Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Assembly of FN-silk with laminin-521 to integrate hPSCs into a three-dimensional culture for neural differentiation†
Carolina
Åstrand
ab,
Veronique
Chotteau
cd,
Anna
Falk
e and
My
Hedhammar
 *a
*a
aDept. of Protein Science, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH-Royal Institute of Technology, SE-10691, Stockholm, Sweden. E-mail: myh@kth.se
bSpiber Technology AB, Stockholm, Sweden
cDept. of Industrial Biotechnology, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH-Royal Institute of Technology, SE-10691, Stockholm, Sweden
dAdBIOPRO Competence Centre for Advanced BioProduction by Continuous Processing, Sweden
eDept. of Neuroscience, Karolinska Institutet, SE-17177, Stockholm, Sweden
First published on 18th March 2020
Abstract
Three-dimensional (3D) neural tissue cultures recapitulate the basic concepts during development and disease better than what can be obtained using conventional two-dimensional cultures. Here, we use a recombinant spider silk protein functionalized with a cell binding motif from fibronectin (FN-silk) in combination with a human recombinant laminin 521 (LN-521) to create a fully defined stem cell niche in 3D. A novel method to assemble silk blended with LN-521 together with human pluripotent stem cells (hPSC) is used to create centimeter-sized foams, which upon cultivation develop into 3D cell constructs supported by a microfibrillar network. After initial cell expansion, neural differentiation was induced to form a homogenous layer of continuous neuroectodermal tissue that allows further differentiation into neuronal subtypes. The silk-supported 3D cell constructs could then be detached from the bottom of the well and cultured as floating entities, where cells appeared in distinctive radial organization resembling early neural tube. This shows that the neural progenitors retain their cellular self-organization ability in the FN-silk/LN-521-supported 3D culture. Calcium imaging demonstrated spontaneous activity, which is important for the formation of neuronal networks. Together, the results show that hPSCs integrated into FN-silk/LN-521 foam develop into neural progenitors and that these stay viable during long-term differentiations. FN-silk/LN-521 also supports morphogenesis mimicking the human brain development and can serve as base for engineering of hPSC-derived neural tissue.
1. Introduction
Human pluripotent stem (hPSC) cells, including human embryonic stem cells (hESC) and human induced pluripotent stem cells (hiPS), have the ability to generate any specialized cell in the body. hPSCs have been used for engineering of a wide variety of human tissue, including models of neurogenesis.1 Three-dimensional (3D) cultures formed by self-organization of hPSCs have been able to partly recapitulate the cellular organization of cortical-,2 retinal-,3 and cerebral tissue.4 Such 3D cultures are currently used for studying human brain development, function and disease. They are also explored for drug screening purposes and for the development of cell replacement therapies.1 Recent advances to differentiate spheroid-based hPSC cultures into cerebral organoids4 have given new insights in neural disease by e.g. modelling of the exposure of Zika virus to the fetal brain5 and autism spectrum disorders.6 An approach to expand the organoid culture to generate laminated slices mimicking cortex-like structures has also been reported.7 However, these systems that are largely based on intrinsic cellular self-organization share limitations such as; (i) little control of the cell type fates, (ii) low reproducibility and (iii) difficulty to scale up.8In contrast to culture systems based on cellular self-assembly, methods using supporting scaffolds/matrices generally give a higher degree of control of the microenvironment.9 Several criteria have to be fulfilled for a matrix to be able to support the important extrinsic signals that regulate cell behavior. Primarily, the chemical composition and morphology of the matrix have to support hPSC attachment and promote survival.10 Neural progenitors have been grown on matrices made of a range of proteins such as collagen, fibrin, fibronectin and laminin,11 but of these only a few have the ability to also support pluripotent cells. Any ill-defined components reduce predictability and reproducibility of tissue organization.12 Recently, a hybrid hydrogel with natural (gelatin) and synthetic (PEG) components were shown to stimulate cell differentiation (in this case mesenchymal stem cells).13 Permeability of the matrix is crucial to allow exchange of nutrients and waste products. Furthermore, the stiffness of the material is known to be important for cellular behavior.14 By screening synthetic matrices for a variety of mechanical and biochemical properties it was seen that the matrix surrounding the cells affected cell orientation during neural tube formation.15 In line with this, synthetic microfilaments have been shown able to aid the organization of cells into a cortical plate.12
Due to its favorable mechanical properties and biocompatibility, silk spun by spiders have successfully been used for peripheral nerve regeneration.16,17 Pre-made scaffolds of recombinant spider silk protein have been shown to support neural stem cell culture.18 Furthermore, silk scaffolds combined with gels consisting of extracellular matrix proteins have been used for mimicking the modularity of cortical tissue.19 The recombinant spider silk protein 4RepCT has previously been functionalized with cell binding motifs from fibronectin (FN-silk)20 and vitronectin (VN-silk).21 A coating of VN-silk was shown able to support long-term expansion of hPSCs grown as monolayers22 and premade scaffolds of VN-silk could serve as a substrate supporting differentiation into neuroectoderm.21 FN-silk has been found to promote attachment and proliferation of human primary cells.20 We recently reported that the FN-silk protein can assemble in the presence of cells under physiological-like conditions into a 3D network of microfibers, rendering evenly distributed cells.23
In this study, we investigated how FN-silk can be used to create a 3D culture system suitable for expansion of hPSCs and following neural differentiation. Recombinant human laminin 521 (LN-521) has been shown advantageous in promoting self-renewal and pluripotency of hPSCs.24 It has also been shown beneficial to combine recombinant laminins with other matrices (fibrin gels) to promote expansion of cells (epithelial progenitor).25 Therefore, we herein investigated the option to combine FN-silk, which enables self-assembly into a 3D environment, with LN-521, which offers biologically relevant cues for the sensitive hPSCs. The method allows integration of hPSCs into an extracellular matrix-like microfibrillar network of fully defined FN-silk/LN-521. We analyzed proliferation, functionality and morphogenesis during expansion and neural differentiation within these cultures. The use of FN-silk/LN-521 allows instant integration of hPSCs that can be expanded into shapeable, macro-sized 3D constructs for neuronal differentiation.
