The nanostructured secretome†
S.
Busatto
 abc,
A.
Zendrini
abc,
A.
Zendrini
 d,
A.
Radeghieri
d,
A.
Radeghieri
 ac,
L.
Paolini
ac,
L.
Paolini
 ac,
M.
Romano
ac,
M.
Romano
 a,
M.
Presta
a,
M.
Presta
 ae and
P.
Bergese
ae and
P.
Bergese
 *ace
*ace
aDepartment of Molecular and Translational Medicine, University of Brescia, 25123 Brescia, Italy. E-mail: paolo.bergese@unibs.it
bDepartment of Transplantation Medicine; Department of Physiology and Biomedical Engineering, Mayo Clinic, Jacksonville, FL 32224, USA
cCSGI, Research Center for Colloids and Nanoscience, 50019 Florence, Italy
dDepartment of Animal Science, Food and Nutrition – Università Cattolica del Sacro Cuore, 29122 Piacenza, Italy
eINSTM, National Inter-university Consortium of Materials Science and Technology, Via G. Giusti 9, 50121 Firenze, Italy
First published on 6th November 2019
Abstract
The term secretome, which traditionally strictly refers to single proteins, should be expanded to also include the great variety of nanoparticles secreted by cells (secNPs) into the extracellular space, which ranges from high-density lipoproteins of a few nanometers to extracellular vesicles and fat globules of hundreds of nanometers. Widening the definition is urged by the ever-increasing understanding of the role of secNPs as regulators/mediators of key physiological and pathological processes, which also puts them in the running as breakthrough cell-free therapeutics and diagnostics. “Made by cells for cells”, secNPs are envisioned as a sweeping paradigm shift in nanomedicine, promising to overcome the limitations of synthetic nanoparticles by unsurpassed circulation and targeting abilities, precision and sustainability. From a longer/wider perspective, advanced manipulation would possibly make secNPs available as building blocks for future “biogenic” nanotechnology. However, the current knowledge is fragmented and sectorial (the majority of the studies being focused on a specific biological and/or medical aspect of a given secNP class or subclass), the understanding of the nanoscale and interfacial properties is limited and the development of bioprocesses and regulatory initiatives is in the early days. We believe that new multidisciplinary competencies and synergistic efforts need to be attracted and augmented to move forward. This review will contribute to the effort by attempting for the first time to rationally gather and elaborate secNPs and their traits into a unique concise framework – from biogenesis to colloidal properties, engineering and clinical translation – disclosing the overall view and easing comparative analysis and future exploitation.
1. Introduction: the secretome nanoparticles
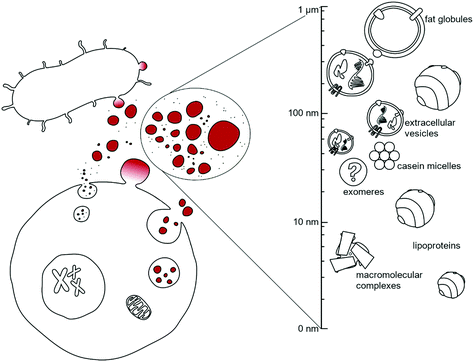
Cells, under both physiological and pathological conditions, secrete a great variety of nanoparticles (secNPs) with different compositions, structures and functions. SecNPs include macromolecular complexes (such as ferritin1 and RNA binding proteins),2 membranous particles (such as extracellular vesicles (EVs)3 and fat globules),4 and micellar structures (such as lipoproteins5 and casein micelles),6 with sizes that range from a few to hundreds of nanometers (Fig. 1). SecNPs are released into the extracellular space together with single molecules, with which they constitute an asset of all biological fluids – e.g. blood, cerebrospinal fluid, saliva, urine, and milk –the biological function of secNPs is shaped by both their molecular and nanoscale (colloidal) properties,7 that is, by their composition, size, structure, surface charge, energetic stability, etc.In the last few years it has becoming more and more evident that cell-to-cell sharing of macromolecular information also occurs via secNPs, “nanolines” of communication that complement “classical” paracrine signalling of single proteins (often referred to as soluble factors), such as cytokines, hormones and growth factors.8,9 In this way, secNPs participate in the regulation of normal physiological processes, such as stem cell maintenance or immune surveillance, and in the pathology underlying diseases.
For these reasons, in the last few years, secNPs have been gaining exponential interest from both academia and biotech companies. Massive production of high grade secNPs is expected to foster a sweeping paradigm shift in nanomedicine, by moving from the synthetic mainstream to natural and hybrid nanosystems “made by cells for cells” with unsurpassed circulation and targeting abilities, personalization and sustainability. Their medical translation might encompass oncology, immunology, tissue regeneration, neurodegenerative disorders and infectious and parasitic diseases. In addition, advanced understanding and manipulation would make secNPs available as effective building blocks for future “biogenic” nanotechnology.
However, the current knowledge of the secNP palette is fragmented, sectorial (research studies and reports are primarily focused on a specific biological and/or therapeutic function of a given kind of secNP) and incomplete (understanding of the nanoscale, colloidal and interfacial properties is poor). On the other hand, the current conception of the secretome misses these nanosized components.
As a first contribution to overcome these hurdles, this review will rationally frame and describe all the secNPs into a unique and simple framework, disclosing the overall view and easing comparative analysis. The traits of SecNPs, which in the present specialized literature are scattered and/or ‘buried’, have been thoroughly reported and elaborated in the sections below, i.e. compositional, structural and colloidal properties (section 2), concentration and separation methods, tweaking and engineering (section 3). This is followed by an effective summary of secNP applications in nanomedicine, including drug delivery, vaccines, regenerative therapeutics and diagnostics (section 4). The review ends with a discussion on the future exploitation and perspectives of secNPs (section 5).
Finally, it is worth noting that this review focuses on biogenic nanoparticles spontaneously secreted by cells. Therefore, nanosized cytoplasmic organelles, viruses10 and fully artificial nanoparticles such as synthetic nanovesicles11 and DNA nanocages12 will not be considered.
2. Physicochemical and biological properties
2.1. Nanosized macromolecular complexes
If not shielded by transport proteins, ions, RNAs, and hormones would be rapidly degraded by enzymes that populate biological fluids. Albumin, ferritin, transferrin and argonaute-2 (Ago2) are just a few examples of proteins that work as transient or regular carriers. Many of them are highly evolutionarily ubiquitous1 and they all can be considered as secNPs naturally equipped with subnano- and/or nano-cages and pockets.Albumin is the most abundant protein found in the plasma. It is a small globular protein (66 kDa) and has a high affinity for metals, fatty acids, amino acids, metabolites and lipophilic xenobiotics.13 The three-dimensional structure comprises three homologous domains that assemble to form a heart-shaped molecule. Each domain is a product of two subdomains and ligand binding regions are located in hydrophobic cavities in subdomains IIA and IIIA.14 The most important physiological roles of albumin are: to bring solutes in the bloodstream to their target organs, and to maintain the pH and osmotic pressure of the plasma.15,16
Ferritin plays a key role in iron sequestration and its protein cages store excess cellular iron for future use by the cell.1,17 Ferritin consists of 24 subunits, typically comprised of different ratios of the H and L chain subunits. The different subunits have divergent functions – H-ferritin utilizes ferroxidase activity for the oxidation of ferrous (Fe2+) to ferric (Fe3+) iron while L-ferritin contains acidic residues on the surface cavity that facilitate ferroxidase turnover and are crucial for the nucleation of Fe3+ within the core protein. Iron-free ferritin, named apoferritin (APOFe), consists of a round-shaped hollow structure loadable with exogenous molecules; indeed, ferritin can be disassembled at very acidic (pH 2–3) or very basic (pH 11–12) pH and it self-reassembles at neutral pH. These assembly properties have been used to load different compounds within the ferritin core. Recent studies demonstrated the association of H-ferritin with exosomes,18,19 another class of secNPs described hereafter, indicating that the intracellular trafficking and secretion of different secNPs can converge.
Transferrin is another important iron carrier that controls the toxic and insoluble characteristics of iron. Both apo- and holotransferrin are single-chain glycoproteins, with 670–690 amino acid residues and a molecular weight of ∼80 kDa. The transferrin molecule consists of two homologous lobes, termed N- and C-lobe. Each lobe consists of two domains that are connected by a flexible hinge, and each lobe can independently bind a Fe3+ ion. The highly specific binding site for Fe3+ is created when the two domains of a lobe close around iron, whereas iron release requires the two domains to open up.20
Argonaute-2 (Ago2) is a ribonucleoprotein complex that serves as a carrier of circulating miRNAs in the plasma and regulates small RNA guided gene silencing processes.21 It is composed of four major domains: N, PAZ, MID and PIWI, each with a specific function. The four domains array into a bi-lobe format consisting of N-PAZ and MID-PIWI connected by Linker 1/2 proteins. The two lobes form a gap which accommodates the guide small RNAs and their complementary fragments in the middle.22
The key physicochemical properties of nanosized macromolecular complexes are summarized in Fig. 2.
