a platform to investigate the interplay of collagen I concentration, soluble cues, and ovarian cancer proliferation.133 These studies demonstrated that increasing collagen density sensitized ovarian cancer cells to soluble cues, resulting in increased proliferation. Other platforms that have been used to investigate ovarian cancer proliferation include PLGA-PEG-PLGA tri-block copolymer hydrogels62 and PEG-based hydrogels.35 Both of these studies evaluated the role of ECM molecules on ovarian cancer cell proliferation, concluding that cell-integrin engagement drives responsiveness to ECM cues. Taken together, these in vitro models reveal significant insights into the progression of ovarian cancer.
The influence of ECM cues in fallopian tube epithelial cell invasion was investigated in a 3D model that captured the size and shape of cortical inclusion cysts,139 which are thought to play a role in the initiation of ovarian cancer. Varying hydrogel concentrations of collagen I, collagen III, or both in a microfluidic lumen system suggested that increases in collagen III promote fallopian tube epithelial cell invasion.140 The role of cortical inclusion cyst curvature was explored in a modified model in which the radius of curvature was varied, demonstrating a link between fallopian tube invasion and substrate curvature.139
Fallopian tube secretory cells are considered the cell of origin in the majority of ovarian cancers. When compared to 2D cultures, 3D spheroid cultures better mimicked the in vivo tissue and cell behavior.92 Results showed that the 3D in vitro model offered a better approximation, compared to the 2D culture, of the biology of healthy tissue and malignant transformation to carcinoma.92 In another study, 3D organoid culture was used to model the fallopian tube epithelium function. The fallopian tube is a dynamic tissue that is continuously changing in response to the stimulus of hormones, such as progesterone. Epithelial cells were embedded in Matrigel, resulting in a 3D model that closely mimicked the in vivo tissue and tested its ability to respond to oestradiol and progesterone, two hormones that fluctuate during the menstrual cycle.141Ex vivo models of fallopian tube epithelium have also used either collagen or alginate matrices.142,143 A co-culture system of ciliated and secretory cells in an ex vivo model studied the behavior of these cells and their response to DNA damage, an initiating step in the progression of carcinoma. The model recapitulated the in vivo environment of the fimbria epithelium and identified a response to mutagenic injury that was confirmed with pathological samples.142 Another ex vivo 3D model used human fallopian fimbriae to study the influence of specific ovulatory factors such as estradiol, oxidative stress mimetic (H2O2), and insulin on cell proliferation. These factors are regulators of healthy tissue physiology and were hypothesized to contribute to carcinogenesis.143 Results showed that the alginate matrix maintained the tissue structure up to 7 days, and the presence of H2O2 and insulin influenced cell proliferation. These tissue models provided valuable information about the response of the fallopian tubes to the exposure to hormonal changes and ovulatory factors. Furthermore, they may provide insights into the understanding of epithelial biology and oncogenesis.
3.2. Uterine muscle
3D cell culture platforms can better model tissue contractility compared with 2D models and is easier to interrogate in vitro than in animal models. Magnetic bioprinting of myometrial cells into hollow rings was used to study uterine contractility. Abnormal uterine contractility is related to common pathological disorders, including irregular menstrual cycle, infertility, and preterm labor.104 For the first time, this bioprinted 3D in vitro model of primary human uterine smooth muscle cells obtained from patients during a cesarean section showed a method for the personalization of therapies for uterine contractility disorders. Furthermore, the bioprinted rings exhibited different responses to contractility inhibitors such as indomethacin and nifedipine. In all experimental conditions, inhibitors slowed or stopped the contraction of the in vitro model.104 However, modeling the human myometrium can be challenging as it is difficult to obtain primary cells from uterine biopsies, and those that are obtained often fail to proliferate in standard culture conditions. Therefore, opportunities to model human myometrium in in vitro culture systems are dependent on finding alternative cell sources of smooth muscle cells or developing more robust methods of expanding human myometrial cells in vitro.
Potential biomaterials used to design 3D in vitro models of the uterus provide new insights on the mechanism of cell behavior and uterus function. A recent study used a PGLA coated PGA scaffold seeded with autologous myometrial and endometrial cells from rabbits to bioengineer uterine tissue.146 Rabbits have been frequently used in reproductive studies as they have a relatively larger uterus with a similar structure to human tissue compared with other animal models. A biodegradable cell-laden scaffold resulted in native tissue structure prior to implantation and upon implantation restored uterine function, resulting in live births. Furthermore, rabbits that received a cell-seeded scaffold were capable of responding to mechanical strains that occurred during pregnancy and developed all of the uterine tissue layers, including the myometrium and endometrium.147 PGLA and PGA are effective scaffolds for tissue regeneration due to their high porosity. Also, these polymers are biocompatible and have tunable mechanical and chemical properties.147 Another study used a combination of fibrinogen with PEG and PVA to build a 3D scaffold with smooth muscle cells. This modified biomaterial was biodegradable, biocompatible, had good cell adhesion, and facilitated the development of myometrial 3D structures (Table 3).105 Overall, the development of these tissue-engineered approaches has led to a deeper understanding of dynamic mechanical and chemical cues on uterine function.
Uterine fibroids (leiomyomas or myomas) are the most common benign yet often painful tumors of the myometrium. In the U.S., they account for approximately 30% of all hysterectomies among women between 18–44 years old.148,149 Associated symptoms include pelvic pain, excessive uterine bleeding, infertility, and pregnancy complications. Despite the frequency of these tumors, there is limited information and understanding of their mechanism and treatment. Studies suggested that the inflammatory events caused by physiological injuries in the uterus are associated with an increase in the production of ECM that results in the formation of leiomyomas.150 Collagens are the most abundant components of the ECM in uterine fibroids (Fig. 2E) and are responsible for producing the rigid structure of leiomyomas. Furthermore, myomas present higher levels of metalloproteinases at the different stages of growth.150,151 Therefore, a guiding principle when developing in vitro models of uterine fibroids is to include collagen and enable ECM variation and degradation.
Fibroid formation is dependent on the deposition of collagen I and III as well as dysregulation of signaling processes such as the mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K) pathways. An in vitro model of leiomyoma muscle cells embedded in collagen hydrogels evaluated their response to growth factors, including platelet-derived growth factor (PDGF), which is associated with fibroid formation and growth. This demonstrated differences in cell morphology, proliferation, and interactions with monomeric and fibrillar collagen in the presence or absence of PDGF.152 Results showed that leiomyoma smooth muscle cells had distinct morphologies on the different collagen matrices and that proliferation depended on overall collagen concentration. Another study used collagen I to model uterine fibroids and myometrial cells to study the changes in cell behavior and gene expression in response to potential therapies. The model maintained the molecular phenotype of in vivo tissue and cell–ECM interaction. Furthermore, this culture system assessed the mechanism of abnormal ECM formation and the effectiveness of potential therapeutic agents.153 Although uterine myomas are persistent tumors and often painful, it is still an understudied research area. In vitro 3D models have the potential to improve and understand the biology and cell behavior of uterine leiomyomas, particularly the importance of ECM in the formation of fibroids.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 women in the U.S154 and comprising only 3–5% of all uterine cancers.8 These tumors are considered highly aggressive, resulting in poor patient prognosis. Uterine sarcomas are classified as leiomyosarcomas, endometrial stromal sarcomas, low grade or high-grade sarcomas, undifferentiated uterine sarcomas, or other.8,155 Leiomyosarcomas are mesenchymal tumors and are the most common uterine sarcoma arising within the myometrium; they are malignant smooth muscle tumors and typically present with prominent necrosis.50 Adenosarcomas are a mixture of benign epithelial cells and malignant mesenchymal cells.48 These distinctions in cell type are critical to building 3D models that replicate the tumor microenvironment.