2. Materials and methods
2.1 Recombinant spider silk/laminin
The recombinant spider silk protein 4RepCT and functional variants thereof (VN-silk, FN-silk)20,21,26 were supplied by Spiber Technologies AB (Sweden). Recombinant human laminin-521 was purchased from Biolamina (Sweden) and added to the silk protein solution at a final concentration of 10 μg ml−1. The FN-silk/LN-521 mixture was incubated 5–10 min at RT before usage.2.2 hPSC culture
hESC line HS980 with hPSCreg name Kle033-A, derived on LN-521,24 was provided by Dr Hovatta, Karolinska University Hospital, Sweden. hiPS cell line C5, derived on fibroblasts and adapted to LN-521 as previously described,21 was provided by Karolinska Institutet, Sweden, Falk lab. Both lines were used in agreements with ethical vetting by the Regional Ethics Board, Stockholm (Dnr 2015/824-39). The hPSCs were maintained at 37 °C, 5% CO2 on LN-521 coated plates in Essential 8™ (ThermoFisher) with daily medium change. The cells were passaged with TrypLE (ThermoFisher) and counted manually before integration into silk scaffolds. Passages 11–25 were used in this study. Cell lines were routinely tested for mycoplasma 2–5 times a year.2.3 Integration of hPSCs into silk foam and analysis of foam stability
Silk foam with integrated hPSCs were prepared in hydrophobic plates of 24-well format (Sarstedt cat no 83.3922.500). Porous foam was formed by rapidly pipetting air (2–3 strokes per s) using a pipette set at 40 μl into a droplet of 20 μl FN-silk/LN-521 placed at the bottom of a well and spread to a diameter of 1 cm. 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 hPSCs resuspended in culture medium (typically 10
000 hPSCs resuspended in culture medium (typically 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–15
000–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per μl) in presence of 10 μg ml−1 Rock inhibitor Y27632 (VWR) were added directly into the foam and distributed by 5 additional strokes with the pipette. The foams were stabilized for 15 min at 37 °C, 5% CO2 in a cell incubator before submerged with 0.7 ml Essential 8™ (ThermoFisher) supplemented with 10 μg ml−1 Y27632. Rock inhibitor was omitted in the medium from 16–20 hours after seeding. For evaluation of foam stability, 12 foams per condition were monitored by bright field microscopy 2 days after foam formation. Stability was graded as % of intact foam attached to the bottom of the well. After the air-bubbles have burst, the foam maintains at least 100 μm height, which, if assuming around 5 cell layers, a similar cell density as the 2D control (14 × 103 cells per cm2).
000 cells per μl) in presence of 10 μg ml−1 Rock inhibitor Y27632 (VWR) were added directly into the foam and distributed by 5 additional strokes with the pipette. The foams were stabilized for 15 min at 37 °C, 5% CO2 in a cell incubator before submerged with 0.7 ml Essential 8™ (ThermoFisher) supplemented with 10 μg ml−1 Y27632. Rock inhibitor was omitted in the medium from 16–20 hours after seeding. For evaluation of foam stability, 12 foams per condition were monitored by bright field microscopy 2 days after foam formation. Stability was graded as % of intact foam attached to the bottom of the well. After the air-bubbles have burst, the foam maintains at least 100 μm height, which, if assuming around 5 cell layers, a similar cell density as the 2D control (14 × 103 cells per cm2).
2.4 Metabolic activity and proliferation
The metabolic activity, as a relative measurement of cell growth, was monitored by Alamar Blue cell viability assay (Invitrogen) at 48 h after cell-integration. Supernatants from cultures were measured after 2 h incubation with the Alamar blue reagent using a fluorescence plate reader (CLARIOstar, BMG Labtech). Fluorescence intensities from four cultures per condition were analyzed. For cell proliferation, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells from HS980 and C5 were dissociated with TryLE and counted manually before seeding and at the endpoint (72 h). Proliferation is presented as fold change. For each experiment, cells from three different passages were analyzed.
000 cells from HS980 and C5 were dissociated with TryLE and counted manually before seeding and at the endpoint (72 h). Proliferation is presented as fold change. For each experiment, cells from three different passages were analyzed.
2.5 Live/dead assay
Silk foams with integrated cells were washed once in pre-warmed PBS before 30 min incubation with calcein and EthD-1 (live/dead viability kit, Invitrogen). Imaging was performed on a Leica DMI6000 B. Images were processed using NIH Image J software.2.6 Immunocytochemistry
Silk foams with integrated cells were washed once in PBS and fixed in 4% paraformaldehyde for 15 min at RT followed by three washes in PBS. For sectioning; fixed samples were incubated in 20% sucrose at 4 °C, over night before embedding in Tissue-Tek (Sakura) and sectioned using a Leica cryostat CM 1950. Permeabilization was done at RT for 15 min in PBS with 0.1% Triton X-100 followed by blocking for 1 h at RT with 10% donkey serum (Jackson ImmunoResearch). Primary antibodies were incubated overnight at 4 °C in PBS with 0.1% Tween-20 (PBST) and 5% donkey serum. Secondary antibodies were incubated for 1 h at RT in PBST with 5% serum. The samples were washed three times with PBST between each incubation. Primary antibodies: anti-OCT3/4 (R&D, #962649, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50), anti-NANOG (R&D, #963488, 1
50), anti-NANOG (R&D, #963488, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50), anti-Laminin (Abcam, ab11575, 1
50), anti-Laminin (Abcam, ab11575, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), anti-PAX6 (Abcam, ab5790, 1
200), anti-PAX6 (Abcam, ab5790, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100), anti-Nestin (R&D, #196908, 1
100), anti-Nestin (R&D, #196908, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), anti-BIII-tubulin (Sigma, #SDL3D10, 1
200), anti-BIII-tubulin (Sigma, #SDL3D10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), anti-SOX2 (Santa Cruz, sc-17320, 1
500), anti-SOX2 (Santa Cruz, sc-17320, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50), anti-KI67 (Abcam, ab92742, 1
50), anti-KI67 (Abcam, ab92742, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100), anti-N-cadherin (Abcam, ab12221, 1
100), anti-N-cadherin (Abcam, ab12221, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), anti-DCX (Abcam, ab18723, 1
200), anti-DCX (Abcam, ab18723, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), anti-MAP2 (Sigma, #AP-20, 1
200), anti-MAP2 (Sigma, #AP-20, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), anti-GAD67 (Millipore, #1G102, 1
500), anti-GAD67 (Millipore, #1G102, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200). Secondary antibodies: anti-rabbit 647 (Abcam, ab150075, 1
200). Secondary antibodies: anti-rabbit 647 (Abcam, ab150075, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-goat 647 (Abcam, ab15031, 1
1000), anti-goat 647 (Abcam, ab15031, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-goat 488 (Jackson ImmunoResearch, 705545147, 1
1000), anti-goat 488 (Jackson ImmunoResearch, 705545147, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-mouse 488 (Jackson ImmunoResearch, 715545150, 1
1000), anti-mouse 488 (Jackson ImmunoResearch, 715545150, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000). Nuclei were visualized with DAPI (Sigma). Slides were mounted in fluorescence mounting medium (DAKO). Fluorescence was analyzed on a Leica DMI6000 B, or for maximum intensity projection, a Zeiss Light Sheet Z.1 with 10×/0.5NA detection objective was used. Images were processed using NIH Image J software.
1000). Nuclei were visualized with DAPI (Sigma). Slides were mounted in fluorescence mounting medium (DAKO). Fluorescence was analyzed on a Leica DMI6000 B, or for maximum intensity projection, a Zeiss Light Sheet Z.1 with 10×/0.5NA detection objective was used. Images were processed using NIH Image J software.