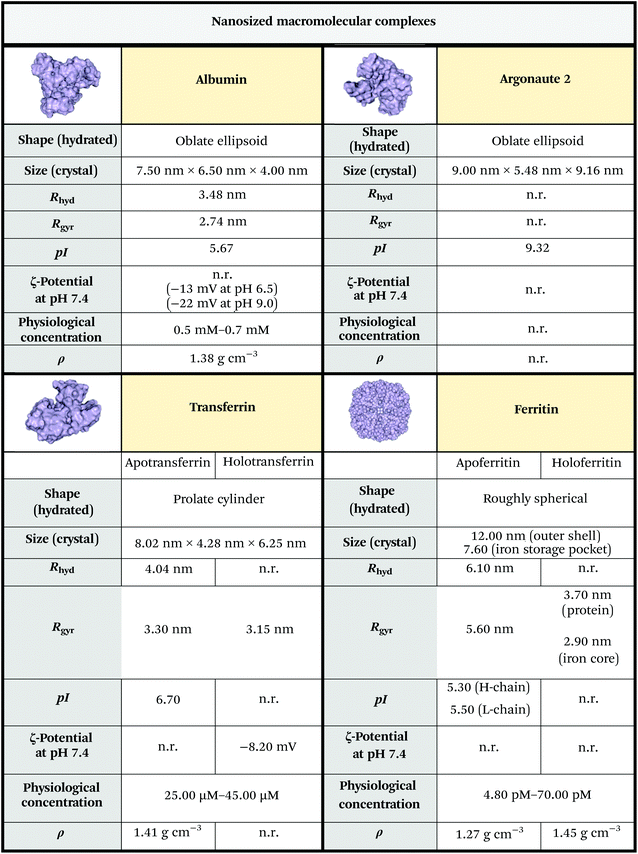 | ||
| Fig. 2 Physicochemical properties of primary macromolecular complex secNPs. The size was estimated from RSCB PDB files 1AO6,153QYT,232FHA24 and 5JS2.25 Albumin, apotransferrin, apoferritin and Ago2 isoelectric points were predicted using the ExPASy compute pI/Mw tool (https://web.expasy.org/compute_pi/). Other data were obtained from ref. 26–31. Unknown parameters are marked as “n.r.” (not reported). Notes: (i) The reported protein concentrations refer to human adult males; the concentrations relative to adult females and infants are slightly lower; (ii) some of the parameters reported refer to the bovine or horse counterpart (which anyway closely resembles the human variant in terms of structure and sequence); (iii) mean protein density is experimentally considered to be 1.35 g cm−3, while theoretical calculation performed on standard proteins led to a mean density of 1.47 g cm−3 or 1.43 g cm−3, in regard to the algorithm used to perform the analysis.32,33 Legend: Rhyd = hydrodynamic radius; Rgyr = gyration radius; pI = isoelectric point; ρ = density. | ||
2.2. Extracellular vesicles
Extracellular vesicles are soft nanoparticles made up of a lipid membrane which encloses proteins, nucleic acids and metabolites (Fig. 3 and 4). They function in cell–cell and cell–microenvironment communication,34,35 emerging as universal agents in intra- and cross-organism communication for mammals, viruses,10 bacteria, archaea, microbes, parasites,36 fungi and plants.37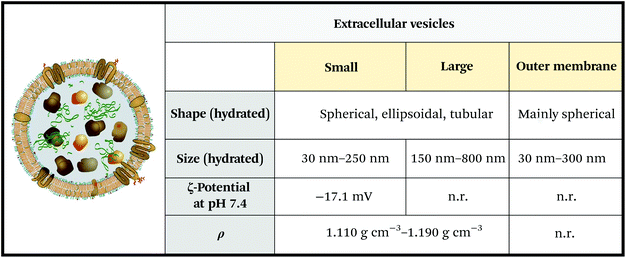 | ||
| Fig. 3 Physicochemical properties of extracellular vesicles (EVs). Note that the reported ζ-potential refers to serum EVs. ζ-Potential varies significantly depending on EV media (e.g. saliva, urine, blood, and cerebrospinal fluid), and oscillates between −15 mV and −34 mV. Unknown parameters are marked as “n.r.” (not reported). Figure adapted and reproduced from ref. 3 with permission from Elsevier, copyright 2012. Data were obtained from ref. 44–46. Legend: ρ = density. | ||
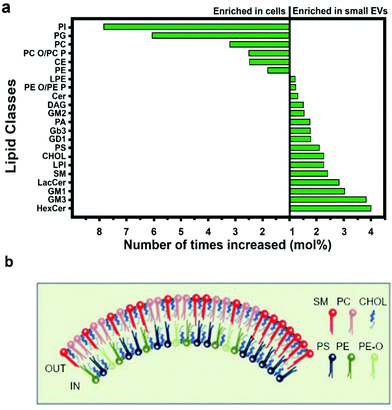 | ||
| Fig. 4 Lipid composition of small EVs. (a) Small EVs and cell membrane are differentially rich in lipid moieties. (b) Composition of the inner and outer leaflets of the lipid bilayer surrounding small EVs. Figures adapted from ref. 47. Legend: PI = phosphatidyl-inositol; PG = phosphatidyl-glycerol; PC = phosphatidyl-choline; PC-O = alkyl-ether substituted phosphatidyl-choline; PC-P = alkenyl-ether substituted phosphatidyl-choline; CE = cholesteryl ester; PE = phosphatidyl-ethanolamine; LPE = lysophosphatidyl-ethanolamine; PE-O = alkyl-ether substituted phosphatidyl-ethanolamine; PE-P = alkenyl-ether substituted phosphatidyl-ethanolamine; Cer = ceramide; DAG = diacyl-glycerol; GM2 = ganglioside GM2; PA = phosphatidic acid; Gb3 = ceramide trihexoside; GD1 = ganglioside GD1; PS = phosphatidyl-serine; CHOL = cholesterol; LPI = lysophosphatidyl-inositol; SM = sphingomyelin; LacCer = lactosyl-ceramide; GM1 = ganglioside GM1; GM3 = ganglioside GM3; HexCer = hexosyl-ceramide. | ||
EVs are emerging as the third way for cell communication other than direct intercellular physical stimuli and the paracrine secretion of active molecules.35 EVs act as vehicles for bioactive cargoes preventing their degradation and delivering them to target cells. Due to their structure and function, EVs actively participate in different physiological processes, such as coagulation and immune system activation.3,35 On the flip side, EVs contribute to the effective maintenance and diffusion of several pathological processes that rely on cell-to-cell communication. For instance, it has been demonstrated that malignant tumours exploit EV-based strategies to initiate the pre-metastatic niche and colonize other healthy organs.41
EVs can be found in every biological fluid derived from common cell secretory pathways. Due to their biogenesis and structure, EVs are representative of the releasing cell phenotype, also possessing the originating cell functions (a “window into the cellular world”).37 On the other hand, EVs feature specific compositional enrichment, starting from lipids (Fig. 4) and proteins (section 4.4.2). For example, EVs isolated from mesenchymal stem cells (MSCs) reproduce part of the biological effects, particularly those responsible for their regenerative potential. MSCs actively secrete multiple growth factors and cytokines encased in EVs that stimulate the repair of different tissues and organs (for further details, see section 4.3.2).42
2.3. Lipoproteins
Lipoproteins are a class of secNPs found in the plasma and tissues, whose primary assignment is lipid transport and delivery.48 They are nanosized complexes composed of a central hydrophobic core of non-polar lipids (Fig. 5, bright orange), enveloped in a mono-layered amphiphilic membrane made of phospholipids, cholesterol (Fig. 5, blue and green) and apolipoproteins (Apo, Fig. 5 grey). The lipid and protein content of a lipoprotein is heterogeneous and evolves during its life and circulation time.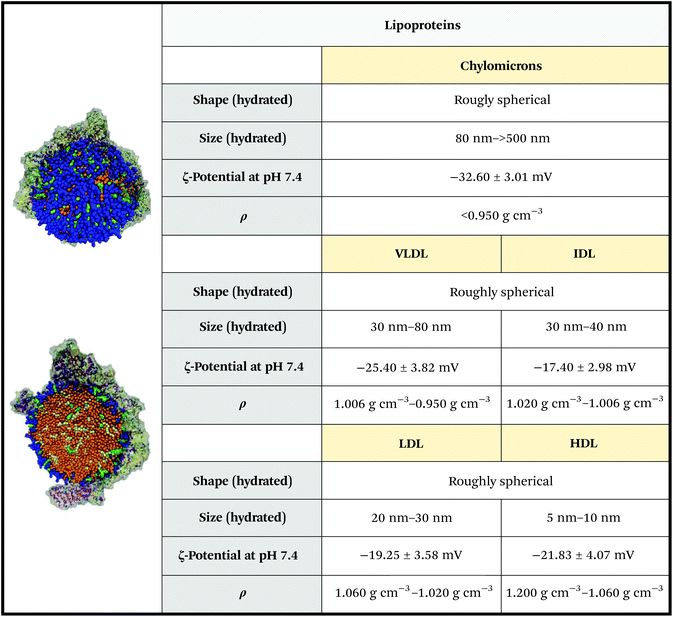 | ||
| Fig. 5 Physicochemical properties of lipoproteins. The ζ-potential of chylomicrons is relative to those of artificial chylomicrons reconstituted via thin layer hydration. Figures adapted and reproduced from ref. 52 with permission from The Royal Society of Chemistry, copyright 2011. Data obtained from ref. 44, 53 and 54. | ||
Five classes of lipoproteins exist. Chylomicrons form the biggest and less dense subclass. They are mainly composed of triacylglycerols and in a minor part of phospholipids, cholesterol, and cholesteryl esters and ApoB-48. Chylomicrons are assembled inside enterocytes (intestine cells) during dietary lipid absorption. Chylomicrons avoid liver passage and transport dietary lipids to storage tissues (adipose and skeletal muscle tissues) and to the cardiac tissue. After the transfer of triacylglycerols to the peripheral tissues, chylomicrons collapse and are catabolized by the liver.49 Very Low-Density Lipoproteins (VLDLs) are synthesized by hepatocytes (liver cells) and are composed of triacylglycerols and cholesteryl esters assembled with apolipoproteins, preferably ApoB-100.49 VLDLs are secreted with a size of 30–80 nm,44 but are rapidly degraded by cleaving of their fatty acid content into the smaller and denser Low-Density Lipoproteins (LDLs) – note: the transformation passes through the so-called Intermediate-Density Lipoproteins (IDLs) – VLDLs and LDLs circulate and transport lipids to peripheral tissues, in particular, LDLs can be taken up by atherosclerotic plaque endothelium (thus representing a high-risk factor).49 High density lipoproteins (HDLs) are smaller and denser than LDLs, with a diameter below 10 nm and a density of 1.060–1.200 g cm−3. They are rich in cholesterol and ApoA-I, A-II, ApoE and ApoC, and are widely studied due to their protective role against cardiovascular diseases and cardiac acute events. The biosynthesis of HDLs begins with the synthesis of ApoA-I in the liver or intestine. HDLs are then formed through subsequent lipidation of ApoA-I, which gives rise to nascent, discoidal particles. Spherical HDLs are then obtained by internalization of cholesterol esters into the core of HDL particles, a step catalysed by lecithin cholesterol acyltransferase. HDLs remove the excess cholesterol from the tissue, which is then transported back to the liver and steroidogenic organs to be recycled.49 HDLs have the additional peculiarity to also carry other molecules than apolipoproteins and lipids, such as small non-coding genetic materials, mainly micro RNAs (miRNAs),50 and other proteins, such as metalloproteinase.51
The salient physicochemical properties of lipoproteins are summarized in Fig. 5 and Table 1.