000 women in the U.S154 and comprising only 3–5% of all uterine cancers.8 These tumors are considered highly aggressive, resulting in poor patient prognosis. Uterine sarcomas are classified as leiomyosarcomas, endometrial stromal sarcomas, low grade or high-grade sarcomas, undifferentiated uterine sarcomas, or other.8,155 Leiomyosarcomas are mesenchymal tumors and are the most common uterine sarcoma arising within the myometrium; they are malignant smooth muscle tumors and typically present with prominent necrosis.50 Adenosarcomas are a mixture of benign epithelial cells and malignant mesenchymal cells.48 These distinctions in cell type are critical to building 3D models that replicate the tumor microenvironment.
The only treatment available for uterine sarcomas is surgery and radiotherapy.8 The stromal sarcomas have characteristic translocations; meanwhile, leiomyosarcomas have complex karyotypes. These characteristics make them difficult to treat and would benefit from the development of targeted therapies. There are no current 3D in vitro models with biomaterials for this type of tumor. However, a 2D in vitro model was used to interrogate the underlying molecular biology and identify biomarkers for potential treatment strategies.156 This study evaluated a possible treatment of uterine sarcomas with PGJ2 and dasatinib, which are considered potential cancer treatments. Results showed that the combined treatment produced a synergistic effect on inhibiting cancer cells proliferation by negatively regulating the MAPK pathway. Furthermore, 3D in vitro models could identify potential molecular targets or evaluate currently available treatment options.
3.3. Endometrial lining
Endometrial lining is one of the most challenging tissues to recapitulate in vitro as it is highly dynamic in response to hormones produced by the ovary during the menstrual cycle. During the first days of the menstrual cycle, glandular and stromal cells proliferate due to the rising estrogen levels produced by the follicle.98 Following ovulation, cell differentiation occurs as the corpus luteum, a dynamic endocrine gland of the ovary, synthesizes and increases progesterone and estradiol.159 If fertilization does not occur, the corpus luteum relapses, which decreases the levels of progesterone and leads to glandular and stromal breakdown (menstruation).99 If fertilization does occur, the changes in the endometrial blood vessels play an important role in the implantation of the blastocyst, as vascularization contributes to uterine receptivity.160,161 Endometrium remodeling depends on different hormone changes. Thus, the guiding principle in designing 3D in vitro models of the endometrium is to provide scaffolding and biophysical cues such that the cells respond to stimulation with hormones.
3D in vitro models have been successfully developed and used to study the underlying physiology of the endometrium. The two main types of endometrial models are organoids and collagen scaffolds. Long-term expandable and stable culture organoids were achieved with the combination of Matrigel, growth and signaling factors such as EGF and WNT activators, and either human or mouse endometrial cells. This culture system showed the capacity to respond in a physiological manner to hormones and specific biomarkers in cell proliferation and maturation.74,120 3D endometrial organoid models allow cells to self-organize in a structure similar to stratifications observed in vivo and demonstrate similar molecular signatures to the in vivo tissue.20 Collagen scaffolds have also been used to model the layers of endometrial epithelial cells for long term expansion.97 A model using collagen I scaffolds with endometrial epithelial cells was responsive to stimulation with hormones, resulting in epithelial differentiation and stromal decidualization.97 Both organoids and scaffolds present good approximations of the in vivo tissue as they recapitulate the original tissue (healthy or pathological) and provide a powerful tool for modeling and deciphering tissue development. By manipulating their structure, these models can be a potential tool to integrate other types of cells, such as immune or trophoblast cells, in order to gain a better understanding of the interactions between cells and their response to hormone stimulation.
Collagen and Matrigel can also be combined in order to harness the basement membrane cues from Matrigel and the physical cues from collagen. A collagen–Matrigel blend was used to create a tissue-engineered model that analyzed the communication between endometrial stromal and epithelial cells in response to endocrine signaling.162 Using this in vitro model, the authors were able to elucidate the effects of cytokines present during the proliferative phase of the human endometrium. This hydrogel allowed cells to respond to endocrine effects and demonstrated that the stromal tissue did not proliferate more in response to increasing concentrations of collagen I.162 Gelatin hydrogels, made of denatured collagen, have been used to model angiogenesis and trophoblast invasion in the endometrium, as gelatin is not fibrillar and allows for easier matrix remodeling and cell attachment compared to fibrillar collagen.28 Trophoblast cells drove the process of the non-pathological angiogenesis, which occurs regularly in the endometrium to rebuild the vascular bed.163 Chemically modified gelatin hydrogels have also been used to explore the relationship between biophysical, biochemical, and cellular signals from the endometrium. Due to its crosslinking properties, methacrylamide-functionalized gelatin (GelMA) provides matrix stiffnesses relevant to the in vivo tissue and similar structure to the in vivo environment. It also promotes cell function, endometrial angiogenesis, and responsiveness to hormones. The attachment of methacrylamide groups allows gelatin to be U.V. light polymerized and relatively homogenous in composition and structure.28 Taken together these studies further our understanding of cell behavior in the endometrium. Additionally, these in vitro models capture the complexity of cell–cell and cell–ECM interactions in the human endometrium.
3D in vitro models have investigated the mechanism of action for endometriosis in order to identify therapeutic targets. Fibrin hydrogels were used to culture cells from a fragment of human endometrium. Fibrin is a major component of blood clots and reflects the environment established in the peritoneal surface as a result of retrograde menstruation.168 Endometrial cells proliferated and invaded the fibrin matrix, generating new glands, stroma, and vessels similar to the in vivo tissue. Furthermore, the authors demonstrated that endometrial cell proliferation and angiogenesis were related to the expression of vimentin. Vimentin is an intermediate filament that crosslinks other cytoskeletal proteins and has an important role in the dynamic of mesenchymal cells.
A 3D functional co-culture model of epithelial cells with stromal cells simulated the endometrium physiology.100 The synthetic 3D matrix of modified PEG hydrogels analyzed the response of the cells to hormones and cell–ECM and cell–cell interactions. The PEG hydrogels were modified to incorporate integrin-binding peptides and ECM-binding peptides to simulate the in vivo tissue. This promoted cell attachment, cell viability, remodeling of the 3D matrix, and hormone-mediated cell communication over two weeks in culture. Furthermore, this synthetic ECM model demonstrated phenotypic differences between co-culture models of patient-derived primary cells and endometrial cell lines. Another study by the same group used a modified PEG hydrogel to investigate the kinetics of gel dissolution as a function of change in concentration of enzyme and substrate as well as crosslinking parameters.169
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 380 women in the United States, predominantly affecting postmenopausal women.170 There are two types of endometrial cancer tumors: low grade (type I) and high grade (type II). Type I tumors are related to unopposed estrogen stimulation and may be associated with other gynecological diseases such as polycystic ovarian syndrome.43 Type II tumors are usually high grade and present at higher stages with associated poor prognosis.43 Early-stage endometrial carcinoma, especially low grade, is highly curable, but advanced stage endometrial carcinomas have low survival rates.171In vitro models can lead to a better understanding of the invasion and progression of these tumors and future treatment strategies (Table 3).