2.7 RNA isolation and gene expression analysis
Extraction of total RNA was carried out using a mini RNA purification kit (Qiagen). The RNA was reverse-transcribed using maxima first strand cDNA synthesis kit (ThermoFisher). Real-time qPCR was performed by TaqMan probe-based detection in a C1000™ Thermal Cycler (Bio-Rad) according to the manufacturer's instructions. TaqMan gene expression assays (Life Technologies) were used for: human NANOG ID Hs 04260366_g1, human POU5F1 ID Hs 04260367_gH, human B2M ID 4333766, human FOXG1 ID Hs 01850784_s1, human TUB3 ID Hs 00801390_s1, human PAX6 ID 00240871_m1, human ZIC1 ID 00602749_m1, human T ID 00610080_m1. PrimePCR assays (Bio-Rad) GAD1 Hsa, MAP2 Hsa, B2M Hsa were analyzed with SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad). B2M was used as an endogenous control and expression was quantified by 2−ΔΔCt.27 Results from real time qPCR measurements were expressed as mean fold change ± standard deviation (n = 5) or as stated.2.8 Quantification of cell growth
The cell expansion in foam was quantified using Hoechst-based DNA quantification (Sigma Aldrich). Foams with integrated HS980 cells were prepared as previously described and transferred to sterile nylon nets fitted in a 24-well culture plate to enable any cell debris to be easily removed from the culture. Whole foams with cells were harvested at indicated timepoints by freezing in PBE buffer (100 mM Na2HPO, 5 mM Na2EDTA at pH 7.5). Thawn samples were supplemented with 10 mM L-cysteine and 125 μg ml−1 papain enzyme (Sigma Aldrich) and digested at 60 °C over night. The DNA was quantified in a 96-well plate in 2 μg ml−1 Hoechst 33258 and 1× fluorescent assay buffer using a CLARIOstar microplate reader (BMG Labtech). Three samples per timepoint was plotted as average cell number ± standard deviation (n = 3).2.9 Neural differentiation
Neural induction was initiated three days after integration into silk foam using a modified version of a dual-SMAD inhibition protoc.28,29 The cells were cultured in neural induction medium (NIM) consisting of 50% Neurobasal medium (ThermoFisher) and 50% DMEM/F12 (ThermoFisher) supplemented with 0.5× B-27 without vitamin A (ThermoFisher), 0.5× N2 (ThermoFisher), 100 μM 2-mercaptoethanol (Sigma) and GlutaMAX (Life Technologies) with the addition of recombinant human Noggin, 500 ng ml−1 (R&D) and SB431542, 10 μM (Tocris) with daily medium change. After seven days, NIM was replaced by neuronal progenitor medium (NPDM); 50% neurobasal medium (ThermoFisher Scientific) and 50% DMEM/F12 (ThermoFisher) supplemented with 0.5× B-27 without vitamin A (ThermoFisher), 0.5× N2 (ThermoFisher), 100 μM 2-mercaptoethanol (Life Technologies) and GlutaMAX (Life Technologies). Medium was changed every second or third day for the remaining culture time.2.10 Calcium imaging analysis
Calcium imaging using cell permeant Fluo-4, AM (Life Technologies) was performed according to manufacturer's protocol at day 40 and day 60. The silk foams with integrated cells were incubated in neural progenitor differentiation medium with Fluo-4 for 30 min. Images of fluorescence intensity over time were acquired with a Leica DMI6000 B at room temperature and analyzed using NIH Image J software. Calcium releases demonstrating spontaneous activity were visualized by spectra. Top view images using a ×10 objective of day 60 cultures, typically revealed up to 10% cells giving spontaneous calcium surges during a 3 min analysis period.2.11 Statistical analysis
Data are presented as mean ± standard deviations. Statistical analyses were performed with Student t-test (preceded by One-way ANOVA when comparing multiple groups). Significant p values or non-significant values (n.s.) are reported for each figure in the respective figure legend. For all tests a p-value <0.05 was considered statistically significant.3. Results
3.1 A combination of FN-silk and laminin 521 support 3D culture of evenly integrated human pluripotent stem cells
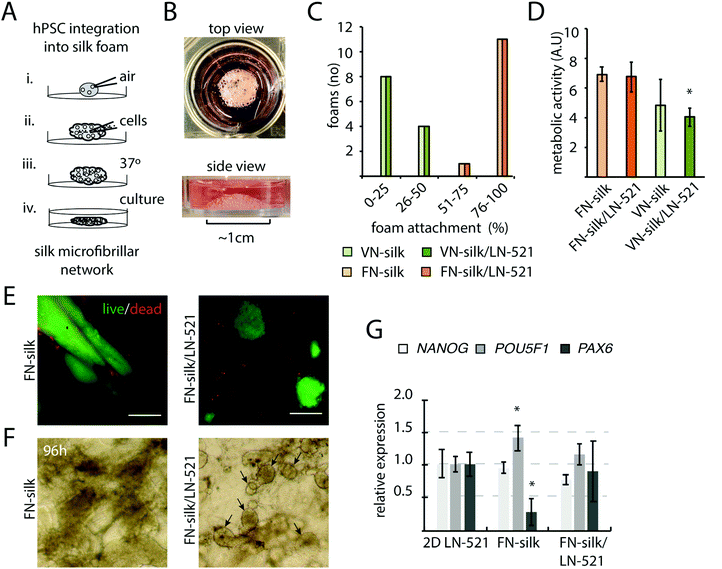
Pre-made scaffolds of the recombinant spider silk proteins FN-silk and VN-silk has previously been found to promote cellular attachment and proliferation of cells seeded on top.20,21 Here, we explored the possibility to integrate hPSCs into a 3D silk foam by allowing cells to be present during the silk-assembly, as described in Fig. 1A. A droplet of soluble silk protein is placed in the well of a hydrophobic culture plate. To transform the silk solution into a foam, air is repeatedly pipetted into the droplet at a speed of 2–3 strokes per s (i). Cells are prepared as a concentrated single-cell suspension in culture medium and added directly into the center of the foam (ii). To obtain a homogenous cell distribution, some additional strokes with the pipette are applied. The foam with integrated cells is then stabilized for 15 min in the cell incubator (iii) before addition of culture medium (iv. and Fig. 1B). Due to the pipetting, each air bubble within the foam is surrounded by a thin silk film. Within 1–3 days of culture, these films will burst, thereby transforming the foam into a network of microfibrillar silk with evenly integrated cells.23In the early embryo, specific isoforms of laminin are expressed, including those with side chains α5β2γ1.30 In culture, coatings of recombinant human LN-521 have been shown to promote survival, self-renewal and pluripotency.24 To create a 3D niche suitable for hPSCs, a fraction of LN-521 was added to the silk solution before silk assembly. Cells of the human embryonic stem cell line, HS980, initially derived and cultivated on LN-521![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31 were then added. First, we investigated the stability of different variants of silk/laminin foams with integrated cells. FN-silk gave rise to stable foam structures, whereas the majority of the foams made of VN-silk were completely or partly disintegrated and detached from the bottom after a 48 h culture period (Fig. 1C). Addition of LN-521 showed no effect on foam stability nor on metabolic activity of the integrated cells (Fig. 1D), although the proliferation rate was lower in VN-silk/LN521, likely due to instability of the VN-silk itself. When comparing viability and colony morphology of cells seeded within FN-silk with and without LN-521, we noticed that cells formed dense and irregular aggregates without LN-521, while cells within FN-silk with LN-521 appeared in well-distributed and round colonies (Fig. 1E), which became more evident under the microscope after four days of culture (Fig. 1F). To further investigate cell behavior during expansion in FN-silk, gene expression analysis revealed slight variations of pluripotency marker POU5f1 and the neural marker PAX6 as compared to cells cultured on a tissue culture plate coated with LN-521 (Fig. 1G). However, the gene expression in cells cultured in FN-silk foam with LN-521 did not reveal this variation. Taken together, due to the irregular colony morphology and decrease of PAX6 in FN-silk without LN-521, we decided to continue further studies to promote neural differentiation in 3D using FN-silk with addition of LN-521.