| Surface components (mol%) | Core lipids (mol%) | |||||
|---|---|---|---|---|---|---|
| Protein components | Protein | Phospholipid | Cholesterol | Cholesterol ester | Triglycerides | |
| Chylomicron | ApoB-48 | 2 | 63 | 35 | 5 | 95 |
| VLDL | ApoB-100 | 2 | 55 | 43 | 24 | 76 |
| LDL | ApoB-100 | 2 | 58 | 42 | 19 | 81 |
| HDL | ApoA-I, A-II, E, C | 2 | 72 | 23 | 82 | 18 |
2.4. Exomeres
Exomeres are a recently discovered class of secNPs. They are small (∼35 nm), non-membranous particles rich in specific enzymes and nucleic acids. Exomeres were first separated and characterized by asymmetrical flow field-flow fractionation (AF4). They possess unique biophysical features, such as mildly negative ζ-potential and higher stiffness compared to EVs.56 Currently, very little is known about the traits and biology of exomeres, except their ability to shuttle functional, bioactive cargoes and to modify the metabolism of recipient cells.572.5. Milk proteins, casein micelles and fat globules
Milk is a mammary gland secretion widely used as a processed food product and as a primary food for newborns of many animal species. Milk is not only able to supply nutrients through proteins and fat-made nanoparticles, but also regulates immune defense and interacts with the gut microbiome.58Milk is an emulsion composed of fat globules, lactose, and soluble proteins, mainly caseins (Fig. 6a). Triglycerides constitute more than 95% of the lipids in milk. Fatty acids used for milk fat globule synthesis are derived from circulating lipids, particularly from chylomicrons and very low-density lipoproteins, or newly synthesized from the mammary gland.59
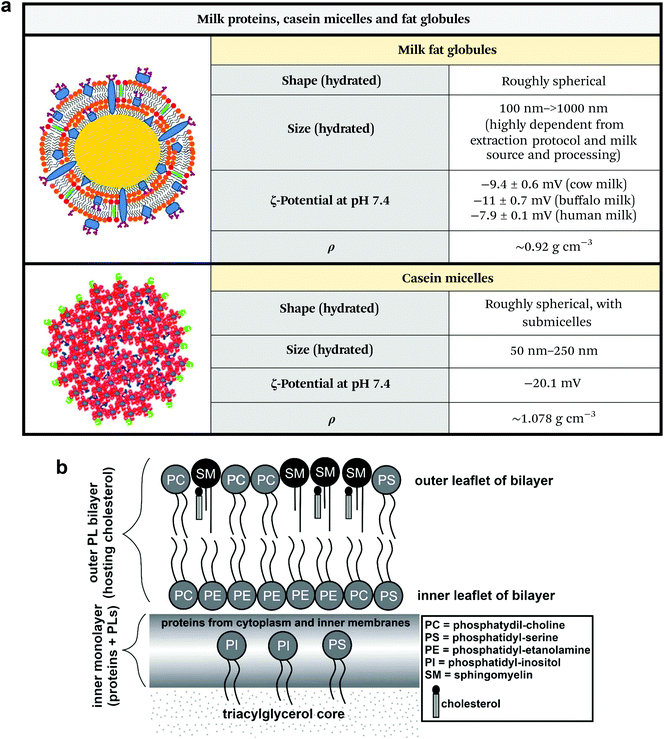 | ||
| Fig. 6 (a) Physicochemical properties of milk fat globules and casein micelles. Unknown parameters are marked as “n.r.” (not reported). Note that the ζ-potential of milk fat globules greatly changes in regard to sample homogenization and source. Figures adapted and reproduced from ref. 4 and 6, with permission from Elsevier, copyright 2017 and 2019. Data extrapolated from ref. 62–66. Legend: ρ = density. (b) Structure and composition of milk fat globules. Figure adapted and reproduced from ref. 67, with permission from Elsevier, copyright 2017. | ||
Fat globules are secNPs with a size ranging from hundreds of nanometres to tens of micrometres (Fig. 6a).59 Fat globules together with lactose and milk proteins originate from epithelial lactating cells that constitute mammal glands. Fat droplets originate in the cell cytoplasm and are already delimited by a single-layer lipid coat that exists prior to secretion. After maturation, the lipid droplet moves into the cell apical region and buds from the plasma membrane acquiring another cell-derived external bilayer that is superimposed during secretion and directly faces the surrounding aqueous environment. The multi-layered membrane is functionalized with multiple proteins and mediate the biological effects of the majority of milk fat globules that are not only related to lipid metabolism but also to the newborn's immune system and to the intestine physiology (Fig. 6b).
Mainly due to their fragile membrane that works as a physical interface between their fat content and the surrounding aqueous environment, fat globules are not present in manufactured food products because of the processing temperature, reagents and timings. In fact, to preserve their native structure, fat globules require dedicated separation protocols. Probably for this reason, their therapeutic potential is not yet fully appreciated.60,61
Caseins are regular components of milks of various species (e.g. cow, goat, rat, mouse and human) and constitute the large majority of proteins in milk. Casein self-assembles to form micelles with a size range of 50 to 250 nm that contain water, calcium and phosphorous salts, and associated enzymes and exist as colloidal particles (Fig. 6a). The specific function and precise internal structure of casein micelles are currently under debate.
2.6. secNP zoo: (buoyant) density versus size
Organizing/visualizing secNP classes and subclasses based on their physicochemical properties may be particularly useful for their rational use, starting from the design of separation protocols (section 3.1). In Fig. 7 we propose as an example the plot obtained by arranging the whole secNP zoo for density versus size. Macromolecular complexes are the densest and smallest, followed by HDL, whose composition is dominated by proteins over lipids. Contrarily, LDLs, VLDLs, and chylomicrons are composed by a growing lipid over protein ratio resulting in increased size and density. As expected from their composition and structure, all the EV populations are characterized by similar densities, shared with the smallest HDL, while their size spans about two orders of magnitude. Milk-derived secNPs slightly overlap in size but differ in density. Specifically, protein-based casein micelles are densest compared to the lipid-based fat globules.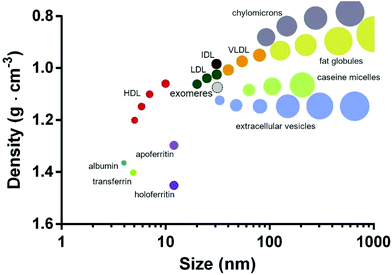 | ||
| Fig. 7 secNPs plotted for density versus size. Exomeres are contoured by a dashed line, since their recent discovery and ongoing physicochemical characterization. The plot was built with the data reported in the previous subsections. The proposed schematic depicts all the classes and subclasses of secNPs discussed in this review integrating the graph previously proposed by Karimi et al.44 | ||
3. Separation, characterization, tweaking, and engineering
3.1. Separation and characterization
Each SecNP class is characterized by unique features, which include protein/lipid ratio, surface charge, size, and density, and is produced through different secretion pathways. Accordingly, native secNPs can be found as mixed constituents of various biological matrices and require dedicated protocols in order to be concentrated or separated.38,49,59,68,69 Concentration (or enrichment) aims to collect in small volumes a large number of a given class of secNPs according to one of their distinct physico-chemical properties, such as size or surface charge. Usually, concentration methods allow the processing of large sample volumes under standardized conditions but conceive the presence in the final formulation of a variable amount of exogenous materials other than solely a specific secNP.70 The separation (or purification) of a class of secNPs instead, aims to purely isolate and retain specific secNPs according to two or more chemico-physical distinctive traits.40 Usually, separation procedures require time-consuming protocols, frequently characterized by small processable sample volumes and a limited yield.71,72 Moreover, an efficient separation is usually hampered by the presence of other classes or subclasses of secNPs in the starting samples that overlap the chosen secNP in size, density, surface biomarkers, etc.71Commonly applied concentration methods exploit the size and density of secNPs (Fig. 7) – e.g. ultracentrifugation (UC) and ultrafiltration (UF) – and/or their surface composition and properties – e.g. immuno-based separation (IBS) assays and polymer-based precipitation protocols (PPs).73 Using UC speeds above or equal to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for a time dependent on the sample volume at a controlled temperature, it is possible to concentrate nanosized materials.74 However, the high speeds at which UC is performed affect the stability and integrity of the secNPs, compromising their final number and properties.
000g for a time dependent on the sample volume at a controlled temperature, it is possible to concentrate nanosized materials.74 However, the high speeds at which UC is performed affect the stability and integrity of the secNPs, compromising their final number and properties.
UF techniques combine controlled pressure flow with filter membranes designed with pores with a determined size and can be performed following different arrangements. For instance, tangential flow filtration devices apply a flow tangential to the filter obtaining a gentle particle separation able to preserve the integrity of the delicate secNP structure.70 Furthermore, UF systems can process large sample volumes under sterile conditions resulting in time saving protocols characterized by a considerable secNP yield.70,75,76
SecNPs expose specific proteins on their external leaflet and, therefore, are eligible for IBS assays.77 IBS usually consists of an immunocapture step, to select secNPs accordingly to the proteins on their surface, followed by a detection method.77 IBS applies both regular and secNP-dedicated detectors, mainly flow cytometers, resulting in a highly specific technique able to enrich a specific secNP subpopulation.78 However, IBS protocols are usually expensive and not scalable to large volumes.78
Polymer-based PPs are applied mostly for EVs, OMVs and HDLs.79 PPs exploit unspecific interactions between secNPs and synthetic polymers that results in a net that precipitates at low-speed centrifugation (<5000g) better preserving the secNP structure.80 However, due to the presence of a protocol-derived matrix that frequently coprecipitates with secNPs, PPs highly impact the amount and purity of secNPs.81 Therefore, PPs are usually used to conduct studies about secNP composition instead of in vitro or in vivo functional studies.79,82
Isolation protocols often combine a first concentration step followed by one or more separation steps based on the physical features of secNPs.83 In particular, density gradients and size exclusion chromatography84 separate secNPs based on their floating density and size and are usually preceded by UC concentration.81 However, in the case secNPs completely overlap in density or size (Fig. 7), these methods have the chance to generate a mixed formulation.
Isolation methods, especially if used in combination, ensure a negligible presence of unwanted components in the final formulations.81 However, laborious and time-consuming protocols may compromise both the structure and yield of the final secNPs.85 Notably, all isolation methods are known to impact to some extent the final secNP formulation and are difficult to standardize.40,61 Therefore, an appropriate approach should be chosen considering the volume, number, and evaluability of the starting biological samples, and also based on the next applications envisioned from the study.40,85
Even though many efforts have been made to introduce and promote guidelines regarding handling and processing of samples recently, still there are differences in almost all the published scientific literature that hamper the sharing of a common practice.85 Microfluidic-based technology represents the latest attempt to combine secNP isolation methods and secNP analysis in scaled-down devices. Dimension and microfluidics enable these devices to couple high throughput and recovery of secNPs at the nanoscale.86
A good isolation practice is always followed by detailed characterization to confirm the presence of the desired secNPs in the final formulation. Characterization usually combines imaging, and biochemical techniques and quantification assays to assess the morphology, size, protein-enrichment, and number of secNPs, respectively.40 Morphology and size range can be analysed by high-resolution imaging techniques, such as atomic force microscopy81 and regular or cryogenic electron microscopy.87 Size distribution and particle quantification techniques are based on the detection of secNP physical properties. Specifically, the secNPs ability to scatter the light (nanoparticle tracking analysis,88 dynamic light scattering,89etc.), to oppose to the flow of an electric current (resistive pulse sensing),90 to interact with metallic nanoparticles (colorimetric nanoplasmonic assay)91 or with fluorescent lipophilic probes (fluorescence correlation spectroscopy89 and flow cytrometry based on micro- and nanofluidics).92 The protein content of secNP formulations can be determined with some of the commonly used protein quantification assays, such as bicinchoninic acid, and Coomassie blue assay. In contrast, the detection of specific proteins has to be performed using more sensitive methods, often based on antigen–antibody reactions and able to detect specific proteins enriched on the secNP surface. However, each protein is present at extremely low concentrations, requiring a high particle number (usually >106) to perform a single analysis with conventional methods, such as western blot,81 immunocapture-based flow cytometry93 or mass spectrometry.94 For this reason, in recent years, intense effort has been made to improve the performance of antibody-based techniques, mainly by coupling them to biosensors and microfluidic devices. New analytical platforms include surface plasmon resonance spectroscopy, surface enhanced Raman spectroscopy, micro-nuclear magnetic resonance, nano-plasmonic sensors, integrated magnetic-electrochemical sensors, etc.94–96
Less commonly characterized components of secNPs are lipids and glycans which can be analysed, for instance, by total reflection Fourier-transform infrared spectroscopy97 and high-resolution glycomics,98 respectively.
Finally, it is important to emphasize that all secNPs share at least one physicochemical feature, making univocal separation between classes or subclasses very difficult to achieve. Therefore, careful characterization of the secNP formulation is fundamental to assess or exclude contamination between subpopulations and to evaluate to which extent it could condition the experimental results.40,44,81
3.2 Tweaking and engineering
The difference between tweaking and engineering is subtle but important and may be profitably applied for better understanding of the plethora of secNP modifications and their intended applications. Cells have been tweaked at the molecular scale for decades. A crucial breakthrough came with the demonstration that an exogenous cDNA derived from an Aequorea victoria gene, encoding for a green fluorescent protein (GFP), produces a fluorescent product when expressed in prokaryotic (E. coli) or eukaryotic (C. elegans) cells without the need for substrates or co-factors.99,100 We will see that this strategy has been largely applied to produce modified secNPs, which for example overexpress a particular targeting ligand.Engineering can be instead interpreted as something different: to design the modified secNP in mechanistic detail, with precise knowledge of all the component parts. This may imply the use of various synthetic components and synthesis techniques, which range from the exogenous loading of drug molecules through electroporation to the production of extracellular vesicles by direct cell extrusion. In secNP modification, very often tweaking and engineering overlap and/or combine,101 as we will concisely present in the next subsections.