380 women in the United States, predominantly affecting postmenopausal women.170 There are two types of endometrial cancer tumors: low grade (type I) and high grade (type II). Type I tumors are related to unopposed estrogen stimulation and may be associated with other gynecological diseases such as polycystic ovarian syndrome.43 Type II tumors are usually high grade and present at higher stages with associated poor prognosis.43 Early-stage endometrial carcinoma, especially low grade, is highly curable, but advanced stage endometrial carcinomas have low survival rates.171In vitro models can lead to a better understanding of the invasion and progression of these tumors and future treatment strategies (Table 3).
The guiding principle for the design of 3D in vitro models for endometrial cancer is to capture the cancer-stromal cell interactions. A blend of collagen and Matrigel matrices was used to support endometrial epithelial cells cultured in a 3D in vitro model to understand endometrial cancer invasion. Similar to the model used to study endometriosis,162 a mixture of collagen I and Matrigel provided a structure to the in vivo tissue. Collagen provided a stromal matrix and Matrigel provided an artificial basement membrane.76 Other 3D in vitro models have been used to target potential treatment alternatives but did not include a biomaterial on their structure. Endometrial cancer patient-derived organoids have been used to study drug sensitivity to endocrine treatments.130 Additionally, a study with organoids models compared 3D multicellular structures with 2D monolayer culture models to study endometrial cancer resistance to various drug treatments. This study used doxorubicin and cisplatin, which are chemotherapeutic regimes for endometrial cancer.172 The authors demonstrated that 3D culture models displayed significant decreases in responsiveness to chemotherapies compared to 2D cell models. Overall, 3D models are a promising tool for drug screening in endometrial cancer.
3.4. Cervical tissue
The ECM plays an important role in the dynamic nature of cervical tissue (Fig. 2D).110 3D in vitro models of this tissue have been used to evaluate cell–ECM interactions. Mechanical and biochemical effects of progesterone on cervical tissue were studied in a 3D in vitro model with cervical fibroblasts seeded on collagen scaffolds. Using this model, the authors investigated the effects of progesterone on cervical shortening. By measuring collagen content and crosslinking in the scaffolds after stimulation with progesterone, they demonstrated that fibroblasts changed their morphology and decreased their production of collagen, leading to a decrease in cervical tissue stiffness.110 An in vitro model commonly used to study skin has also been employed to study cervical cancer: organotypic raft cultures. Organotypic raft cultures are multilayered models of collagen, fibroblasts, and keratinocytes that are maintained at an air–liquid interface. Organotypic raft culture was used to observe differences between neoplastic and healthy cervical tissue in response to acetic acid, which is commonly used in clinical treatments for cervical lesions.174 A separate study used 3D organotypic cultures with human stromal and epithelial cells from each cervical region to evaluate the invasion of epithelial cells and their interaction with stromal cells.109 Increasing the concentration of stromal cells increased the invasion of epithelial cells from each cervical region, suggesting that stromal–epithelial interactions play an essential role in cervical tissue homeostasis.109 The establishment of organotypic raft cultures of cervical cells represents a crucial step forward in understanding cervix biology and function.
There are limited 3D in vitro models of cervical cancer and the underlying mechanism by which cervical epithelium respond to HPV infection is still under investigation. A 3D printed model with a cervical cancer cell line and a hydrogel compromised of gelatin, alginate, and fibrinogen simulated the tumor microenvironment. Chemoresistance was evaluated, and cells within the 3D in vitro model demonstrated increased cell viability and proliferation in the 3D model compared to the conventional 2D culture model. Furthermore, cells within the 3D printed model formed spheroids, while in the monolayer culture, cells formed sheets.178 Similar to healthy cervix, organotypic raft cultures are useful models to study lesions of the cervix caused by HPV.179 For example, a co-cultured organotypic raft culture was used to simulate the infiltration of human lymphocytes into HPV and cervical lesions.180 Overall, in vitro models used to evaluate the progression of cervical cancer provide new insights and are a promising tool to identify potential therapies.
4. Conclusions
Tissue engineering has the potential to transform the landscape of women's health. Studies using in vitro models of gynecological tissues have created hope for women suffering from gynecological diseases. Furthermore, understanding the anatomy, function, and architecture of the tissue is critical for building in vitro models. Specifically, the concentrations and temporal nature of soluble signals such as growth factors, hormones, and small molecules should be evaluated; the scaffold supporting the cells, including ECM components, stiffness, and overall architecture, should be examined; and the cells present, importantly the types of cells, their densities, locations, and dynamics, should be assessed. These properties are summarized in Fig. 3. The biophysical cues present in native tissue can then serve guiding principles to develop 3D in vitro models using biomaterials, cells, and exogenous stimuli.Building tissue-engineered constructs should be done in collaboration with clinicians, pathologists, and cell biologists. Techniques used to evaluate patient biopsies and animal models have a strong overlap with how tissue engineers strive to evaluate in vitro constructs. Engineering biological tissue of the female reproductive system relies on the use of biomaterials, as 3D in vitro models enable us to understand cell–cell and cell–matrix interactions. The in vitro models and biomaterials presented in this review reveal guiding principles and capture complex interactions of different gynecological tissues. There are still many questions to be answered regarding which biomaterial should be used for each model. However, the development of new models opens opportunities to better understand gynecological diseases, identify underlying signaling networks, and evaluate potential treatment strategies.
Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
The authors would like to thank Dr Marian Hettiaratchi for her valuable feedback on this article.Notes and references
- S. Xiao, J. R. Coppeta, H. B. Rogers, B. C. Isenberg, J. Zhu, S. A. Olalekan, K. E. McKinnon, D. Dokic, A. S. Rashedi, D. J. Haisenleder, S. S. Malpani, C. A. Arnold-Murray, K. Chen, M. Jiang, L. Bai, C. T. Nguyen, J. Zhang, M. M. Laronda, T. J. Hope, K. P. Maniar, M. E. Pavone, M. J. Avram, E. C. Sefton, S. Getsios, J. E. Burdette, J. J. Kim, J. T. Borenstein and T. K. Woodruff, Nat. Commun., 2017, 8, 14584 CrossRef CAS.
- A. Fawole and D. Awonuga, Ann. Ib Postgrad. Med., 2007, 5(1), 12–20 Search PubMed.
- M. K. Whiteman, E. Kuklina, D. J. Jamieson, S. D. Hillis and P. A. Marchbanks, Am. J. Obstet. Gynecol., 2010, 202, 541 CrossRef.
- M. L. Pfieffer, Nursing2020, 2019, 49, 34–40 CrossRef.
- A. J. T. Pedersen, T. B. Stage, D. Glintborg, M. Andersen and M. M. H. Christensen, Basic Clin. Pharmacol. Toxicol., 2018, 122, 239–244 CrossRef CAS.
- L. Ibáñez, S. E. Oberfield, S. Witchel, R. J. Auchus, R. J. Chang, E. Codner, P. Dabadghao, F. Darendeliler, N. S. Elbarbary, A. Gambineri, C. Garcia Rudaz, K. M. Hoeger, A. López-Bermejo, K. Ong, A. S. Peña, T. Reinehr, N. Santoro, M. Tena-Sempere, R. Tao, B. O. Yildiz, H. Alkhayyat, A. Deeb, D. Joel, R. Horikawa, F. de Zegher and P. A. Lee, Horm. Res. Paediatr., 2017, 88, 371–395 CrossRef.
- M. D. Pietro, N. Pascuali, F. Parborell and D. Abramovich, Reproduction, 2018, 155, R199–R209 CAS.
- H. Kobayashi, C. Uekuri, J. Akasaka, F. Ito, A. Shigemitsu, N. Koike and H. Shigetomi, Mol. Clin. Oncol., 2013, 1, 599–609 CrossRef.