31 were then added. First, we investigated the stability of different variants of silk/laminin foams with integrated cells. FN-silk gave rise to stable foam structures, whereas the majority of the foams made of VN-silk were completely or partly disintegrated and detached from the bottom after a 48 h culture period (Fig. 1C). Addition of LN-521 showed no effect on foam stability nor on metabolic activity of the integrated cells (Fig. 1D), although the proliferation rate was lower in VN-silk/LN521, likely due to instability of the VN-silk itself. When comparing viability and colony morphology of cells seeded within FN-silk with and without LN-521, we noticed that cells formed dense and irregular aggregates without LN-521, while cells within FN-silk with LN-521 appeared in well-distributed and round colonies (Fig. 1E), which became more evident under the microscope after four days of culture (Fig. 1F). To further investigate cell behavior during expansion in FN-silk, gene expression analysis revealed slight variations of pluripotency marker POU5f1 and the neural marker PAX6 as compared to cells cultured on a tissue culture plate coated with LN-521 (Fig. 1G). However, the gene expression in cells cultured in FN-silk foam with LN-521 did not reveal this variation. Taken together, due to the irregular colony morphology and decrease of PAX6 in FN-silk without LN-521, we decided to continue further studies to promote neural differentiation in 3D using FN-silk with addition of LN-521.
3.2 Expanding hPSCs express pluripotency markers after integration into FN-silk/LN-521 foam
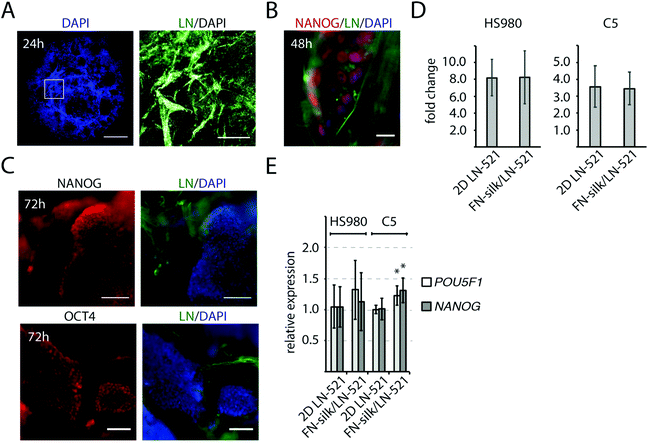
At initiation of a differentiation protocol, a homogenous starting cell population as well as proper cell densities are important factors,32,33 and we therefore characterized the initial cell growth and pluripotency of cells seeded within the FN-silk/LN-521 foam. The introduction of air bubbles into the silk droplet enlarged its original size ∼three times, resulting in a hemisphere-shaped foam with a diameter of 1 cm. One day after cell integration, the vast majority of the silk films surrounding the air-bubbles had burst, leaving evenly distributed cells attached to a network of silk/laminin microfibrils (Fig. 2A). After two days, the initially dispersed cells had formed small 3D colonies of NANOG+ cells (Fig. 2B). The distance between the cell colonies were directly dependent on the volume of the silk solution and the number of cells integrated. When analyzing the cell proliferation for HS980 and a human induced pluripotent cell line (hiPS, C5)21 after three days in FN-silk/LN-521 and compared with 2D control, no difference could be detected (Fig. 2D). At this stage, the majority of the cells had reached their logarithmic growth phase and the cell colonies covered the majority of the space within the microfibrillar network (ESI Fig. S1A†). Immunofluorescence analysis of the pluripotency markers NANOG and OCT4 revealed positive staining in all visible nuclei. The cell colonies had grown considerably in size compared to the time of cell integration (compare Fig. 1E, 2B with C). Gene expression analysis of the two cell lines cultured in FN-silk/LN-521 revealed similar levels of pluripotency markers (even slight increase for C5), when compared to cells grown in 2D on conventional tissue culture plates coated with LN-521 (Fig. 2E).3.3 Induction of early neural progenitors in FN-silk/LN-521 foam
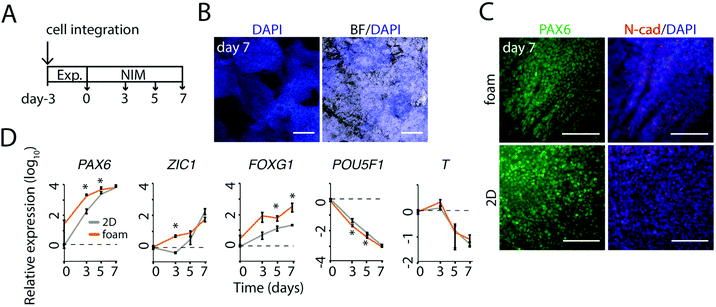
To test in situ differentiation towards neural linages in silk foam, we performed a series of trials using a general dual-SMAD inhibition protocol.32 As previously described, pluripotent cells integrated into foams made of FN-silk/LN-521 can be expanded for 3 days to gain sufficient density for initiation of neural differentiation (Fig. 2). Neural differentiation was induced with a medium (NIM) including Noggin and SB431542 (Fig. 3A). After seven days, the colonies of neural progenitors had expanded throughout the foam (Fig. 3B). Immunofluorescence analysis of cells integrated in the foam revealed a homogenous distribution of PAX6+ and symmetrically distributed N-CAD staining, at this stage similar to cells grown as monolayer on LN-521 coatings (Fig. 3C).Temporal gene expression analysis confirmed rapid induction of early neural markers PAX6, ZIC1 and FOXG1 in cells integrated in foam (Fig. 3D). Notably, in early induction up to day 5, the expression of the three neural markers were found equal to or slightly higher for cells integrated in FN-silk/LN-521 foam than in controls on LN-521 coatings. This was mirrored by a somewhat quicker drop in pluripotency marker POU5f1. At day 7, no major differences in loss of pluripotency marker POU5f1 or T-box transcription factor, a marker for epithelial-to-mesenchymal transition, were detected. Taken together, the abundancy of early neural markers and a rapid loss of pluripotency indicated that the combination of FN-silk/LN-521 would be a suitable platform for neural differentiation in a 3D culture.