SecNPs such as lipoproteins104 and casein micelles105 are unique examples of NPs that can be reassembled or synthesized by mixing their constitutive components. Synthetic or reconstituted secNPs efficiently mimic the physicochemical and biological properties characteristic of their native counterparts and have been widely applied and studied mainly as delivery vehicles for molecular therapeutics.104
Diverse engineering approaches have been applied to secNPs, in order to produce formulations with known, completed, or enhanced abilities.106 The majority of the applied engineering strategies aim at the modification of secNPs with organic molecules, such as peptides, antibodies, lipids, and nucleic acids, resulting in homogeneous systems with new features improving the translational potential of the secNPs.68,102,106–109 Interestingly, the chemico-physical properties of secNPs promote a high-system flexibility opening to different reagents, timings and techniques. Some approaches, usually referred to as endogenous engineering, modify the secreting cells in order to obtain altered secNPs, whereas other strategies called exogenous engineering are based on physico-chemical techniques applied to secNPs after their isolation or during their assembly. SecNPs, mainly nanosized macromolecular complexes,107 lipoproteins, and EVs,106 are biomaterials suitable to undergo different strategies of passive (incubation) or active (fusion peptides) targeting and passive (mixing) or active (sonication, electroporation, thaw–freeze cycles, etc.) drug loading.
Ferritin-L-chain (FTL) fused with activator protein 1 (AP-1) resulted in protein-based nanocages targeting interleukin-4 receptor (IL-4R) overexpressing cells.110 Notably, engineered ferritin nanocages expressing AP-1 peptides were able to selectively bind IL-4R expressing lung tumor cells blocking the pro-inflammatory pathways activated by IL-4 signalling.110 AP-1 ferritin nanocages were also effective when tested as a treatment for in vivo murine models of induced asthma. Compared to the bare ones, AP-1 ferritin nanocages promoted an efficient reduction of inflammation causing an overall inhibitory effect on allergic asthma symptoms.110
Active targeting strategies have been applied also to natural LDLs. In particular, engineered LDLs exposing on their surface the transferrin receptor ligand and loaded with chemotherapeutics were used as a treatment for brain cancer.111 Transferrin receptors are overexpressed in both the blood–brain barrier (BBB) and brain tumor cells. Therefore, engineered LDLs loaded with chemotherapeutics were able to pass through the BBB and accumulate in the brain tissue showing a dose-dependent anti-tumor activity towards glioma cells.111 Interestingly, glioma-bearing mice repeatedly treated with intravenous injection of engineered LDLs showed significant tumor growth inhibition compared to saline, free drug, and native loaded LDLs promoting the highest animal survival rate.111
The scientific literature describes a multitude of different approaches intended to manipulate and engineer both eukaryotic and prokaryotic EVs that range from passive or active drug loading, active targeting (see section 4), click chemistry, to pH responsive EVs.
EVs have been introduced to click chemistry as it represents a modification technique with limited impact on the EV structure and function. EVs were modified using surface chemistry to expose on their outer leaflet proteins functionalized with alkaline groups and successively linked to fluorescent azide molecules. Interestingly, post-modification EVs did not show any change in size and uptake from parent cells compared to native EVs.112
Additionally, Lee et al. designed and produced pH-responsive EVs that efficiently incorporated Dox and hyaluronic acid (HA) grafted with 3-(diethylamino)propylamine (DEAP) into their membrane. In particular, HA mediated the targeting of cancer cells overexpressing cluster of differentiation 44 (CD44), whereas DEAP led to EVs responsive to an acidic tumor microenvironment. Interestingly, engineered HDEA-EVs exhibited high tumor toxicity both in vitro incubated with cancer cells and in vivo after intravenous administration in mice bearing subcutaneous colon carcinoma.113
Lastly, Chen et al. applied for the first time two separate strategies to obtain OMVs simultaneously functionalized for biosensing and bioimaging. This innovative approach consisted in the modification of two membrane-bound bacterial proteins, namely the periplasm-oriented outer membrane lipoprotein SlyB (SlyB) and the transmembrane ice nucleation protein (INP). The bacterial cells were genetically modified to overexpress SlyB fused with nano-luciferase (nLuc) and INP linked to a protein scaffold ending with antibody binding Z domains.68 Consequently, the nLuc was stably anchored into the inner leaflet of the OMV membrane whereas the INP antibody binding domains were protruding out of the OMV surface. Interestingly, the co-expression of SlyB and INP did not affect the nLuc activity or the Z domain binding, resulting in customized secNPs suitable for simultaneous bioimaging and biosensing.68
In conclusion, the majority of engineering approaches assemble secNPs characterized by enhanced features and able to overcome drawbacks typical of therapeutic small molecules, namely poor stability and affinity. However, there are many hurdles in their translation into clinical nanomedicine due to the lack of protocol standardization and scalability.
Ideal in vivo imaging methods should be unequivocally associated with the secNP structure, have a high signal-to-noise ratio, and mirror the secNP half-life.114 To date, different types of molecular imaging techniques have been proposed for non-invasive secNP tracking68,108,115 following both direct and indirect approaches.115 Direct labelling techniques entail secNP association with trackable molecules through weak, unspecific bonds or by other loading techniques, namely electroporation and sonication. Indirect labelling instead starts with the modification of the secreting cells in order to obtain secNPs endowed with imaging molecules or NPs.115
In particular, fluorescence and bioluminescence are the widely applied techniques to label secNPs directly and indirectly.116 Fluorescent lipophilic probes spontaneously intercalate into the secNP structure by passive incubation protocols.114 However, they require specific post-labelling expedients in order to eliminate unbound fluorescent molecules (washing steps or size exclusion chromatography protocols) that could compromise the final EV integrity and yield.114 Additionally, lipophilic fluorescent probes can spontaneously dissociate from the secNP structure generating an unspecific signal not ascribable to EV distribution or uptake.114 Other alternative approaches use fluorescent probes that selectively label DNA or RNA shuttled or loaded into EVs,117 HDLs118 and OMVs.119 The main drawback of these labelling techniques is represented by an inadequate sensitivity connected to the limited and varied secNP content of small genetic materials and proteins.
Bioluminescence is characterized by a high signal/noise ratio since animal tissues do not have any intrinsic bioluminescent activity.116 Cells stably transfected with the luciferase gene translate it into a functional bioluminescent reporter protein naturally uploaded into secNPs.116
Contrary to lipoproteins and albumin, EVs and OMVs are characterized by a limited half-life that usually ranges from 30 minutes to 12 hours. Accordingly, highly stable fluorescent probes that generate signals up to 48–72 hours after their administration are not suitable to provide reliable information about the half-life of EVs and OMVs. In a study published in 2015 by Lai et al., tumor derived EVs were visualized and tracked in vivo using a combination of indirect labelling techniques based on fluorescence and bioluminescence. Secreting cells were simultaneously transfected with two plasmids encoding for the Palmtd Tomato fluorescent plasma membrane protein tagged with a repeated sequence binding MS2 RNA and for the bacteriophage MS2 coat protein fused with GFP.116 This engineering approach allowed one to first detect EV budding and mRNA cargo sorting in vitro and to subsequently monitor EV tumour targeting, mRNA delivery and translation in vivo.116
Other secNP labelling techniques use radionucleotides and ultramagnetic NPs. Contrary to ultramagnetic NPs, radionucleotides benefit from high in vivo sensitivity provided by the tissue permeability of gamma rays. However, radionucleotides are expensive and difficult to handle and require dedicated detection systems. Isaac-Olivè et al. used reconstituted HDLs (rHDLs) as carriers for radio-imaging agents. Given the high demand of cholesterol for membrane synthesis, tumor cells frequently overexpress SR-B1 receptors that mediate rHDL uptake. Therefore, rHDLs loaded with a modified hydrophobic radiopharmaceutical have been used as vehicles to target SR-B1 overexpressing cancer cells.120 Interestingly, rHDLs loaded with hydrophobic radio-imaging agents retained the intrinsic tumor targeting properties both in vitro and in vivo, exhibiting great potential as a radiopharmaceutical transporter for the imaging and diagnosis of SR-B1 overexpressing tumors.120
4. Clinical opportunities
4.1. Drug delivery
Drug delivery nanoscale vehicles able to reach and accumulate in the target site without damaging other organs promise to significantly improve treatment results and patient outcome. After two decades of intense efforts and funding, the use of inorganic NPs for the task has been recently severely questioned, with reasons which include low circulating stability, poor targeting ability and toxicity.121 Accordingly, the advancement of synthetic NPs to the clinic has been far below expectations.122Contrarily, since their discovery all the classes of secNPs, mainly lipoproteins and EVs, have been envisioned as natural delivery vehicles for molecular therapeutics. In fact, secNPs are inherently provided with superior biological/biomedical properties compared to even the most advanced synthetic NPs, including improved targeting, and circulation performance combined with increased bioavailability, personalization and sustainability. The state of the art in this particular field, which also takes advantage of several tweaking and engineering strategies outlined in section 3.2, will be summarized in the following subsections.
Noteworthily, Table 2 lists some review papers that discuss techniques, applications, and future perspectives of secNPs as drug delivery vehicles.
| Nanosized macromolecular complexes | Extracellular vesicles | Lipoproteins | Milk proteins, casein micelles and fat globules | |
|---|---|---|---|---|
| Drug delivery | 17, 97–99 | 103–106 | 82, 141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 142 142 |
146 |
| Vaccines | 149–152 | 43, 153–156 | 157–158 | — |
| Regenerative therapeutics | 159 | 162 | 171, 172 | — |
| Multiplexed biomarkers | — | 7, 185, 186 | 209–212 | — |
One of the first examples of clinically approved NPs for cancer treatment is represented by Abraxane that consists of albumin bound to paclitaxel through hydrophobic chemical interactions. However, after systemic administration albumin and paclitaxel dissociate after a short circulation time entailing limited therapeutic effects.