- L. Doherty, L. Mutlu, D. Sinclair and H. Taylor, Reprod. Sci., 2014, 21, 1067–1092 CrossRef.
- M. Kim, D. H. Suh, K. H. Lee, K. Y. Eom, J. Y. Lee, Y. Y. Lee, H. F. Hansen, M. R. Mirza and J. W. Kim, J. Gynecol. Oncol., 2020, 31, e48 CrossRef.
- S. Ayehunie, A. Islam, C. Cannon, T. Landry, J. Pudney, M. Klausner and D. J. Anderson, Reprod. Sci., 2015, 22, 980–990 CrossRef CAS.
- T. H. Kim and J.-W. Jeong, in Encyclopedia of Reproduction (Second Edition), ed. M. K. Skinner, Academic Press, Oxford, 2018, pp. 305–311 Search PubMed.
- M. Emoto, K. Yano, B. Choijamts, S. Sakai, S. Hirasawa, S. Wakamori, M. Aizawa, K. Nabeshima, K. Tachibana and N. Kanomata, Anticancer Res., 2015, 35, 2739–2746 CAS.
- R. Edmondson, J. J. Broglie, A. F. Adcock and L. Yang, Assay Drug Dev. Technol., 2014, 12, 207–218 CrossRef CAS.
- Y. Wang, Z. Chen, F. Bian, L. Shang, K. Zhu and Y. Zhao, Expert Opin. Drug Discovery, 2020, 1–11 Search PubMed.
- T. Salo, M. Sutinen, E. Hoque Apu, E. Sundquist, N. K. Cervigne, C. E. de Oliveira, S. U. Akram, S. Ohlmeier, F. Suomi, L. Eklund, P. Juusela, P. Åström, C. C. Bitu, M. Santala, K. Savolainen, J. Korvala, A. F. Paes Leme and R. D. Coletta, BMC Cancer, 2015, 15, 981 CrossRef.
- Y. Maru, N. Tanaka, M. Itami and Y. Hippo, Gynecol. Oncol., 2019, 154, 189–198 CrossRef CAS.
- T. K. Woodruff, Reproduction, 2019, 158, F113–F126 Search PubMed.
- E. S. Gargus, H. B. Rogers, K. E. McKinnon, M. E. Edmonds and T. K. Woodruff, Nat. Biomed. Eng., 2020, 4, 381–393 CrossRef.
- L. Alzamil, K. Nikolakopoulou and M. Y. Turco, Cell Death Differ., 2020 DOI:10.1038/s41418-020-0565-5.
- D. C. Fernandes, R. F. Canadas, R. L. Reis and J. M. Oliveira, in Biomaterials- and Microfluidics-Based Tissue Engineered 3D Models, ed. J. M. Oliveira and R. L. Reis, Springer International Publishing, Cham, 2020, pp. 137–159 Search PubMed.
- M. Kapałczyńska, T. Kolenda, W. Przybyła, M. Zajączkowska, A. Teresiak, V. Filas, M. Ibbs, R. Bliźniak, Ł. Łuczewski and K. Lamperska, Arch. Med. Sci., 2018, 14, 910–919 Search PubMed.
- J. M. Aamodt and D. W. Grainger, Biomaterials, 2016, 86, 68–82 CrossRef CAS.
- K. J. Ornell, K. S. Mistretta, E. Newman, C. Q. Ralston and J. M. Coburn, ACS Biomater. Sci. Eng., 2019, 5, 6742–6754 CrossRef CAS.
- T. Wiwatpanit, A. R. Murphy, Z. Lu, M. Urbanek, J. E. Burdette, T. K. Woodruff and J. J. Kim, J. Clin. Endocrinol. Metab., 2020, 105, 769–780 CrossRef.
- E. A. Brooks, M. F. Gencoglu, D. C. Corbett, K. R. Stevens and S. R. Peyton, APL Bioeng., 2019, 3, 026106 CrossRef.
- A. E. G. Baker, L. C. Bahlmann, R. Y. Tam, J. C. Liu, A. N. Ganesh, N. Mitrousis, R. Marcellus, M. Spears, J. M. S. Bartlett, D. W. Cescon, G. D. Bader and M. S. Shoichet, Adv. Mater., 2019, 31, 1901166 CrossRef.
- S. G. Zambuto, K. B. H. Clancy and B. A. C. Harley, Interface Focus, 2019, 9(5) DOI:10.1098/rsfs.2019.0016.
- N. Wang, J. Wang, X. Meng, Y. Bao, S. Wang and T. Li, Sci. Rep., 2018, 8, 12285 CrossRef.
- F. Di Modugno, C. Colosi, P. Trono, G. Antonacci, G. Ruocco and P. Nisticò, J. Exp. Clin. Cancer Res., 2019, 38, 117 CrossRef.
- V. Poltavets, M. Kochetkova, S. M. Pitson and M. S. Samuel, Front. Oncol., 2018, 8 DOI:10.3389/fonc.2018.00431.
- J. Sapudom and T. Pompe, Biomater. Sci., 2018, 6, 2009–2024 RSC.
- K. Duval, H. Grover, L.-H. Han, Y. Mou, A. F. Pegoraro, J. Fredberg and Z. Chen, Physiology, 2017, 32, 266–277 CrossRef CAS.
- G. Benton, I. Arnaoutova, J. George, H. K. Kleinman and J. Koblinski, Adv. Drug Delivery Rev., 2014, 7980, 3–18 CrossRef.
- D. Loessner, K. S. Stok, M. P. Lutolf, D. W. Hutmacher, J. A. Clements and S. C. Rizzi, Biomaterials, 2010, 31, 8494–8506 CrossRef CAS.
- A. Nyga, U. Cheema and M. Loizidou, J. Cell Commun. Signaling, 2011, 5, 239–248 CrossRef.
- F. R. Nezhat, R. Apostol, C. Nezhat and T. Pejovic, Am. J. Obstet. Gynecol., 2015, 213, 262–267 CrossRef CAS.
- R. C. Bast, U. A. Matulonis, A. K. Sood, A. A. Ahmed, A. E. Amobi, F. R. Balkwill, M. Wielgos-Bonvallet, D. D. L. Bowtell, J. D. Brenton, J. S. Brugge, R. L. Coleman, G. F. Draetta, K. Doberstein, R. I. Drapkin, M. A. Eckert, R. P. Edwards, K. M. Elias, D. Ennis, A. Futreal, D. M. Gershenson, R. A. Greenberg, D. G. Huntsman, J. X. Y. Ji, E. C. Kohn, C. Iavarone, E. R. Lengyel, D. A. Levine, C. J. Lord, Z. Lu, G. B. Mills, F. Modugno, B. H. Nelson, K. Odunsi, J. A. Pilsworth, R. K. Rottapel, D. J. Powell, L. Shen, I.-M. Shih, D. R. Spriggs, J. Walton, K. Zhang, R. Zhang and L. Zou, Cancer, 2019, 125, 1963–1972 CrossRef.
- P. Harter, J. Sehouli, D. Lorusso, A. Reuss, I. Vergote, C. Marth, J.-W. Kim, F. Raspagliesi, B. Lampe, G. Aletti, W. Meier, D. Cibula, A. Mustea, S. Mahner, I. B. Runnebaum, B. Schmalfeldt, A. Burges, R. Kimmig, G. Scambia, S. Greggi, F. Hilpert, A. Hasenburg, P. Hillemanns, G. Giorda, I. von Leffern, C. Schade-Brittinger, U. Wagner and A. du Bois, N. Engl. J. Med., 2019, 380, 822–832 CrossRef.