3.4 Formation of continuous neuroectodermal 3D structures in FN-silk/LN-521 foam
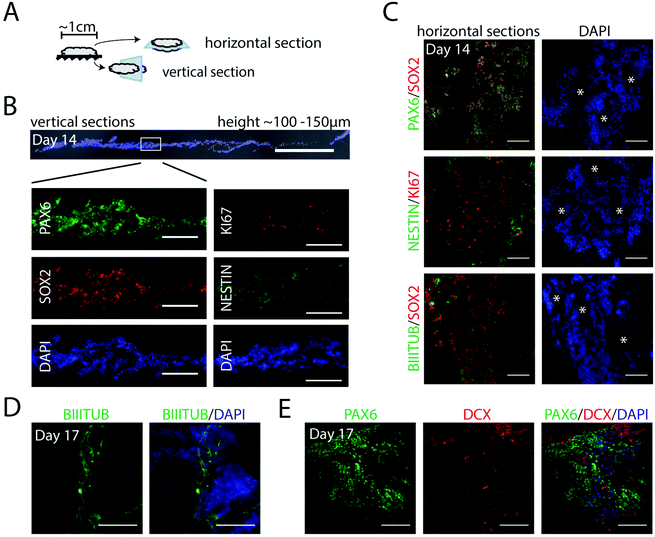
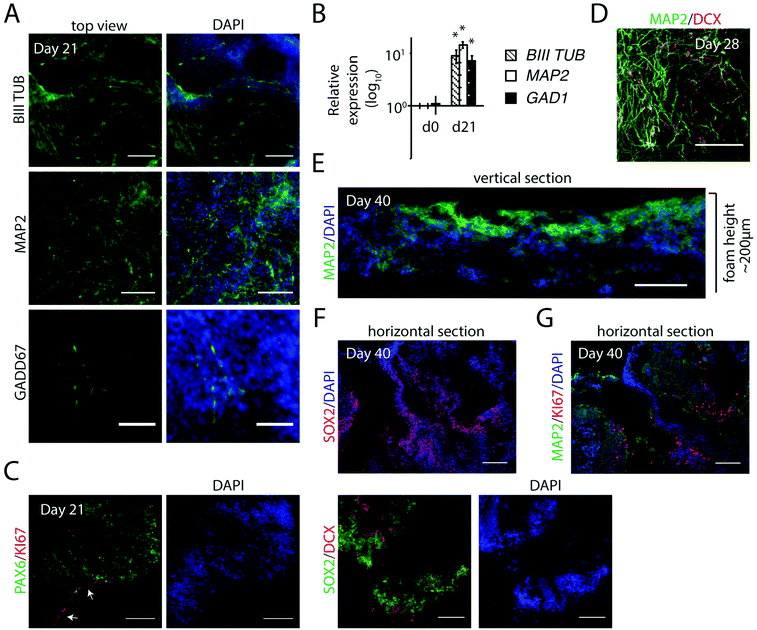
With the aim to form controlled, homogenous 3D populations of neural progenitors for further neuronal differentiation, we assessed the first steps of neurogenesis in defined x, y, z axis planes of foams. Cell integration, expansion and neural induction was initiated as described above (including 7 days in NIM medium), before switching to a neural progenitor medium without the inhibitors (ESI Fig. S2A†). Foams with integrated cells were fixed at day 14 and sectioned as indicated in Fig. 4A. The foams, 1 cm in diameter, contained cells arranged in layers with a total approximate height of 100–150 μm (Fig. 4B). At this stage, we observed a homogenous cell distribution throughout the entire section with strongly expressed early neural markers PAX6 and NESTIN, neural stem cell marker SOX2 and the proliferation marker KI67 (Fig. 4B), indicating dividing progenitors throughout the vertical plane. From analyses of multiple horizontal sections, we noticed that the pores of the fibrillar silk network had developed into channels throughout the 3D culture (Fig. 4C, white asterisks). These channels facilitate access of media for also the innermost cells inside the silk network. Cells co-expressing PAX6 and SOX2, as well as uniformly distributed KI67+ cells, were found throughout the cultures in all horizontal and vertical sections analyzed (Fig. 4B and C). Furthermore, cells positive for neural marker NESTIN were prominent, as well as cells displaying the neuronal marker BIII-TUBULIN, confirming differentiation into neuroectoderm (Fig. 4C and D). Gene expression analysis showed induction of PAX6 and FOXG1, while the pluripotency marker POU5f1 was markedly reduced (ESI Fig. S2B†). At day 17, also DCX, a marker of newborn neurons, was clearly visible along with the PAX6+ cells (Fig. 4E). Together, these results show an effective and uniform differentiation of hPSCs towards a neuroepithelial fate in the centimeter-sized FN-silk/LN-521 network.3.5 Generation of neuronal progenitors in FN-silk/LN-521 foam
To evaluate the potential of FN-silk/LN-521 silk to support development of neurons, we extended the cultivation period in neuronal progenitor medium for a general differentiation without additional patterning factors. The appearance of neuronal markers was analyzed after 3 and 6 weeks. An intricate 3D network of a mixture of cells positive for BIII-TUBULIN and for MAP2 (more mature neurons), was observed at day 21 (Fig. 5A) which was further developed one week later (Fig. 5D). In addition, GADD67+ cells were found integrated within the silk microfibrils, indicative of development of inhibitory GABAergic interneurons. Transcription analysis showed strong expression of BIII-TUBULIN and MAP2 (Fig. 5B). Up-regulation of GAD1, an additional marker for GABAergic interneurons, confirmed the presence of cells of interneuronal subtypes.In contrast to the uniform distribution of cells expressing markers for cycling progenitors found at day 14 (see Fig. 4B and C), analysis of horizontal sections at day 21 showed a more heterogeneous pattern of KI67+ cells, preferentially residing at the walls of the channels (Fig. 5C). This was further visualized by the abundance of neural stem cell marker SOX2, indicating that many of the progenitors within the 3D culture remained in their early state of differentiation.
After a culture period extended to 40 days, analysis of vertical sections revealed MAP2+ cells throughout the foam structure, but especially on the upper side (Fig. 5E). At this stage, the total construct of cells within silk had typically grown to a height of >200 μm. Horizontal sections revealed loose structures of cells at different stages of neurogenesis. No necrotic areas were found, even when analyzing the inner parts. Partial expression of neural stem cell markers SOX2 and PAX6 remained within the 3D culture, preferentially in areas of lower cell density (ESI Fig. S3A†), or next to the channels (Fig. 5F). When analyzing sequential sections, KI67 staining confirmed the presence of cycling cells, flanked by more mature neurons (MAP2+) (Fig. 5F and G). To summarize, generation of hPSC-derived neuronal progenitors integrated in the FN-silk/LN-521 microfibrillar network is a simple way to efficiently obtain a continuous 3D distribution of the cell types that constitute the foundation of the developing brain.