In order to overcome these limitations, endogenous albumin has been engineered by using active targeting strategies and re-arranging it into nanosized macromolecular complexes with enhanced stability and targeting abilities. In particular, albumin-based macromolecular complexes used to treat tumor-bearing mice resulted in an enhanced antitumor effect, and limited off-target delivery to secondary organs.126
Globular nanosized proteins were also exploited as biogenic vehicles for therapeutic molecules, such as doxorubicin, that if systemically administered cause multiple long-term side effects. Apoferritin-based NPs functionalized with antibodies against prostate specific membrane antigens and loaded with doxorubicin (APOFe-Dox-α) have been applied as drug delivery vehicles for prostatic cancer treatment.127 Notably, APOFe-Dox-α active targeting mediated tumor specific drug delivery both in vitro and in vivo in mice bearing subcutaneous xenografts of human prostatic cancer cells.127 Specifically, APOFe-Dox-α targeted and accumulated in subcutaneous prostatic tumors showing a negligible liver, kidney and heart uptake, thus reducing Dox-mediated toxicity to secondary organs.127
Ago proteins are largely known to bind and mediate the intercellular transfer of hydrophobic and poorly stable non-coding genetic materials, specifically silencing RNAs (siRNAs) and miRNAs.125
siRNAs regulate cell metabolism through gene silencing and recently emerged as a novel therapeutic strategy for a broad range of life-threatening pathologies. However, siRNA application is severely hampered by their poor stability. Interestingly, Ago2 and siRNA complexes coated with cationic polymers resulted in stable platforms able to perform gene silencing both in vitro and in vivo.128 Cationic polymers not only improved the Ago2-siRNA complex stability but also mediated an efficient endo-lysosomal escape that resulted in enhanced gene silencing. Accordingly, Ago2-siRNA NPs coated with polyamines caused significant tumor growth inhibition in melanoma mouse models increasing their survival rate.128
One of the first pioneering papers proposing EVs as vehicles for therapeutic siRNAs was published in 2011 by Alvarez Erviti et al. They produced and isolated engineered EVs exposing a brain-targeting fusion peptide on their surface and successively loaded with a siRNA targeting a gene involved in Alzheimer's disease pathogenesis.138 Notably, engineered and loaded EVs intravenously injected into mouse models were able to efficiently silence target gene expression in different brain areas, proving their efficacy as therapeutic vehicles.138
Compared to other secNPs, EVs are delimited by a functionalized and decorated cellular membrane which confers specific EV targeting abilities both in vitro and in vivo. For example, engineered ferritin nanocages and EVs resulted in different tumor targeting efficiencies mainly due to structural differences. Ferritin NPs and EVs displaying tumor targeting ligands on their surface were able to bind to cancer cells avoiding their interaction with phagocytes and consequently blocking the harmful crosstalk between tumor and immune cells. However, if compared with ferritin NPs, EVs resulted in an improved binding to cultured cancer cells inducing an efficient immune-mediated antitumor response in vitro.139 Furthermore, contrary to ferritin NPs, engineered EVs repeatedly administered in vivo efficiently impaired the growth of subcutaneous tumors in both immunocompetent and immunocompromised mice.139
Lately, EVs isolated from mouse blood and loaded with dopamine (dopEVs) have been studied as a treatment for Parkinson's disease.140 Interestingly, in this work no pre- or post-isolation engineering strategy was applied to EVs. In fact, transferrin – an abundant blood protein often associated with the structure of blood EVs – directly mediated EV passage across the BBB.140 DopEVs associated with transferrin molecules were used as a treatment in murine models of Parkinson's disease, resulting in the delivery of a large quantity of dop into the mouse brain, striatum, and substantia nigra and in an overall pathological phenotype improvement.140
OMVs work as natural nanocarriers for highly immunogenic antigens, such as membrane-associated proteins and proteinaceous virulence factors, that induce systemic immune-protection against pathogens43 (see section 4.2). OMVs also offer an effective support structure for enzyme immobilization or transportation. In particular, OMV enzyme immobilization is naturally modulated by bacterial metabolism, outperforming in quality and efficiency conventional artificial methods based on resins.43
Recently, chitosan NPs loaded with Dox and functionalized with LDLs encasing therapeutic siRNA have been applied for the treatment of multidrug resistant liver cancer. LDL-based chitosan NPs successfully combined the stability of chitosan NPs and the targeting ability of LDLs exhibiting dose-dependent cytotoxicity towards a Dox resistant liver cancer cell line and effective gene silencing.143 Furthermore, LDL-based chitosan NPs efficiently escaped immune system clearance and accumulated in subcutaneous hepatic tumors, avoiding the healthy hepatic tissue, and reducing multi-drug resistant hepatic cancer growth.143
HDLs are actively taken up by cells through scavenger receptor class B type 1 (SR-BI) proteins which are typically overexpressed in hepatocytes and some types of cancer cells. Due to their physical properties and functional role (see section 2.2 and Fig. 3), native HDLs or rHDLs have been conceived as carriers of small molecular therapeutics and siRNAs for the treatment and prevention of atherosclerosis and tumors. rHDLs loaded with siRNAs targeting the Pokemon proto-oncogene (siRNA-HDLs) resulted in stable complexes applicable as a treatment for human hepatocellular carcinoma.144 Notably, siRNA-HDLs efficiently targeted cancer cells resulting in negligible cytotoxicity in vitro, and a time-dependent antitumor response in vivo achieved through Pokemon gene transcription inhibition.144
Considering natural plasmatic HDLs, they can be isolated and engineered with ligands mediating both active targeting and transport. For example, engineered HDLs loaded with chemotherapeutics and exposing on their surface a transferrin receptor ligand and a glioma-homing peptide were able to overcome the BBB and target glioma tumors.145 In particular, HDLs engineered with the transferrin receptor ligand were able to overcome the BBB leading to the highest animal survival time compared to HDLs engineered with the glioma targeting peptide alone, bare HDLs, free drug, and saline.
In conclusion, both natural and reconstituted lipoproteins alone or coupled with other NPs are suitable as stable carriers for small molecules, which include hydrophobic chemotherapeutics and siRNAs, resulting in targeted intratumoral delivery.144 To the best of our knowledge, the application of chylomicrons and VLDLs as drug-delivery vehicles has been overlooked.
To the best of our knowledge, the application of milk fat globules as drug delivery vehicles has so far been overlooked.
4.2 Vaccines
Vaccines represent one of the greatest successes of modern medicine for reducing infectious diseases and are estimated to save millions of lives globally each year. Nevertheless, many diseases are not yet preventable by vaccination and some current vaccines could be improved for high efficacy and safety. This large unmet medical need demands further research which has already succeeded in developing modern vaccines. Compared to the 19th and early 20th century vaccines that were made of killed, inactivated, or live-attenuated pathogens, modern vaccines contain isolated, highly purified antigenic protein subunits which are safer but tend to induce lower levels of immunization.An effective way to address these limitations has gradually emerged through studies of structural vaccinology and nanoparticle research by developing antigen nanoparticles, which can mediate multi-copy antigen display, thereby mimicking the repetitive surface architecture of a natural microbe.148 Please refer to Table 2 to find examples of the scientific literature discussing the application of the different subclasses of secNPs for immunostimulatory purposes.
Among these, albumin (used for years as a stabilizer in different vaccine formulations) has been recently employed in the development of novel vaccine nanocomplexes. By conjugating specific antigens with Evans blue (EB) into albumin-binding vaccines (AlbiVax), Zhu et al. developed clinically promising albumin/AlbiVax nanocomplexes that self-assemble in vivo from AlbiVax and endogenous albumin for efficient vaccine delivery and potent cancer immunotherapy.149
Ferritin self-assembles into nanoparticles with robust thermal and chemical stability that are potentially suited to carry and expose immunogens. As an example, a ferritin nanocage-based influenza vaccine has been developed in an eukaryotic cell and could elicit broad neutralizing antibodies against a variety of influenza viruses.150 A ferritin-based peptide epitope display system has also been successfully developed in prokaryotic cells achieving a 100% passive protection rate after immunization.151 Ferritin nanoparticles have finally been applied in personalized tumor immunotherapy by building a SpyTag/SpyCatcher-enabled click vaccine platform.152
The major limitations of the applications of OMVs in vivo concern their safety and post-administration stability. Therefore, ongoing investigations are aimed to determine their compositions and alter their contents to improve these issues.43,154
Apart from OMVs, EVs from other pathogens such as helminths can potentially elicit an immune response due to the high immunogenicity of EV proteins. Different research approaches are gathering information about the composition of helminth EVs for their future exploitation as vaccines.155
Finally, also EVs isolated from animals previously infected with viruses (e.g. porcine respiratory and reproductive virus) containing viral proteins but free of virus particles can be recognized by the immune system and represent an alternative vaccination strategy.156
Finally, LDLs conjugated with fluorescent ovalbumin (OVA) and lactobionic acid resulted in a substantially increased uptake of OVA by murine macrophage-like ANA1 cells, and by primary peritoneal macrophages due to uptake by a G-protein coupled receptor.230 This antigen delivery system could be used as an alternative way of delivering drugs or vaccines directly to macrophages.
4.3. Regenerative therapeutics
Regenerative medicine takes advantage of the capability of stem cells to induce repair pathways in damaged tissues and diseased organs. Since the advent of the first cell-based regenerative techniques, significant progress has been made and several therapies have begun to enter the clinic. Many stem cell-secreted secNPs exhibit restorative potential comparable to parental cells, laying the groundwork for cell-free therapeutics. This approach presents considerably lower risks in regard to their cell-based counterparts (e.g. neoplastic transformation, poor immune compatibility, and difficulties in handling and storage).Albumin possesses features that fall outside its transport duties. Findings suggest that albumin plays an active role in bone proliferation, although a precise mechanism has not been identified. Therefore, several studies examined albumin employment in regenerative medicine, especially in bone reconstruction. Albumin coating enhances mesenchymal stem cell adhesion, proliferation and long-term survival in bone allografts, which have been successfully used in human bone reconstruction.160 Bone allografts represent a good alternative to autologous bone, which has excellent regenerative properties but suffers from drawbacks, such as scarce availability and donor site morbidity. On the other hand, standard allografts present less osteogenic and osteoinductive properties, due to destruction of bone cells and denaturation of proteins occurring during scaffold preparation. Functionalization with albumin led to the creation of more performant allografts; for instance, albumin-doped bone allografts led to faster and better outcomes in patients who had undergone anterior cruciate ligament reconstruction.160 The same research group functionalized suture materials with albumin, creating bio-active sutures for surgical stem cell transplantation.161 In this case, albumin was freeze-dried on the surface of a polyfilament absorbable thread, which was subsequently seeded with rat bone marrow mesenchymal stem cells and used to suture a wound located in the gluteal and tricepts surae muscles of the rats. Suture-transplanted stem cells maintained their vitality after surgery and readily migrated in the damaged tissue, supporting the regenerative process.
MSC-EVs exert their action through the delivery of encapsulated bioactive molecules (including enzymes and various non-coding RNAs) to the cells of the tissue microenvironment, modulating many signalling pathways. Indeed, the MSC-EV cargo is functionally complex and comprises molecules involved in the regulation of metabolism, angiogenesis, inflammation and others. As a direct result, MSC-EVs have been successfully employed in cell-free treatment of many injury models. For instance, EVs produced by hypoxic MSCs ameliorated the conditions of APP/PS1 mice, an Alzheimer's disease model.163 After systemic administration of EVs, mice showed reduced levels of β-amyloid and deposition of Aβ plaques in the brain, and reduced activation of pro-inflammatory pathways of STAT3 and NF-κB. Moreover, mice exhibited lower expression of pro-inflammatory cytokines IL-1β and TNF-α and higher expression of anti-inflammatory molecules, such as IL-4 and IL-10, in the brain tissue. An overall improvement of cognition and memory was also observed. The beneficial effects were ascribable to the raising of brain levels of miR-21, a small non-coding nucleic acid known to be directly involved in the downregulation of inflammation processes. Similar results were obtained in peripheral nerve injury animal models. Ma et al.164 observed motor function restoration in rats, after sciatic nerve transection. Systemic injection of EVs produced by human umbilical cord MSCs (hUCMSC-EVs) promoted extensive Schwann cell growth, axonal regeneration and myelination, and consequently reduced muscular atrophy. Interestingly, near the site of injury the aggregation of hUCMSC-EVs was observed, suggesting that hUCMSC-EVs promoted nerve regeneration establishing a favourable environment for tissue reconstruction. In particular, hUCMSC-EVs locally lowered the expression of pro-inflammatory molecules, in a similar way also observed by Cui et al.163 Many other organs were treated with MSC-EVs (e.g. skin, cartilage, liver and lung), highlighting their broad spectrum of healing modalities. A few ongoing early clinical trials are testing the regenerative potential of MSC-EVs in patients with acute ischemic stroke (NCT03384433), intractable cutaneous ulcers (NCT02565264) and large eye macular holes (NCT03437759).