- M. R. Mirza, B. J. Monk, J. Herrstedt, A. M. Oza, S. Mahner, A. Redondo, M. Fabbro, J. A. Ledermann, D. Lorusso, I. Vergote, N. E. Ben-Baruch, C. Marth, R. Mądry, R. D. Christensen, J. S. Berek, A. Dørum, A. V. Tinker, A. du Bois, A. González-Martín, P. Follana, B. Benigno, P. Rosenberg, L. Gilbert, B. J. Rimel, J. Buscema, J. P. Balser, S. Agarwal and U. A. Matulonis, Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer, www.ncbi.nlm.nih.gov, (accessed July 3, 2020).
- E. Rolla, F1000Research, 2019, 8 DOI:10.12688/f1000research.14817.1.
- A. Daniilidis, K. Chatzistamatiou and E. Assimakopoulos, Minerva Ginecol., 2017, 69, 488–503 Search PubMed.
- M. M. Braun, Endometrial Cancer, 2016, 93, 7 Search PubMed.
- E. Kalampokas, F. Payne, A. Nomikos and M. Gurumurthy, Gynecol. Oncol., 2018, 150, 378–386 CrossRef CAS.
- M. E. Randall, V. Filiaci, D. S. McMeekin, V. von Gruenigen, H. Huang, C. M. Yashar, R. S. Mannel, J.-W. Kim, R. Salani, P. A. DiSilvestro, J. J. Burke, T. Rutherford, N. M. Spirtos, K. Terada, P. R. Anderson, W. R. Brewster, W. Small, C. A. Aghajanian and D. S. Miller, J. Clin. Oncol., 2019, 37, 1810–1818 CrossRef CAS.
- P. Morice, A. Leary, C. Creutzberg, N. Abu-Rustum and E. Darai, Lancet, 2016, 387, 1094–1108 CrossRef.
- R. R. Cui, J. D. Wright and J. Y. Hou, BJOG, 2017, 124, 1028–1037 CrossRef CAS.
- I. M. E. Desar, P. B. Ottevanger, C. Benson and W. T. A. van der Graaf, Crit. Rev. Oncol. Hematol., 2018, 122, 10–20 CrossRef CAS.
- C. Parra-Herran and B. E. Howitt, Surg. Pathol. Clin., 2019, 12, 363–396 CrossRef.
- G. Fernandez, S. M. i Borràs, V. N. Pérez and F. Guedea, Rep. Pract. Oncol. Radiother., 2013, 18, 153–158 CrossRef.
- H. C. Chung, W. Ros, J.-P. Delord, R. Perets, A. Italiano, R. Shapira-Frommer, L. Manzuk, S. A. Piha-Paul, L. Xu, S. Zeigenfuss, S. K. Pruitt and A. Leary, J. Clin. Oncol., 2019, 37, 1470–1478 CrossRef CAS.
- H. Li, X. Wu and X. Cheng, J. Gynecol. Oncol., 2016, 27, e43 CrossRef.
- Y. Liu, Y. Zhang, R. Cheng, S. Liu, F. Qu, X. Yin, Q. Wang, B. Xiao and Z. Ye, J. Magn. Reson. Imaging, 2019, 49, 280–290 CrossRef.
- W.-J. Koh, N. R. Abu-Rustum, S. Bean, K. Bradley, S. M. Campos, K. R. Cho, H. S. Chon, C. Chu, R. Clark, D. Cohn, M. A. Crispens, S. Damast, O. Dorigo, P. J. Eifel, C. M. Fisher, P. Frederick, D. K. Gaffney, E. Han, W. K. Huh, J. R. Lurain, A. Mariani, D. Mutch, C. Nagel, L. Nekhlyudov, A. N. Fader, S. W. Remmenga, R. K. Reynolds, T. Tillmanns, S. Ueda, E. Wyse, C. M. Yashar, N. R. McMillian and J. L. Scavone, J. Natl. Compr. Cancer Network, 2019, 17, 64–84 CrossRef CAS.
- N. Bhatla, D. Aoki, D. N. Sharma and R. Sankaranarayanan, Int. J. Gynecol. Obstet., 2018, 143, 22–36 CrossRef.
- A. Tamadon, K.-H. Park, Y. Y. Kim, B.-C. Kang and S.-Y. Ku, Tissue Eng. Regener. Med., 2016, 13, 447–454 CrossRef CAS.
- Z. Yang, H. Xu and X. Zhao, Adv. Sci., 2020, 7(9) DOI:10.1002/advs.201903718.
- J. Vanacker and C. A. Amorim, Ann. Biomed. Eng., 2017, 45, 1633–1649 CrossRef.
- K. Wolf, S. Alexander, V. Schacht, L. M. Coussens, U. H. von Andrian, J. van Rheenen, E. Deryugina and P. Friedl, Semin. Cell Dev. Biol., 2009, 20, 931–941 CrossRef CAS.
- E. A. Aisenbrey and W. L. Murphy, Nat. Rev. Mater., 2020, 1–13 Search PubMed.
- S. Pradhan, J. M. Clary, D. Seliktar and E. A. Lipke, Biomaterials, 2017, 115, 141–154 CrossRef CAS.
- N. Zhou, K. Hu, Z. Guo, Q. Zhang, J. Chen, T. Zhang and N. Gu, J. Nanosci. Nanotechnol., 2018, 18, 5252–5255 CrossRef CAS.
- L. Gu and D. J. Mooney, Nat. Rev. Cancer, 2016, 16, 56–66 CrossRef CAS.
- M. Egeblad, M. G. Rasch and V. M. Weaver, Curr. Opin. Cell Biol., 2010, 22, 697–706 CrossRef CAS.
- S. Zhu, Q. Yuan, T. Yin, J. You, Z. Gu, S. Xiong and Y. Hu, J. Mater. Chem. B, 2018, 6, 2650–2676 RSC.
- R. G. Rowe and S. J. Weiss, Trends Cell Biol., 2008, 18, 560–574 CrossRef CAS.
- K. Wolf, S. Alexander, V. Schacht, L. M. Coussens, U. H. von Andrian, J. van Rheenen, E. Deryugina and P. Friedl, Semin. Cell Dev. Biol., 2009, 20, 931–941 CrossRef CAS.
- C. Liverani, L. Mercatali, L. Cristofolini, E. Giordano, S. Minardi, G. D. Porta, A. De Vita, G. Miserocchi, C. Spadazzi, E. Tasciotti, D. Amadori and T. Ibrahim, Cell. Mol. Bioeng., 2017, 10, 223–234 CrossRef CAS.
- K. L. Sodek, K. J. Murphy, T. J. Brown and M. J. Ringuette, Cancer Metastasis Rev., 2012, 31, 397–414 CrossRef CAS.
- T. Hoshiba and T. Yamaoka, Decellularized Extracellular Matrix: Characterization, Fabrication and Applications, 2019, pp. 1–14, 10.1039/9781788015998-00001.
- H. Hopfer, C. A. Rinehart, G. Vollmer and D. G. Kaufman, Pathobiology, 1994, 62, 104–108 CrossRef CAS.
- A. Nyga, U. Cheema and M. Loizidou, J. Cell Commun. Signaling., 2011, 5, 239 CrossRef.
- J. Wang, J. Ou, Y. Guo, T. Dai, X. Li, J. Liu, M. Xia, L. Liu and M. He, Br. J. Cancer, 2014, 111, 112–124 CrossRef CAS.