3.6 Neural progenitors retain self-organizing ability in FN-silk/LN-521
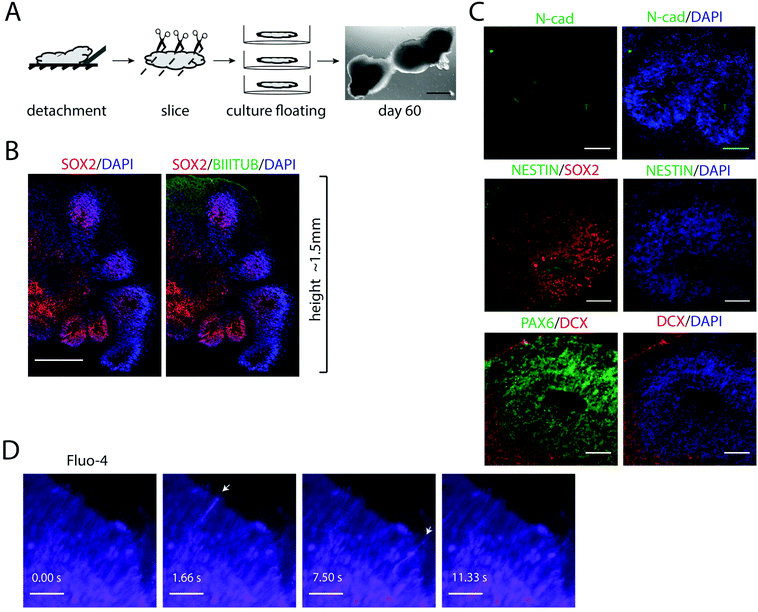
Pluripotent stem cells and early progenitors have a strong self-organizing ability to regulate the development, which can be utilized to obtain organ-like units in vitro.34 After longer cultivations in FN-silk/LN-521, zones of cycling progenitor cells were found organized into polarized cellular organizations (ESI Fig. S3†). These findings encouraged us to further investigate whether FN-silk/LN-521 3D cultures would provide a suitable microenvironment for supporting self-organization during differentiation.We reasoned that cellular self-organization would be facilitated by increasing the flexibility of the silk construct. To test this, we first used the protocol described above to generate homogeneous layers of neuronal progenitors in FN-silk/LN-521. The silk foams with integrated progenitors were then manually detached from the bottom of the wells, cut into slices of ≈2 mm thickness and transferred to low-attachment culture plates where they were kept as floating entities (Fig. 6A). By further extending the cultivation time, this allowed the cells to spontaneously reorganize within the silk network and the cellular constructs to grow in size.
Immunofluorescence analysis of 3D cultures sectioned 60 days after initiation of differentiation revealed multiple regions of SOX2+ cell populations radially organized around the channels and surrounded by layers of cells positive for BIII-TUBULIN (Fig. 6B). The internal structures are an indicative of neural stem cell organization and highly reminiscent of the ventricular zones formed during early neural tube development, which has previously been modeled in cerebral organoids.4 Looking more closely within the 3D culture, multiple columnar shaped organizations of SOX2+ and NESTIN+ cells surrounding ventricle-like cavities with apical localization of the neural specific N-cadherin were found (Fig. 6C). Many of these regions also contained layers of PAX6+ cells, surrounded by newborn neurons marked by DCX+ cells at the basal surface.
3.7 FN-silk/LN-521 supports the generation of cells displaying spontaneous calcium oscillations
To test whether the progenitors that had formed within the FN-silk/LN-521 cultures were functionally active, we imaged the cells after treatment with the calcium indicator Fluo-4. At day 40, widespread spikes in intracellular calcium concentration [Ca2+] were seen, indicating spontaneous activity (ESI Fig. S3B†). In addition, areas of sequential activity involving several cells were observed. Similar spontaneous calcium activity was also noticed at day 60 (Fig. 6D, n = 3), and ESI Movie 1.† Together, this demonstrates the ability of hPSC-derived neural progenitors to spontaneously form cerebral structures with active, signaling progenitors, thereby recapitulating the early neurodevelopment.4. Discussion
In this study we developed a fully defined 3D culture setup for neural differentiation of human pluripotent stem cells using a functionalized recombinant silk protein, FN-silk20 in combination with recombinant laminin, LN-521.24 The procedure to instantly integrate pluripotent stem cells during silk assembly resulted in centimeter-sized 3D constructs, suitable for differentiation and long-term cell survival. We found that this culture set-up provides an easy method for engineering of constructs such as layers of continuous neuroectoderm, network with neuronal activity and self-organizing neural tube-like structures. The method described here constitutes a viable base also for the development of a variety of neuronal differentiation protocols for hPSCs in 3D formats.Cell density is an important factor for neural differentiation outcome32 and at the same time a challenge for any 3D culture set up. The procedure for integration of cells into FN-silk/LN-521 foam involves a mild dispersion of single cells before stabilization, resulting in an even distribution of cells within a network of flexible silk microfibers and laminin. By adjusting the number of hPSCs at integration and the time of cell expansion, different densities of integrated cells are easily obtained. Herein, we used 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pluripotent cells during seeding, rendering ∼14 × 103 cells per cm2 silk foam if assuming 4–5 cell layers. The fact that the hPSCs survive the integration process and stay pluripotent indicates that the silk/laminin is able to provide supportive cell-matrix contact points. After an initial culture period in neural induction medium, a defined and homogenous neural population forms a uniform layer of SOX2+ and PAX6+ neuroectoderm (Fig. 4). Advantages of using a biomaterial-based approach for engineering 3D constructs are the higher degree of control of the microenvironment and the possibility to obtain scalable constructs.8 With this silk-based culture system it would be feasible to create combinations and/or gradients of e.g. different laminin isoforms or other extra cellular matrix proteins for the study of cell fate in 3D.
000 pluripotent cells during seeding, rendering ∼14 × 103 cells per cm2 silk foam if assuming 4–5 cell layers. The fact that the hPSCs survive the integration process and stay pluripotent indicates that the silk/laminin is able to provide supportive cell-matrix contact points. After an initial culture period in neural induction medium, a defined and homogenous neural population forms a uniform layer of SOX2+ and PAX6+ neuroectoderm (Fig. 4). Advantages of using a biomaterial-based approach for engineering 3D constructs are the higher degree of control of the microenvironment and the possibility to obtain scalable constructs.8 With this silk-based culture system it would be feasible to create combinations and/or gradients of e.g. different laminin isoforms or other extra cellular matrix proteins for the study of cell fate in 3D.
Many applications for neural modelling using hPSCs involve long-term differentiation protocols to obtain functionality of a specific cell type, such as oligodendrocytes,35 or the maturation of higher order self-organizing brain structures, as for example formation of the cortical plate.12 Herein, we show that during culture of cells supported by silk, channels are formed within the network of silk microfibers. These channels facilitate access of oxygen and nutrients throughout the construct which enable long term (>month-long) differentiations to be performed also without applying a flow. Analyses of sections from the inner part of the constructs confirmed that cells maintain viable and dividing progenitor cells were found also after a longer culture period, indicating sufficient permeability throughout the construct (Fig. 5F and G).
Pluripotent stem cells and early progenitors have a strong ability to self-organize into layered structures of cortical tissue, closely resembling the human fetal brain.36 Establishing robust protocols for such complex 3D cultures with reduced variations and using defined materials is important to be able to efficiently model the neural development and disease.37 The silk/laminin microfibrillar network was found to be highly flexible and able to support extensive cellular remodeling during differentiation. Increasing the flexibility further by detaching the cell-silk construct from its support enhanced spontaneous reorganization into polarized neuroectoderm, allowing development of neural-tube like structures (Fig. 6). By this, we showed that the key steps during early neural development can be recapitulated in FN-silk/LN-521 matrix in similar ways as in spheroid-based cerebral organoids.4,5
The ability to create oriented and multi-layered cultures is attractive for mimicking the complex in vivo situation of tissue.38 Combining several foams with integrated cells next to or on top of each other would be a feasible approach for the study of interactions between brain regions, similar to what has been demonstrated using fused cerebral organoids.39 We recently described the possibility to integrate multiple cell types in FN-silk, including endothelial cells for in vitro vascularization.23 Previously, this type of recombinant spider silk was shown to be degraded within 3 months after transplantation.40 The 3D culture approach described herein could thus potentially be used for in vivo applications.