Notably, MSCs represent the most common but not the exclusive source of staminal EVs. Indeed, even induced pluripotent stem cells165 and cardiac,166 amniotic fluid,167 and embryonic168 stem cells release EVs. It is likely that other stem cell EVs operate on the injured tissue in the very same way MSC-EVs do. Although the use of natural, unmodified EVs is prominent in regenerative medicine, a few examples of EV engineering of stem cells and non-stem cells are reported in the literature. Such approaches are mainly intended to enhance natural EV healing properties by loading EVs with endogenous molecules, or by augmenting EV targeting capabilities by means of surface modifications. For instance, Vandergriff et al. decorated cardiac-stem cell derived EVs using a cardiac homing peptide and tested them in a mice model of heart ischemic/reperfusion injury. Such modification allows one to circumvent dangerous EV intracardiac injections in favour of more safe EV intravenous administration with a minimal loss of treatment efficacy and negligible off-target effects. In vitro, ex vivo and in vivo experiments were performed, pointing out that engineered cardiac-stem cells derived EVs truly possess infarct-targeting features and regenerative potential: indeed, the hearts of the treated mice showed lowered cardiac fibrosis and enhanced cell proliferation and angiogenesis when compared to the control animals. Finally, it is worth noting that EV engineering specifically intended for regenerative purposes has been to date far less represented than other topics (e.g. cancer and neurodegenerative disorders), suggesting that this field is still in its infancy.
HDLs reduce palmitate-induced apoptosis in cardiomyocytes.173 Palmitate cause lipotoxicity via downregulation of AMP-activated protein kinase (AMPK). HDL treatment of palmitate-fed cardiomyocytes resulted in the improvement of several pro-apoptotic parameters (e.g. Annexin-V levels and Bcl2/Bax ratio), and in enhanced expression of genes involved in cell contractility and fatty acid oxidation. Interestingly, such genes are under the control of AMPK-dependent pathways.173 Thus, HDLs are effective contrasting lipoapoptosis caused by the accumulation of free fatty acids in cells or organs (steatosis). Steatosis of non-adipose tissues is typical in patients affected by metabolic syndrome and obesity. HDLs could be implemented as a treatment for such conditions, besides traditional approaches (e.g. physical exercise, anti-steatotic drugs, and caloric restriction). However, in clinics, the application of natural HDLs is clearly limited. Synthetic rHDLs (see section 4.1.3 for further details) are used instead.174 rHDLs have been tested in vitro and in vivo, and their benefits resemble almost perfectly those of natural HDLs.175–177 For instance, in ref. 176, C57BL/6N mice were fed with a coconut oil (CC) diet for 6 months, in order to cause heart dysfunction. High fat feeding induced several cardiac impairments, such as ventricular hypertrophy, increased interstitial fibrosis, decreased stroke volume and lower capillary density, compared to the control mice (which were fed with a standard diet). Strikingly, periodic intraperitoneal administration of rHDLs (8 infusions, with an interval of 48 h) after a CC diet reversed pathological heart remodelling that occurred in mice and restored cardiac function to normalcy. Moreover, the treatment had no side effects on healthy control mice. It is worth noting that the rHDL formulation used in this study was based on the so-called “Milano” isoform of Apo-A1, a mutated variant of Apo-A which presents enhanced activity in regard to the wild type form. Current clinical trials are testing Milano Apo-A1 efficacy in the treatment of cardiovascular diseases (e.g. NCT02678923).
4.4. Diagnostics
Biomarkers in both basic, and clinical research and clinical practice are to date necessary to monitor the physiology and disease onset. Indeed, parameters with high predictive and prognostic values are mandatory to define therapy effectiveness and clinical outcomes. The nanostructured secretome represents an immense, easily accessible reservoir of such information. SecNPs are inherently multiplexed and very sensible to changes occurring during disease onset and evolution. Some of the secNPs treated in the present section are long-established biomarkers, and their use is well documented. Other categories of secNPs are not yet implemented in clinics but conceal great potential.EV molecular content is fostered as a diagnostic and prognostic tool also for non-cancer diseases, such as auto-immune187 and cardiovascular diseases (CVD).196 In the latter field, efforts have been made to relate the levels of subpopulations of EVs positive to specific proteins or miRNAs with the most diffused CVDs, e.g. heart failure (e.g. CD144+ or mir-192+ EVs),197,198 coronary artery disease (e.g. CD3+/CD45+ or α-SMA+ EVs),199 and stroke (e.g. mir-9+ or mir-124+ positive EVs).200
The second source of the information of EVs, namely their colloidal properties, has been far less investigated than their compositional counterparts, although it proved to be significant. For instance, discrepancies in particle concentration between pathological versus healthy samples have been registered in multiple myeloma,201 bladder,202 pancreatic,203 and breast cancer studies.204 Particle size is another biophysical factor influencing the biological functions and uptake kinetics of EVs; differences in such parameters are reported in ovarian205 and prostate cancer,206 but also in pre-eclampsia studies.207 The nanomechanical properties of EVs (e.g. membrane stiffness) could have a similar importance for EV–cell interactions, but they have been less investigated compared to other potential physical biomarkers. So far, only one study has examined EV nanomechanics in relation to pathology.208 In this seminal work, the authors compared the biophysical properties of two bladder cancer cell lines and one non-malignant, immortalized cell line. All the three types of EVs exhibited similar sizes and concentrations but marked differences in membrane stiffness as observed through Quantitative Nano-Mapping Atomic Force Microscopy (QNM-AFM). Such a difference was also associated with the ability of malignant EVs to induce local complement activation and augment endothelium permeability and leakiness, facilitating EV trans-endothelial transport.
However, integrated data from nuclear magnetic resonance lipoprotein profile, metabolomics and soluble marker levels were found to be predictive of immunological recovery of HIV patients after anti-retroviral therapy,213 while lipoprotein infrared (IR) spectroscopy has been used for the detection of alcohol abuse.214 Indeed, chronic alcohol consumption causes liver to synthesize Fatty Acid Ethyl Esters (FAEE) from lipids and ethanol. FAEE are primarily bound to albumin and lipoproteins and can be easily detected using IR spectroscopy due to the presence of specific ester C–O–C bonds, which are normally absent in lipids. Moreover, differences in the IR spectrograms of alcoholic people specifically correlate with two other parameters used as alcohol biomarkers: AST/ALT enzymes and Carbohydrate-Deficient Transferrin (CDT) levels. Therefore, the authors claim that IR could represent a potential, non-destructive and costless tool for the detection of alcohol biomarkers, although several obstacles need to be overcome (e.g. short life of non-HDL compared to CDT or enzymes and dysregulation in lipoprotein levels due to drugs or diseases). Lipoproteins also contribute to the biomarker set with the nucleic acids they transport. HDLs carry significant amounts of miRNAs,215 which are envisioned as potential markers for myocardial infarction, stable and unstable angina pectoris,216 hypercholesterolemia and atherosclerosis.217 This is supported by several in vitro studies highlighting the role of HDL-miRNAs in inflammation and atherosclerosis regulation.218,219 miRNA transport by lipoproteins other than HDLs is little investigated, but clues to LDL involvement exist.217
5. Towards nanoparticles 2.0 and biogenic nanotechnology
Nature has been exploiting NPs much longer than we have in our laboratories. The secretome (intended in its wider meaning) carries a variety of secNPs. These nanoparticles, either native/bare or modified, promise to be those “nanoparticles 2.0” able to bring much needed physicochemical heterogeneity and intrinsic precision to nanomedicine, while opening exciting new perspectives.Researchers around the world are beginning to tackle this truly cross-disciplinary field, but the way is still long and scattered with cultural and technical obstacles. Comparative analysis and study of secNPs are very infrequent; the majority of the studies are focused on a specific biological and/or medical aspect of a given secNP class or subclass. In order to consolidate and harness a unified perspective, which also includes compelling physics, chemistry and materials science, will be transformative. This review intends to provide a tool in this endeavor, by gathering and elaborating secNPs and their traits into a unique and concise framework.
A major challenge to secNP translation is posed by the need for sustainable (in ethical, economic, scale and environmental terms) methods for the production of high-grade secNP formulations (see, for example, ref. 5, 220 and 221). Noteworthily, these objectives are actively pursued by two ongoing European projects (http://www.evfoundry.eu and https://ves4us.eu/project). To be brought to the society, sustainable production must be also standardized. The wider scientific community dealing with bionanomaterials is starting to vigorously debate and work on the topic. Recently, as a result of important collective efforts, general guidelines for minimal information reporting in the bio-nano experimental literature222 and specific guidelines for minimal requirements for EV studies40 have been proposed. Sustainable standardized protocols and technology will be foundational towards the large-scale production of secNP formulations which comply with good manufacturing practice guidelines (EU GMP guidelines available at: https://ec.europa.eu/health/documents/eudralex/vol-4_en) and eventually clinical-grade regulations. Standardization and regulatory initiatives will also open up large-scale clinical studies aimed at the validation of secNP formulations.
On the opposite side, scientists are trying to learn about the nature of single cells, as both physiological and pathological processes rely on the fine interplay between different kinds of cells (https://www.nature.com/news/single-cell-biology-1.22241). Current single-cell experiments are focused on genomics, whereas soluble and nanostructured secretome remains overlooked. Adding secNPs to single-cell analyzed components could substantially improve the current picture of both physiological and pathological processes. This seems a feasible objective, as recently demonstrated by Li et al., who by integrating an optofluidic biosensor and a single cell bioreactor could successfully detect in real time the spatial fingerprints of the angiogenetic growth factors secreted by a single cancer cell.223
From a wider alternative perspective, secNPs can be used as naturally pre-assembled building blocks for future “biogenic” surface- and nanotechnology, such as the realization of heterogeneous systems or the synthesis of living cells from scratch.224 For example, the first shall see the combination of synthetic surfaces or nanosystems with secNPs in order to upgrade artificial nanotechnology with the natural features of secNPs;225–228 the latter will see the enrichment with secNPs of the current toolbox used for the bottom-up assembly of synthetic protocells.229
Author contributions
Conceptualization, P.B. and S.B.; writing – original draft preparation, all the authors; writing – review and editing, all the authors; visualization, A.Z.; supervision, P.B.; funding acquisition, P.B. and M.P.; please turn to the CRediT taxonomy for the term explanation.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
This work was supported by the Università degli Studi di Brescia and the Center for Colloid and Surface Science (CSGI) through the evFOUNDRY project, Horizon 2020 – Future and emerging technologies (H2020-FETOPEN), ID: 801367, and the Associazione Italiana per la Ricerca sul Cancro (AIRC) through IG grant ID: 18493. P. B. and M. P. also acknowledge the support by National Inter-university Consortium of Materials Science and Technology (INSTM).References
- Y. Zhang and B. P. Orner, Int. J. Mol. Sci., 2011, 12, 5406–5421 CrossRef CAS PubMed.