- M. Boretto, B. Cox, M. Noben, N. Hendriks, A. Fassbender, H. Roose, F. Amant, D. Timmerman, C. Tomassetti, A. Vanhie, C. Meuleman, M. Ferrante and H. Vankelecom, Development, 2017, 144, 1775–1786 CrossRef CAS.
- M. V. Monteiro, V. M. Gaspar, L. P. Ferreira and J. F. Mano, Biomater. Sci., 2020, 8, 1855–1864 RSC.
- D. W. Park, D. S. Choi, H.-S. Ryu, H. C. Kwon, H. Joo and C. K. Min, Cancer Lett., 2003, 195, 185–192 CrossRef CAS.
- L. D. Shea, T. K. Woodruff and A. Shikanov, Annu. Rev. Biomed. Eng., 2014, 16, 29–52 CrossRef CAS.
- J. C. Pence, K. B. H. Clancy and B. A. C. Harley, Adv. Biosyst., 2017, 1(9) DOI:10.1002/adbi.201700056.
- F. Ahmadi, Z. Oveisi, S. M. Samani and Z. Amoozgar, Res. Pharm. Sci., 2015, 10, 1–16 CAS.
- M. C. Chiti, M. M. Dolmans, J. Donnez and C. A. Amorim, Ann. Biomed. Eng., 2017, 45, 1650–1663 CrossRef CAS.
- B. J. Engel, P. E. Constantinou, L. K. Sablatura, N. J. Doty, D. D. Carson, M. C. Farach-Carson, D. A. Harrington and T. I. Zarembinski, Adv. Healthcare Mater., 2015, 4, 1664–1674 CrossRef CAS.
- J. Zhu, Biomaterials, 2010, 31, 4639–4656 CrossRef CAS.
- S. Pradhan, J. M. Clary, D. Seliktar and E. A. Lipke, Biomaterials, 2017, 115, 141–154 CrossRef CAS.
- E. Cambria, K. Renggli, C. C. Ahrens, C. D. Cook, C. Kroll, A. T. Krueger, B. Imperiali and L. G. Griffith, Biomacromolecules, 2015, 16, 2316–2326 CrossRef CAS.
- C.-C. Lin and K. S. Anseth, Pharm. Res., 2009, 26, 631–643 CrossRef CAS.
- A. Colom, R. Galgoczy, I. Almendros, A. Xaubet, R. Farré and J. Alcaraz, J. Biomed. Mater. Res., Part A, 2014, 102, 2776–2784 CrossRef.
- A. K. Miri, A. Khalilpour, B. Cecen, S. Maharjan, S. R. Shin and A. Khademhosseini, Biomaterials, 2019, 198, 204–216 CrossRef CAS.
- J. Rouwkema, N. C. Rivron and C. A. van Blitterswijk, Trends Biotechnol., 2008, 26, 434–441 CrossRef CAS.
- I. Szmelskyj, L. Aquilina and A. O. Szmelskyj, in Acupuncture for IVF and Assisted Reproduction, ed. I. Szmelskyj, L. Aquilina and A. O. Szmelskyj, Churchill Livingstone, 2015, pp. 23–58 Search PubMed.
- P. K. Kreeger, J. W. Deck, T. K. Woodruff and L. D. Shea, Biomaterials, 2006, 27, 714–723 CrossRef CAS.
- M. Zuccotti, V. Merico, P. Rebuzzini, M. Belli, G. Vigone, F. Mulas, L. Fassina, W. Wruck, J. Adjaye, R. Bellazzi and S. Garagna, Int. J. Dev. Biol., 2015, 59, 211–216 CrossRef.
- K. Lawrenson, M. Notaridou, N. Lee, E. Benjamin, I. J. Jacobs, C. Jones and S. A. Gayther, BMC Cell Biol., 2013, 14, 43 CrossRef CAS.
- C. Godoy-Guzmán, C. Nuñez, P. Orihuela, A. Campos and V. Carriel, J. Anat., 2018, 233, 73–85 CrossRef.
- A. T. Yamada, J. R. Bianco, E. M. O. Lippe, K. Y. Degaki, A. F. Dalmorin, A. K. Edwards, P. D. A. Lima and V. A. Paffaro, in The Guide to Investigation of Mouse Pregnancy, ed. B. A. Croy, A. T. Yamada, F. J. DeMayo and S. L. Adamson, Academic Press, Boston, 2014, pp. 163–173 Search PubMed.
- L. A. Salamonsen and J. Evans, in Encyclopedia of Reproduction (Second Edition), ed. M. K. Skinner, Academic Press, Oxford, 2018, pp. 320–325 Search PubMed.
- H. Zhu, C.-C. Hou, L.-F. Luo, Y.-J. Hu and W.-X. Yang, Gene, 2014, 551, 1–14 CrossRef CAS.
- Y. Abbas, L. G. Brunel, M. S. Hollinshead, R. C. Fernando, L. Gardner, I. Duncan, A. Moffett, S. Best, M. Y. Turco, G. J. Burton and R. E. Cameron, Interface Focus, 2020, 10, 20190079 CrossRef.
- H. N. Jabbour, R. W. Kelly, H. M. Fraser and H. O. D. Critchley, Endocr. Rev., 2006, 27, 17–46 CrossRef CAS.
- S. A. Olalekan, J. E. Burdette, S. Getsios, T. K. Woodruff and J. J. Kim, Biol. Reprod., 2017, 96, 971–981 CrossRef.
- C. D. Cook, A. S. Hill, M. Guo, L. Stockdale, J. P. Papps, K. B. Isaacson, D. A. Lauffenburger and L. G. Griffith, Integr. Biol., 2017, 9, 271–289 CrossRef CAS.
- P. J. Brighton, Y. Maruyama, K. Fishwick, P. Vrljicak, S. Tewary, R. Fujihara, J. Muter, E. S. Lucas, T. Yamada, L. Woods, R. Lucciola, Y. Hou Lee, S. Takeda, S. Ott, M. Hemberger, S. Quenby and J. J. Brosens, eLife, 2017, 6 DOI:10.7554/eLife.31274.
- S. Biswas Shivhare, J. N. Bulmer, B. A. Innes, D. K. Hapangama and G. E. Lash, Hum. Reprod., 2018, 33, 399–410 CrossRef.
- P. G. M. Figueira, M. S. Abrão, G. Krikun and H. Taylor, Ann. N. Y. Acad. Sci., 2011, 1221, 10–17 CrossRef CAS.
- G. R. Souza, H. Tseng, J. A. Gage, A. Mani, P. Desai, F. Leonard, A. Liao, M. Longo, J. S. Refuerzo and B. Godin, Int. J. Mol. Sci., 2017, 18(4) DOI:10.3390/ijms18040683.
- M. Heidari Kani, E.-C. Chan, R. C. Young, T. Butler, R. Smith and J. W. Paul, Ann. Biomed. Eng., 2017, 45, 1746–1757 CrossRef.
- O. Shynlova, J. A. Mitchell, A. Tsampalieros, B. L. Langille and S. J. Lye, Biol. Reprod., 2004, 70, 986–992 CrossRef CAS.
- V. De Gregorio, F. Urciuolo, P. A. Netti and G. Imparato, Cancers, 2020, 12(5) DOI:10.3390/cancers12051150.
- H. Feltovich and L. Carlson, Semin. Perinatol., 2017, 41, 477–484 CrossRef.