5. Conclusions
By assembly of recombinant functionalized silk protein together with laminin 521, we created a defined 3D silk fibrillar network for expansion and long-term differentiation of hPSCs. The mild assembly process of the silk enables an even integration of viable cells, creating a controlled and homogeneous neural population rendering functionally active neurons. Transfer of these flexible constructs into free-floating entities allowed development of neural tube like-tissue including progenitor polarity. Our results suggest that FN-silk/LN521 assembly could be a useful tool for a wide range of 3D culture aiming for neural tissue engineering.Conflicts of interest
M.H. has shares in Spiber Technologies AB, a company that aims to commercialize recombinant spider silk.Acknowledgements
We thank Spiber Technologies AB for providing recombinant spider silk proteins. We are grateful to Dr Steven Edwards at Advanced Light Microscopy Facility at SciLifeLab, Solna and the National Microscopy Infrastructure, NMI (VR-RFI 2016-00968) for providing assistance in microscopy and to the iPS Core at Karolinska Institutet, Stockholm.References
- K. A. Lemke, A. Aghayee and R. S. Ashton, Deriving, regenerating, and engineering CNS tissues using human pluripotent stem cells, Curr. Opin. Biotechnol., 2017, 47, 36–42 CrossRef CAS.
- M. Eiraku, K. Watanabe, M. Matsuo-Takasaki, M. Kawada, S. Yonemura, M. Matsumura, T. Wataya, A. Nishiyama, K. Muguruma and Y. Sasai, Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals, Cell Stem Cell, 2008, 3(5), 519–532 CrossRef CAS PubMed.
- T. Nakano, S. Ando, N. Takata, M. Kawada, K. Muguruma, K. Sekiguchi, K. Saito, S. Yonemura, M. Eiraku and Y. Sasai, Self-Formation of Optic Cups and Storable Stratified Neural Retina from Human ESCs, Cell Stem Cell, 2012, 10(6), 771–785 CrossRef CAS PubMed.
- M. A. Lancaster, M. Renner, C.-A. Martin, D. Wenzel, L. S. Bicknell, M. E. Hurles, T. Homfray, J. M. Penninger, A. P. Jackson and J. A. Knoblich, Cerebral organoids model human brain development and microcephaly, Nature, 2013, 501, 373 CrossRef CAS PubMed.
- X. Qian, Ha N. Nguyen, M. M. Song, C. Hadiono, S. C. Ogden, C. Hammack, B. Yao, G. R. Hamersky, F. Jacob, C. Zhong, K.-j. Yoon, W. Jeang, L. Lin, Y. Li, J. Thakor, D. A. Berg, C. Zhang, E. Kang, M. Chickering, D. Nauen, C.-Y. Ho, Z. Wen, K. M. Christian, P.-Y. Shi, B. J. Maher, H. Wu, P. Jin, H. Tang, H. Song and G.-l. Ming, Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure, Cell, 2016, 165(5), 1238–1254 CrossRef CAS PubMed.
- J. Mariani, G. Coppola, P. Zhang, A. Abyzov, L. Provini, L. Tomasini, M. Amenduni, A. Szekely, D. Palejev, M. Wilson, M. Gerstein, E. L. Grigorenko, K. Chawarska, K. A. Pelphrey, J. R. Howe and F. M. Vaccarino, FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders, Cell, 2015, 162(2), 375–390 CrossRef CAS PubMed.
- A. M. Paşca, S. A. Sloan, L. E. Clarke, Y. Tian, C. D. Makinson, N. Huber, C. H. Kim, J.-Y. Park, N. A. O'Rourke, K. D. Nguyen, S. J. Smith, J. R. Huguenard, D. H. Geschwind, B. A. Barres and S. P. Paşca, Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture, Nat. Methods, 2015, 12, 671 CrossRef PubMed.
- S. B. Shah and A. Singh, Cellular self-assembly and biomaterials-based organoid models of development and diseases, Acta Biomater., 2017, 53, 29–45 CrossRef CAS PubMed.
- S. Knowlton, Y. Cho, X.-J. Li, A. Khademhosseini and S. Tasoglu, Utilizing stem cells for three-dimensional neural tissue engineering, Biomater. Sci., 2016, 4(5), 768–784 RSC.
- R. Peerani, B. M. Rao, C. Bauwens, T. Yin, G. A. Wood, A. Nagy, E. Kumacheva and P. W. Zandstra, Niche-mediated control of human embryonic stem cell self-renewal and differentiation, EMBO J., 2007, 26(22), 4744 CrossRef CAS PubMed.
- Y. Li, M. Liu, Y. Yan and S.-T. Yang, Neural differentiation from pluripotent stem cells: The role of natural and synthetic extracellular matrix, World J. Stem Cells, 2014, 6(1), 11–23 CrossRef PubMed.
- M. A. Lancaster, N. S. Corsini, S. Wolfinger, E. H. Gustafson, A. W. Phillips, T. R. Burkard, T. Otani, F. J. Livesey and J. A. Knoblich, Guided self-organization and cortical plate formation in human brain organoids, Nat. Biotechnol., 2017, 35, 659 CrossRef CAS PubMed.
- B. J. Klotz, L. A. Oosterhoff, L. Utomo, K. S. Lim, Q. Vallmajo-Martin, H. Clevers, T. B. F. Woodfield, A. Rosenberg, J. Malda, M. Ehrbar, B. Spee and D. Gawlitta, A Versatile Biosynthetic Hydrogel Platform for Engineering of Tissue Analogues, Adv. Healthcare Mater., 2019, 8(19), e1900979 CrossRef PubMed.
- K. Saha, A. J. Keung, E. F. Irwin, Y. Li, L. Little, D. V. Schaffer and K. E. Healy, Substrate Modulus Directs Neural Stem Cell Behavior, Biophys. J., 2008, 95(9), 4426–4438 CrossRef CAS PubMed.
- A. Ranga, M. Girgin, A. Meinhardt, D. Eberle, M. Caiazzo, E. M. Tanaka and M. P. Lutolf, Neural tube morphogenesis in synthetic 3D microenvironments, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(44), E6831 CrossRef CAS PubMed.
- C. Allmeling, A. Jokuszies, K. Reimers, S. Kall, C. Y. Choi, G. Brandes, C. Kasper, T. Scheper, M. Guggenheim and P. M. Vogt, Spider silk fibres in artificial nerve constructs promote peripheral nerve regeneration, Cell Proliferation, 2008, 41(3), 408–420 CrossRef CAS PubMed.
- C. Radtke, C. Allmeling, K. H. Waldmann, K. Reimers, K. Thies, H. C. Schenk, A. Hillmer, M. Guggenheim, G. Brandes and P. M. Vogt, Spider silk constructs enhance axonal regeneration and remyelination in long nerve defects in sheep, PLoS One, 2011, 6(2), e16990 CrossRef CAS PubMed.