- K. Li, R. S. Rodosthenous, F. Kashanchi, T. Gingeras, S. J. Gould, L. S. Kuo, P. Kurre, H. Lee, J. N. Leonard, H. Liu, T. B. Lombo, S. Momma, J. P. Nolan, M. J. Ochocinska, D. M. Pegtel, Y. Sadovsky, F. Sanchez-Madrid, K. M. Valdes, K. C. Vickers, A. M. Weaver, K. W. Witwer, Y. Zeng, S. Das, R. L. Raffai and T. K. Howcroft, JCI Insight, 2018, 3(7), e98942 CrossRef PubMed.
- A. V. Vlassov, S. Magdaleno, R. Setterquist and R. Conrad, Biochim. Biophys. Acta, 2012, 1820, 940–948 CrossRef CAS PubMed.
- H. Singh and S. Gallier, Food Hydrocolloids, 2017, 68, 81–89 CrossRef CAS.
- S. Raut, J. L. Dasseux, N. A. Sabnis, L. Mooberry and A. Lacko, Ther. Delivery, 2018, 9, 257–268 CrossRef CAS PubMed.
- F. Rehan, N. Ahemad and M. Gupta, Colloids Surf., B, 2019, 179, 280–292 CrossRef CAS PubMed.
- L. Paolini, A. Zendrini and A. Radeghieri, Biomarkers Med., 2018, 12, 383–391 CrossRef CAS PubMed.
- S. L. N. Maas, X. O. Breakefield and A. M. Weaver, Trends Cell Biol., 2017, 27, 172–188 CrossRef CAS PubMed.
- R. Vescovi, M. Monti, D. Moratto, L. Paolini, F. Consoli, L. Benerini, L. Melocchi, S. Calza, M. Chiudinelli, G. Rossi, M. Bugatti, M. Maio, E. Fonsatti, C. Farisoglio, M. Simbolo, C. Almici, R. Verardi, A. Scarpa, P. Bergese, A. Manganoni, F. Facchetti and W. Vermi, Cancer Immunol. Res., 2019, 7, 12–28 CrossRef PubMed.
- E. N. Nolte- 't Hoen, T. Cremer, R. C. Gallo and L. B. Margolis, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 9155 CrossRef PubMed.
- F. Fernandez-Trillo, L. M. Grover, A. Stephenson-Brown, P. Harrison and P. M. Mendes, Angew. Chem., Int. Ed., 2017, 56, 3142–3160 CrossRef CAS PubMed.
- A. R. Chandrasekaran and O. Levchenko, Chem. Mater., 2016, 28, 5569–5581 CrossRef CAS.
- K. W. Lexa, E. Dolghih and M. P. Jacobson, PLoS One, 2014, 9, e93323 CrossRef PubMed.
- X. M. He and D. C. Carter, Nature, 1992, 358, 209–215 CrossRef CAS PubMed.
- S. Sugio, A. Kashima, S. Mochizuki, M. Noda and K. Kobayashi, Protein Eng., 1999, 12, 439–446 CrossRef CAS PubMed.
- G. J. Quinlan, G. S. Martin and T. W. Evans, Hepatology, 2005, 41, 1211–1219 CrossRef CAS PubMed.
- B. Chiou and J. R. Connor, Pharmaceuticals, 2018, 11(4), 124 CrossRef CAS PubMed.
- O. D. Mrowczynski, A. B. Madhankumar, B. Slagle-Webb, S. Y. Lee, B. E. Zacharia and J. R. Connor, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 1921–1928 CrossRef CAS PubMed.
- M. Truman-Rosentsvit, D. Berenbaum, L. Spektor, L. A. Cohen, S. Belizowsky-Moshe, L. Lifshitz, J. Ma, W. Li, E. Kesselman, I. Abutbul-Ionita, D. Danino, L. Gutierrez, H. Li, K. Li, H. Lou, M. Regoni, M. Poli, F. Glaser, T. A. Rouault and E. G. Meyron-Holtz, Blood, 2018, 131, 342–352 CrossRef CAS PubMed.
- H. M. Baker, B. F. Anderson and E. N. Baker, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 3579–3583 CrossRef CAS PubMed.
- J. D. Arroyo, J. R. Chevillet, E. M. Kroh, I. K. Ruf, C. C. Pritchard, D. F. Gibson, P. S. Mitchell, C. F. Bennett, E. L. Pogosova-Agadjanyan, D. L. Stirewalt, J. F. Tait and M. Tewari, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5003–5008 CrossRef CAS PubMed.
- Z. Ye, H. Jin and Q. Qian, J. Cancer, 2015, 6, 877–882 CrossRef CAS PubMed.
- N. Yang, H. Zhang, M. Wang, Q. Hao and H. Sun, Sci. Rep., 2012, 2, 999 CrossRef PubMed.
- P. D. Hempstead, S. J. Yewdall, A. R. Fernie, D. M. Lawson, P. J. Artymiuk, D. W. Rice, G. C. Ford and P. M. Harrison, J. Mol. Biol., 1997, 268, 424–448 CrossRef CAS PubMed.
- N. T. Schirle, G. A. Kinberger, H. F. Murray, W. F. Lima, T. P. Prakash and I. J. MacRae, J. Am. Chem. Soc., 2016, 138, 8694–8697 CrossRef CAS PubMed.
- B. Jachimska and A. Pajor, Bioelectrochemistry, 2012, 87, 138–146 CrossRef CAS PubMed.
- M. Kabiri, Z. Amiri-Tehranizadeh, A. Baratian, M. R. Saberi and J. Chamani, Molecules, 2012, 17, 3114–3147 CrossRef CAS PubMed.
- I. Moglia, M. Santiago, Á. Olivera-Nappa and M. Soler, J. Inorg. Biochem., 2018, 183, 184–190 CrossRef CAS PubMed.
- H. P. Erickson, Biol. Proced. Online, 2009, 11, 32–51 CrossRef CAS PubMed.
- M. A. Kiselev, I. A. Gryzunov, G. E. Dobretsov and M. N. Komarova, Biofizika, 2001, 46, 423–427 CAS.
- F. Kilar and I. Simon, Biophys. J., 1985, 48, 799–802 CrossRef CAS PubMed.
- M. L. Quillin and B. W. Matthews, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2000, 56, 791–794 CrossRef CAS PubMed.
- H. Fischer, I. Polikarpov and A. F. Craievich, Protein Sci., 2004, 13, 2825–2828 CrossRef CAS PubMed.
- G. van Niel, G. D'Angelo and G. Raposo, Nat. Rev. Mol. Cell Biol., 2018, 19, 213 CrossRef CAS PubMed.
- M. Mathieu, L. Martin-Jaular, G. Lavieu and C. Thery, Nat. Cell Biol., 2019, 21, 9–17 CrossRef CAS PubMed.
- B. L. Deatherage and B. T. Cookson, Infect. Immun., 2012, 80, 1948 CrossRef CAS PubMed.
- M. Yanez-Mo, P. R. Siljander, Z. Andreu, A. B. Zavec, F. E. Borras, E. I. Buzas, K. Buzas, E. Casal, F. Cappello, J. Carvalho, E. Colas, A. Cordeiro-da Silva, S. Fais, J. M. Falcon-Perez, I. M. Ghobrial, B. Giebel, M. Gimona, M. Graner, I. Gursel, M. Gursel, N. H. Heegaard, A. Hendrix, P. Kierulf, K. Kokubun, M. Kosanovic, V. Kralj-Iglic, E. M. Kramer-Albers, S. Laitinen, C. Lasser, T. Lener, E. Ligeti, A. Line, G. Lipps, A. Llorente, J. Lotvall, M. Mancek-Keber, A. Marcilla, M. Mittelbrunn, I. Nazarenko, E. N. Nolte- 't Hoen, T. A. Nyman, L. O'Driscoll, M. Olivan, C. Oliveira, E. Pallinger, H. A. Del Portillo, J. Reventos, M. Rigau, E. Rohde, M. Sammar, F. Sanchez-Madrid, N. Santarem, K. Schallmoser, M. S. Ostenfeld, W. Stoorvogel, R. Stukelj, S. G. Van der Grein, M. H. Vasconcelos, M. H. Wauben and O. De Wever, J. Extracell. Vesicles, 2015, 4, 27066 CrossRef PubMed.
- G. Raposo and W. Stoorvogel, J. Cell Biol., 2013, 200, 373–383 CrossRef CAS PubMed.
- E. Cocucci, G. Racchetti and J. Meldolesi, Trends Cell Biol., 2009, 19, 43–51 CrossRef CAS PubMed.