- H. Deng, S. Mondal, S. Sur and C. D. Woodworth, J. Cell. Physiol., 2019, 234, 7683–7694 CrossRef CAS.
- M. House, J. Kelly, N. Klebanov, K. Yoshida, K. Myers and D. L. Kaplan, Tissue Eng., Part A, 2018, 24, 1765–1774 CrossRef CAS.
- E. Cho, Y. Y. Kim, K. Noh and S.-Y. Ku, J. Tissue Eng. Regener. Med., 2019, 13, 1294–1315 CrossRef CAS.
- N. Desai, A. Alex, F. AbdelHafez, A. Calabro, J. Goldfarb, A. Fleischman and T. Falcone, Reprod. Biol. Endocrinol., 2010, 8, 119 CrossRef.
- C. E. Tomaszewski, E. Constance, M. M. Lemke, H. Zhou, V. Padmanabhan, K. B. Arnold and A. Shikanov, Biomater. Sci., 2019, 7, 571–580 RSC.
- H. Zhou, M. A. Malik, A. Arab, M. T. Hill and A. Shikanov, PLoS One, 2015, 10, e0140205 CrossRef.
- J. I. Ahn, G. A. Kim, H. S. Kwon, J. Y. Ahn, J. A. Hubbell, Y. S. Song, S. T. Lee and J. M. Lim, J. Tissue Eng. Regener. Med., 2015, 9, 319–323 CrossRef CAS.
- Z. Sleiman, E. Karaman, M. Terzic, S. Terzic, G. Falzone and S. Garzon, J. Reprod. Infertil., 2019, 20, 201–208 Search PubMed.
- D. Muzzio, M. L. Foglia, M. F. Desimone and M. Zygmunt, Curr. Pharm. Des., 2017, 23, 3603–3613, DOI:10.2174/1381612823666170509104848.
- H. Zhang, Z. Gao, Y. Zhang, H. Wang and Y. Li, RSC Adv., 2018, 8, 39098–39105 RSC.
- T. Wiwatpanit, A. R. Murphy, Z. Lu, M. Urbanek, J. E. Burdette, T. K. Woodruff and J. J. Kim, J. Clin. Endocrinol. Metab., 2020, 105, 769–780 CrossRef.
- M. Y. Turco, L. Gardner, J. Hughes, T. Cindrova-Davies, M. J. Gomez, L. Farrell, M. Hollinshead, S. G. E. Marsh, J. J. Brosens, H. O. Critchley, B. D. Simons, M. Hemberger, B.-K. Koo, A. Moffett and G. J. Burton, Nat. Cell Biol., 2017, 19, 568–577 CrossRef CAS.
- K. N. Shah and S. S. Patel, Pharm. Biol., 2016, 54, 975–983 CrossRef CAS.
- K. C. Fogg, W. R. Olson, J. N. Miller, A. Khan, C. Renner, I. Hale, P. S. Weisman and P. K. Kreeger, Cancer Lett., 2019, 458, 92–101 CrossRef CAS.
- B. K. Erickson, M. G. Conner and C. N. Landen, Am. J. Obstet. Gynecol., 2013, 209, 409–414 CrossRef.
- T.-L. Yeung, C. S. Leung, K.-P. Yip, C. L. Au Yeung, S. T. C. Wong and S. C. Mok, Am. J. Physiol.: Cell Physiol., 2015, 309, C444–C456 CrossRef CAS.
- T. Jiang, J. Zhao, S. Yu, Z. Mao, C. Gao, Y. Zhu, C. Mao and L. Zheng, Biomaterials, 2019, 188, 130–143 CrossRef CAS.
- A. Hendriks, A. R. Cruz, E. Soldaini, A. G. O. Manetti and F. Bagnoli, Curr. Top. Microbiol. Immunol., 2018, 1–23, DOI:10.1007/82_2018_130.
- S. Dumont, Z. Jan, R. Heremans, T. Van Gorp, I. Vergote and D. Timmerman, J. Ovarian Res., 2019, 12, 105 CrossRef.
- H. A. Kenny, M. Lal-Nag, E. A. White, M. Shen, C.-Y. Chiang, A. K. Mitra, Y. Zhang, M. Curtis, E. M. Schryver, S. Bettis, A. Jadhav, M. B. Boxer, Z. Li, M. Ferrer and E. Lengyel, Nat. Commun., 2015, 6, 6220 CrossRef CAS.
- M. Lu, C. E. Henry, H. Lai, Y. Y. Khine, C. E. Ford and M. H. Stenzel, Biomater. Sci., 2019, 7, 1652–1660 RSC.
- H. Yu, H. Lee, A. Herrmann, R. Buettner and R. Jove, Nat. Rev. Cancer, 2014, 14, 736–746 CrossRef CAS.
- M. J. Carroll, A. Kapur, M. Felder, M. S. Patankar and P. K. Kreeger, Oncotarget, 2016, 7, 86608–86620 CrossRef.
- E. Kaemmerer, F. P. W. Melchels, B. M. Holzapfel, T. Meckel, D. W. Hutmacher and D. Loessner, Acta Biomater., 2014, 10, 2551–2562 CrossRef CAS.
- K. C. Fogg, C. M. Renner, H. Christian, A. Walker, L. Marty-Santos, A. Khan, W. R. Olson, C. Parent, A. O'Shea, D. M. Wellik, P. S. Weisman and P. K. Kreeger, Tissue Eng., Part A, 2020, 26, 747–758 CrossRef CAS.
- I. Briceag, A. Costache, V. Purcarea, R. Cergan, M. Dumitru, I. Briceag, M. Sajin and A. Ispas, J. Med. Life, 2015, 8, 129–131 CAS.
- K. Narang, Z. S. Cope and J. M. Teixeira, in Human Reproductive and Prenatal Genetics, ed. P. C. K. Leung and J. Qiao, Academic Press, 2019, pp. 129–153 Search PubMed.
- D. Y. Paik, D. M. Janzen, A. M. Schafenacker, V. S. Velasco, M. S. Shung, D. Cheng, J. Huang, O. N. Witte and S. Memarzadeh, Stem Cells, 2012, 30, 2487–2497 CrossRef CAS.
- K. Lawrenson, E. Benjamin, M. Turmaine, I. Jacobs, S. Gayther and D. Dafou, Cell Proliferation, 2009, 42, 385–393 CrossRef CAS.
- R. J. Kurman and I.-M. Shih, Am. J. Pathol., 2016, 186, 733–747 CrossRef.
- A. J. Fleszar, A. Walker, P. K. Kreeger and J. Notbohm, Integr. Biol., 2019, 11, 342–352 CrossRef.
- A. J. Fleszar, A. Walker, V. Porubsky, W. Flanigan, D. James, P. J. Campagnola, P. S. Weisman and P. K. Kreeger, APL Bioeng., 2018, 2(3) DOI:10.1063/1.5022595.
- M. Kessler, K. Hoffmann, V. Brinkmann, O. Thieck, S. Jackisch, B. Toelle, H. Berger, H.-J. Mollenkopf, M. Mangler, J. Sehouli, C. Fotopoulou and T. F. Meyer, Nat. Commun., 2015, 6, 8989 CrossRef CAS.
- K. Levanon, V. Ng, H. Y. Piao, Y. Zhang, M. C. Chang, M. H. Roh, D. W. Kindelberger, M. S. Hirsch, C. P. Crum, J. A. Marto and R. Drapkin, Oncogene, 2010, 29, 1103–1113 CrossRef CAS.