- M. Lewicka, O. Hermanson and A. U. Rising, Recombinant spider silk matrices for neural stem cell cultures, Biomaterials, 2012, 33(31), 7712–7717 CrossRef CAS PubMed.
- M. D. Tang-Schomer, J. D. White, L. W. Tien, L. I. Schmitt, T. M. Valentin, D. J. Graziano, A. M. Hopkins, F. G. Omenetto, P. G. Haydon and D. L. Kaplan, Bioengineered functional brain-like cortical tissue, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(38), 13811–13816 CrossRef CAS PubMed.
- M. Widhe, N. D. Shalaly and M. Hedhammar, A fibronectin mimetic motif improves integrin mediated cell biding to recombinant spider silk matrices, Biomaterials, 2016, 74, 256–266 CrossRef CAS PubMed.
- S. Wu, J. Johansson, P. Damdimopoulou, M. Shahsavani, A. Falk, O. Hovatta and A. Rising, Spider silk for xeno-free long-term self-renewal and differentiation of human pluripotent stem cells, Biomaterials, 2014, 35(30), 8496–8502 CrossRef CAS PubMed.
- S. Wu, J. Johansson, O. Hovatta and A. Rising, Efficient passage of human pluripotent stem cells on spider silk matrices under xeno-free conditions, Cell. Mol. Life Sci., 2016, 73(7), 1479–1488 CrossRef CAS PubMed.
- U. Johansson, M. Widhe, N. D. Shalaly, I. L. Arregui, L. Nilebäck, C. P. Tasiopoulos, C. Åstrand, P.-O. Berggren, C. Gasser and M. Hedhammar, Assembly of functionalized silk together with cells to obtain proliferative 3D cultures integrated in a network of ECM-like microfibers, Sci. Rep., 2019, 9(1), 6291 CrossRef PubMed.
- S. Rodin, L. Antonsson, C. Niaudet, O. E. Simonson, E. Salmela, E. M. Hansson, A. Domogatskaya, Z. Xiao, P. Damdimopoulou, M. Sheikhi, J. Inzunza, A.-S. Nilsson, D. Baker, R. Kuiper, Y. Sun, E. Blennow, M. Nordenskjöld, K.-H. Grinnemo, J. Kere, C. Betsholtz, O. Hovatta and K. Tryggvason, Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment, Nat. Commun., 2014, 5, 3195 CrossRef PubMed.
- N. Polisetti, L. Sorokin, N. Okumura, N. Koizumi, S. Kinoshita, F. E. Kruse and U. Schlotzer-Schrehardt, Laminin-511 and -521-based matrices for efficient ex vivo-expansion of human limbal epithelial progenitor cells, Sci. Rep., 2017, 7(1), 5152 CrossRef PubMed.
- M. Widhe, U. Johansson, C.-O. Hillerdahl and M. Hedhammar, Recombinant spider silk with cell binding motifs for specific adherence of cells, Biomaterials, 2013, 34(33), 8223–8234 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method, Methods, 2001, 25(4), 402–408 CrossRef CAS PubMed.
- S. M. Chambers, C. A. Fasano, E. P. Papapetrou, M. Tomishima, M. Sadelain and L. Studer, Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling, Nat. Biotechnol., 2009, 27(3), 275–280 CrossRef CAS PubMed.
- C. Nicoleau, C. Varela, C. Bonnefond, Y. Maury, A. Bugi, L. Aubry, P. Viegas, F. Bourgois-Rocha, M. Peschanski and A. L. Perrier, Embryonic stem cells neural differentiation qualifies the role of Wnt/beta-Catenin signals in human telencephalic specification and regionalization, Stem Cells, 2013, 31(9), 1763–1774 CrossRef CAS PubMed.
- E. Klaffky, R. Williams, C.-C. Yao, B. Ziober, R. Kramer and A. Sutherland, Trophoblast-Specific Expression and Function of the Integrin α7 Subunit in the Peri-implantation Mouse Embryo, Dev. Biol., 2001, 239(1), 161–175 CrossRef CAS PubMed.
- S. Rodin, L. Antonsson, C. Niaudet, O. E. Simonson, E. Salmela, E. M. Hansson, A. Domogatskaya, Z. Xiao, P. Damdimopoulou, M. Sheikhi, J. Inzunza, A. S. Nilsson, D. Baker, R. Kuiper, Y. Sun, E. Blennow, M. Nordenskjold, K. H. Grinnemo, J. Kere, C. Betsholtz, O. Hovatta and K. Tryggvason, Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment, Nat. Commun., 2014, 5, 3195 CrossRef PubMed.
- S. M. Chambers, C. A. Fasano, E. P. Papapetrou, M. Tomishima, M. Sadelain and L. Studer, Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling, Nat. Biotechnol., 2009, 27, 275 CrossRef CAS PubMed.
- G. Lee, S. M. Chambers, M. J. Tomishima and L. Studer, Derivation of neural crest cells from human pluripotent stem cells, Nat. Protoc., 2010, 5, 688 CrossRef CAS PubMed.
- S. J. Arnold and E. J. Robertson, Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo, Nat. Rev. Mol. Cell Biol., 2009, 10, 91 CrossRef CAS PubMed.
- G. M. C. Rodrigues, T. Gaj, M. M. Adil, J. Wahba, A. T. Rao, F. K. Lorbeer, R. U. Kulkarni, M. M. Diogo, J. M. S. Cabral, E. W. Miller, D. Hockemeyer and D. V. Schaffer, Defined and Scalable Differentiation of Human Oligodendrocyte Precursors from Pluripotent Stem Cells in a 3D Culture System, Stem Cell Rep., 2017, 8(6), 1770–1783 CrossRef CAS PubMed.
- J. G. Camp, F. Badsha, M. Florio, S. Kanton, T. Gerber, M. Wilsch-Brauninger, E. Lewitus, A. Sykes, W. Hevers, M. Lancaster, J. A. Knoblich, R. Lachmann, S. Paabo, W. B. Huttner and B. Treutlein, Human cerebral organoids recapitulate gene expression programs of fetal neocortex development, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(51), 15672–15677 CrossRef CAS PubMed.
- C.-T. Lee, R. M. Bendriem, W. W. Wu and R.-F. Shen, 3D brain Organoids derived from pluripotent stem cells: promising experimental models for brain development and neurodegenerative disorders, J. Biomed. Sci., 2017, 24(1), 59 CrossRef PubMed.
- P. Zhuang, A. X. Sun, J. An, C. K. Chua and S. Y. Chew, 3D neural tissue models: From spheroids to bioprinting, Biomaterials, 2018, 154, 113–133 CrossRef CAS PubMed.
- J. A. Bagley, D. Reumann, S. Bian, J. Levi-Strauss and J. A. Knoblich, Fused cerebral organoids model interactions between brain regions, Nat. Methods, 2017, 14(7), 743–751 CrossRef CAS PubMed.
- C. Fredriksson, M. Hedhammar, R. Feinstein, K. Nordling, G. Kratz, J. Johansson, F. Huss and A. Rising, Tissue Response to Subcutaneously Implanted Recombinant Spider Silk: An in Vivo Study, Materials, 2009, 2(4), 1908–1922 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9bm01624d |
| This journal is © The Royal Society of Chemistry 2020 |