- S. L. Eddie, S. M. Quartuccio, J. Zhu, J. A. Shepherd, R. Kothari, J. J. Kim, T. K. Woodruff and J. E. Burdette, Gynecol. Oncol., 2015, 136, 348–354 CrossRef.
- U. Fidan, U. Keskin, M. Ulubay, M. Öztürk and S. Bodur, Clin. Anat., 2017, 30, 404–408 CrossRef.
- J. Rosner, T. Samardzic and M. S. Sarao, in StatPearls, StatPearls Publishing, Treasure Island (FL), 2020 Search PubMed.
- R. S. Magalhaes, J. K. Williams, K. W. Yoo, J. J. Yoo and A. Atala, Nat. Biotechnol., 2020, 1–8 Search PubMed.
- A. Atala, J. Tissue Eng. Regener. Med., 2007, 1, 83–96 CrossRef CAS.
- D. D. Baird, D. B. Dunson, M. C. Hill, D. Cousins and J. M. Schectman, Am. J. Obstet. Gynecol., 2003, 188, 100–107 CrossRef.
- L. A. Wise and S. K. Laughlin-Tommaso, Clin. Obstet. Gynecol., 2016, 59, 2–24 CrossRef.
- M. S. Islam, A. Ciavattini, F. Petraglia, M. Castellucci and P. Ciarmela, Hum. Reprod. Update, 2018, 24, 59–85 CrossRef CAS.
- M. Wolańska, K. Sobolewski, E. Bańkowski and S. Jaworski, Gynecol. Obstet. Invest., 2004, 58(1), 14–18 CrossRef.
- F. Koohestani, A. G. Braundmeier, A. Mahdian, J. Seo, J. Bi and R. A. Nowak, PLoS One, 2013, 8, e75844 CrossRef CAS.
- M. Malik and W. H. Catherino, Fertility Sterility, 2012, 97, 1287–1293 CrossRef.
- A. S. Brohl, L. Li, V. Andikyan, S. G. Običan, A. Cioffi, K. Hao, J. T. Dudley, C. Ascher-Walsh, A. Kasarskis and R. G. Maki, Oncologist, 2015, 20, 433–439 CrossRef.
- B. Choijamts, S. Jimi, T. Kondo, Y. Naganuma, T. Matsumoto, M. Kuroki, H. Iwasaki and M. Emoto, Stem Cells, 2011, 29, 1485–1495 CrossRef CAS.
- T. Kawakita, N. Masato, E. Takiguchi, A. Abe and M. Irahara, Exp. Ther. Med., 2017, 13, 2939–2945 CrossRef CAS.
- D. L. Keefe and K. P. Wright, in General Gynecology, ed. A. I. Sokol and E. R. Sokol, Mosby, Philadelphia, 2007, pp. 21–41 Search PubMed.
- T. J. Colgan and C. Meg McLachlin, in Comprehensive Cytopathology (Third Edition), ed. M. Bibbo and D. Wilbur, W.B. Saunders, Edinburgh, 2008, pp. 247–271 Search PubMed.
- A. R. Baerwald, G. P. Adams and R. A. Pierson, Ultrasound Obstet. Gyneco.l, 2005, 25, 498–507 CrossRef CAS.
- P. a. W. Rogers, Hum. Reprod. Update, 1996, 2, 57–62 CrossRef CAS.
- A. M. Hantak, I. C. Bagchi and M. K. Bagchi, Int. J. Dev. Biol., 2014, 58, 139–146 CrossRef CAS.
- S. C. Schutte, C. O. James, N. Sidell and R. N. Taylor, Reprod. Sci., 2015, 22, 308–315 CrossRef CAS.
- R. Demir, A. Yaba and B. Huppertz, Acta Histochem., 2010, 112, 203–214 CrossRef CAS.
- L. Kuznetsov, K. Dworzynski, M. Davies and C. Overton, Br. Med. J., 2017, 358 DOI:10.1136/bmj.j3935.
- L. Mei, J. Bao, L. Tang, C. Zhang, H. Wang, L. Sun, G. Ma, L. Huang, J. Yang, L. Zhang, K. Liu, C. Song and H. Sun, Eur. J. Pharm. Sci., 2010, 39, 421–427 CrossRef CAS.
- Z. Chen, Y. Dai, Z. Dong, M. Li, X. Mu, R. Zhang, Z. Wang, W. Zhang, J. Lang, J. Leng and X. Jiang, Integr. Biol., 2012, 4, 1090–1095 CrossRef CAS.
- H. Fan, Exp. Ther. Med., 2020, 19, 1617–1625 CAS.
- A. Fasciani, G. Bocci, J. Xu, R. Bielecki, E. Greenblatt, N. Leyland and R. F. Casper, Fertil. Steril., 2003, 80, 1137–1143 CrossRef.
- J. Valdez, C. Cook, C. C. Ahrens, A. J. Wang, A. Brown, M. Kumar, L. Stockdale, D. Rothenberg, K. Renggli, E. Gordon, D. Lauffenburger, F. White and L. Griffith, Biomaterials, 2017, 130, 90–103 CrossRef CAS.
- K. Moore and M. A. Brewer, American Society of Clinical Oncology Educational Book, 2017, pp. 435–442 Search PubMed.
- A. T. Ali, Ceska Gynekol., 2013, 78, 448–459 Search PubMed.
- K. Chitcholtan, P. H. Sykes and J. J. Evans, J. Transl. Med., 2012, 10, 38 CrossRef CAS.
- L. Shi, W. Yao, Y. Gan, L. Y. Zhao, W. Eugene McKee, J. Vink, R. J. Wapner, C. P. Hendon and K. Myers, J. Biomech. Eng., 2019, 141(9) DOI:10.1115/1.4043977.
- S. F. Martin, A. D. Wood, M. M. McRobbie, M. Mazilu, M. P. McDonald, I. D. W. Samuel and C. S. Herrington, Int. J. Cancer, 2007, 120, 1964–1970 CrossRef CAS.
- M. Vu, J. Yu, O. A. Awolude and L. Chuang, Curr. Probl. Cancer, 2018, 42, 457–465 CrossRef.
- S. D. Kang, S. Chatterjee, S. Alam, A. C. Salzberg, J. Milici, S. H. van der Burg and C. Meyers, J. Virol., 2018, 92(20) DOI:10.1128/JVI.01261–18.
- P. Tsikouras, S. Zervoudis, B. Manav, E. Tomara, C. Romanidis, A. Bothou and G. Galazios, J. BUON, 2016212320–325 Search PubMed.
- Y. Zhao, R. Yao, L. Ouyang, H. Ding, T. Zhang, K. Zhang, S. Cheng and W. Sun, Biofabrication, 2014, 6, 035001 CrossRef.
- G. Raikhy, B. L. Woodby, M. L. Scott, G. Shin, J. E. Myers, R. S. Scott and J. M. Bodily, J. Virol., 2019, 93(19) DOI:10.1128/JVI.00458–19.
- N. Jacobs, M. P. Moutschen, E. Franzen-Detrooz, V. Boniver, J. Boniver and P. Delvenne, Virchows Arch., 1998, 432, 323–330 CrossRef CAS.
- A. J. Fleszar, A. Walker, V. Porubsky, W. Flanigan, D. James, P. J. Campagnola, P. S. Weisman and P. K. Kreeger, APL Bioeng., 2018, 2(3) DOI:10.1063/1.5022595.
- A. J. Fleszar, A. Walker, P. K. Kreeger and J. Notbohm, Integr. Biol., 2019, 11, 342–352 CrossRef.
| This journal is © The Royal Society of Chemistry 2021 |