- C. Thery, K. W. Witwer, E. Aikawa, M. J. Alcaraz, J. D. Anderson, R. Andriantsitohaina, A. Antoniou, T. Arab, F. Archer, G. K. Atkin-Smith, D. C. Ayre, J. M. Bach, D. Bachurski, H. Baharvand, L. Balaj, S. Baldacchino, N. N. Bauer, A. A. Baxter, M. Bebawy, C. Beckham, A. Bedina Zavec, A. Benmoussa, A. C. Berardi, P. Bergese, E. Bielska, C. Blenkiron, S. Bobis-Wozowicz, E. Boilard, W. Boireau, A. Bongiovanni, F. E. Borras, S. Bosch, C. M. Boulanger, X. Breakefield, A. M. Breglio, M. A. Brennan, D. R. Brigstock, A. Brisson, M. L. Broekman, J. F. Bromberg, P. Bryl-Gorecka, S. Buch, A. H. Buck, D. Burger, S. Busatto, D. Buschmann, B. Bussolati, E. I. Buzas, J. B. Byrd, G. Camussi, D. R. Carter, S. Caruso, L. W. Chamley, Y. T. Chang, C. Chen, S. Chen, L. Cheng, A. R. Chin, A. Clayton, S. P. Clerici, A. Cocks, E. Cocucci, R. J. Coffey, A. Cordeiro-da-Silva, Y. Couch, F. A. Coumans, B. Coyle, R. Crescitelli, M. F. Criado, C. D'Souza-Schorey, S. Das, A. Datta Chaudhuri, P. de Candia, E. F. De Santana, O. De Wever, H. A. Del Portillo, T. Demaret, S. Deville, A. Devitt, B. Dhondt, D. Di Vizio, L. C. Dieterich, V. Dolo, A. P. Dominguez Rubio, M. Dominici, M. R. Dourado, T. A. Driedonks, F. V. Duarte, H. M. Duncan, R. M. Eichenberger, K. Ekstrom, S. El-Andaloussi, C. Elie-Caille, U. Erdbrugger, J. M. Falcon-Perez, F. Fatima, J. E. Fish, M. Flores-Bellver, A. Forsonits, A. Frelet-Barrand, F. Fricke, G. Fuhrmann, S. Gabrielsson, A. Gamez-Valero, C. Gardiner, K. Gartner, R. Gaudin, Y. S. Gho, B. Giebel, C. Gilbert, M. Gimona, I. Giusti, D. C. Goberdhan, A. Gorgens, S. M. Gorski, D. W. Greening, J. C. Gross, A. Gualerzi, G. N. Gupta, D. Gustafson, A. Handberg, R. A. Haraszti, P. Harrison, H. Hegyesi, A. Hendrix, A. F. Hill, F. H. Hochberg, K. F. Hoffmann, B. Holder, H. Holthofer, B. Hosseinkhani, G. Hu, Y. Huang, V. Huber, S. Hunt, A. G. Ibrahim, T. Ikezu, J. M. Inal, M. Isin, A. Ivanova, H. K. Jackson, S. Jacobsen, S. M. Jay, M. Jayachandran, G. Jenster, L. Jiang, S. M. Johnson, J. C. Jones, A. Jong, T. Jovanovic-Talisman, S. Jung, R. Kalluri, S. I. Kano, S. Kaur, Y. Kawamura, E. T. Keller, D. Khamari, E. Khomyakova, A. Khvorova, P. Kierulf, K. P. Kim, T. Kislinger, M. Klingeborn, D. J. Klinke 2nd, M. Kornek, M. M. Kosanovic, A. F. Kovacs, E. M. Kramer-Albers, S. Krasemann, M. Krause, I. V. Kurochkin, G. D. Kusuma, S. Kuypers, S. Laitinen, S. M. Langevin, L. R. Languino, J. Lannigan, C. Lasser, L. C. Laurent, G. Lavieu, E. Lazaro-Ibanez, S. Le Lay, M. S. Lee, Y. X. F. Lee, D. S. Lemos, M. Lenassi, A. Leszczynska, I. T. Li, K. Liao, S. F. Libregts, E. Ligeti, R. Lim, S. K. Lim, A. Line, K. Linnemannstons, A. Llorente, C. A. Lombard, M. J. Lorenowicz, A. M. Lorincz, J. Lotvall, J. Lovett, M. C. Lowry, X. Loyer, Q. Lu, B. Lukomska, T. R. Lunavat, S. L. Maas, H. Malhi, A. Marcilla, J. Mariani, J. Mariscal, E. S. Martens-Uzunova, L. Martin-Jaular, M. C. Martinez, V. R. Martins, M. Mathieu, S. Mathivanan, M. Maugeri, L. K. McGinnis, M. J. McVey, D. G. Meckes Jr., K. L. Meehan, I. Mertens, V. R. Minciacchi, A. Moller, M. Møller Jørgensen, A. Morales-Kastresana, J. Morhayim, F. Mullier, M. Muraca, L. Musante, V. Mussack, D. C. Muth, K. H. Myburgh, T. Najrana, M. Nawaz, I. Nazarenko, P. Nejsum, C. Neri, T. Neri, R. Nieuwland, L. Nimrichter, J. P. Nolan, E. N. Nolte- 't Hoen, N. Noren Hooten, L. O'Driscoll, T. O'Grady, A. O'Loghlen, T. Ochiya, M. Olivier, A. Ortiz, L. A. Ortiz, X. Osteikoetxea, O. Ostergaard, M. Ostrowski, J. Park, D. M. Pegtel, H. Peinado, F. Perut, M. W. Pfaffl, D. G. Phinney, B. C. Pieters, R. C. Pink, D. S. Pisetsky, E. Pogge von Strandmann, I. Polakovicova, I. K. Poon, B. H. Powell, I. Prada, L. Pulliam, P. Quesenberry, A. Radeghieri, R. L. Raffai, S. Raimondo, J. Rak, M. I. Ramirez, G. Raposo, M. S. Rayyan, N. Regev-Rudzki, F. L. Ricklefs, P. D. Robbins, D. D. Roberts, S. C. Rodrigues, E. Rohde, S. Rome, K. M. Rouschop, A. Rughetti, A. E. Russell, P. Saa, S. Sahoo, E. Salas-Huenuleo, C. Sanchez, J. A. Saugstad, M. J. Saul, R. M. Schiffelers, R. Schneider, T. H. Schoyen, A. Scott, E. Shahaj, S. Sharma, O. Shatnyeva, F. Shekari, G. V. Shelke, A. K. Shetty, K. Shiba, P. R. Siljander, A. M. Silva, A. Skowronek, O. L. Snyder 2nd, R. P. Soares, B. W. Sodar, C. Soekmadji, J. Sotillo, P. D. Stahl, W. Stoorvogel, S. L. Stott, E. F. Strasser, S. Swift, H. Tahara, M. Tewari, K. Timms, S. Tiwari, R. Tixeira, M. Tkach, W. S. Toh, R. Tomasini, A. C. Torrecilhas, J. P. Tosar, V. Toxavidis, L. Urbanelli, P. Vader, B. W. van Balkom, S. G. van der Grein, J. Van Deun, M. J. van Herwijnen, K. Van Keuren-Jensen, G. van Niel, M. E. van Royen, A. J. van Wijnen, M. H. Vasconcelos, I. J. Vechetti Jr., T. D. Veit, L. J. Vella, E. Velot, F. J. Verweij, B. Vestad, J. L. Vinas, T. Visnovitz, K. V. Vukman, J. Wahlgren, D. C. Watson, M. H. Wauben, A. Weaver, J. P. Webber, V. Weber, A. M. Wehman, D. J. Weiss, J. A. Welsh, S. Wendt, A. M. Wheelock, Z. Wiener, L. Witte, J. Wolfram, A. Xagorari, P. Xander, J. Xu, X. Yan, M. Yanez-Mo, H. Yin, Y. Yuana, V. Zappulli, J. Zarubova, V. Zekas, J. Y. Zhang, Z. Zhao, L. Zheng, A. R. Zheutlin, A. M. Zickler, P. Zimmermann, A. M. Zivkovic, D. Zocco and E. K. Zuba-Surma, J. Extracell. Vesicles, 2018, 7, 1535750 CrossRef PubMed.
- R. Xu, A. Rai, M. Chen, W. Suwakulsiri, D. W. Greening and R. J. Simpson, Nat. Rev. Clin. Oncol., 2018, 15, 617–638 CrossRef CAS PubMed.
- Y. Seo, H. S. Kim and I. S. Hong, Stem Cells Int., 2019, 2019, 5126156 Search PubMed.
- K. Watanabe, Appl. Microbiol. Biotechnol., 2016, 100, 9837–9843 CrossRef CAS PubMed.
- N. Karimi, A. Cvjetkovic, S. C. Jang, R. Crescitelli, M. A. Hosseinpour-Feizi, R. Nieuwland, J. Lötvall and C. Lässer, Cell. Mol. Life Sci., 2018, 75, 2873–2886 CrossRef CAS PubMed.
- T. Soares Martins, J. Catita, I. Martins Rosa, O. A. B. da Cruz e Silva and A. G. Henriques, PLoS One, 2018, 13, e0198820 CrossRef PubMed.
- C. Lässer, S. C. Jang and J. Lötvall, Mol. Aspects Med., 2018, 60, 1–14 CrossRef PubMed.
- T. Skotland, K. Sandvig and A. Llorente, Prog. Lipid Res., 2017, 66, 30–41 CrossRef CAS PubMed.
- I. Ramasamy, Clin. Chem. Lab. Med., 2014, 52, 1695–1727 CAS.
- K. R. Feingold and C. Grunfeld, in Endotext, ed. K. R. Feingold, B. Anawalt, A. Boyce, G. Chrousos, K. Dungan, A. Grossman, J. M. Hershman, G. Kaltsas, C. Koch, P. Kopp, M. Korbonits, R. McLachlan, J. E. Morley, M. New, L. Perreault, J. Purnell, R. Rebar, F. Singer, D. L. Trence, A. Vinik and D. P. Wilson, South Dartmouth (MA), 2000 Search PubMed.
- H. Ishikawa, H. Yamada, N. Taromaru, K. Kondo, A. Nagura, M. Yamazaki, Y. Ando, E. Munetsuna, K. Suzuki, K. Ohashi and R. Teradaira, Ann. Clin. Biochem., 2017, 54, 134–142 CrossRef CAS PubMed.
- N. Auge, F. Maupas-Schwalm, M. Elbaz, J. C. Thiers, A. Waysbort, S. Itohara, H. W. Krell, R. Salvayre and A. Negre-Salvayre, Circulation, 2004, 110, 571–578 CrossRef CAS PubMed.
- T. Murtola, T. A. Vuorela, M. T. Hyvönen, S.-J. Marrink, M. Karttunen and I. Vattulainen, Soft Matter, 2011, 7, 8135–8141 RSC.
- M. Rodriguez, M. Guardiola, I. Oliva, J. Carles Vallvè, R. Ferre, L. Masana, S. Parra, J. Ribalta and A. Castro, Int. J. Rheum. Dis., 2019, 22, 480–487 CrossRef CAS PubMed.
- M. A. Elsheikh, Y. S. R. Elnaggar, D. Y. Otify and O. Y. Abdallah, Pharm. Res., 2018, 35, 18 CrossRef PubMed.
- K. K. Ng, J. F. Lovell and G. Zheng, Acc. Chem. Res., 2011, 44, 1105–1113 CrossRef CAS PubMed.
- H. Zhang, D. Freitas, H. S. Kim, K. Fabijanic, Z. Li, H. Chen, M. T. Mark, H. Molina, A. B. Martin, L. Bojmar, J. Fang, S. Rampersaud, A. Hoshino, I. Matei, C. M. Kenific, M. Nakajima, A. P. Mutvei, P. Sansone, W. Buehring, H. Wang, J. P. Jimenez, L. Cohen-Gould, N. Paknejad, M. Brendel, K. Manova-Todorova, A. Magalhães, J. A. Ferreira, H. Osório, A. M. Silva, A. Massey, J. R. Cubillos-Ruiz, G. Galletti, P. Giannakakou, A. M. Cuervo, J. Blenis, R. Schwartz, M. S. Brady, H. Peinado, J. Bromberg, H. Matsui, C. A. Reis and D. Lyden, Nat. Cell Biol., 2018, 20, 332–343 CrossRef CAS PubMed.
- Q. Zhang, J. N. Higginbotham, D. K. Jeppesen, Y.-P. Yang, W. Li, E. T. McKinley, R. Graves-Deal, J. Ping, C. M. Britain, K. A. Dorsett, C. L. Hartman, D. A. Ford, R. M. Allen, K. C. Vickers, Q. Liu, J. L. Franklin, S. L. Bellis and R. J. Coffey, Cell Rep., 2019, 27, 940–954 CrossRef CAS PubMed.
- N. J. Andreas, B. Kampmann and K. Mehring Le-Doare, Early Hum. Dev., 2015, 91, 629–635 CrossRef CAS PubMed.
- N. Argov, D. G. Lemay and J. B. German, Trends Food Sci. Technol., 2008, 19(12), 617–623 CrossRef CAS PubMed.
- C. Lopez, C. Cauty and F. Guyomarc'h, Eur. J. Lipid Sci. Technol., 2019, 121, 1800201 Search PubMed.
- W. Holzmüller and U. Kulozik, Int. Dairy J., 2016, 61, 51–66 CrossRef.
- O. Ménard, S. Ahmad, F. Rousseau, V. Briard-Bion, F. Gaucheron and C. Lopez, Food Chem., 2010, 120, 544–551 CrossRef.
- M.-C. Michalski, F. Michel, D. Sainmont and V. Briard, Colloids Surf., B, 2002, 23, 23–30 CrossRef CAS.
- M. C. Michalski, V. Briard, F. Michel, F. Tasson and P. Poulain, J. Dairy Sci., 2005, 88, 1927–1940 CrossRef CAS PubMed.
- B. Ingham, G. D. Erlangga, A. Smialowska, N. M. Kirby, C. Wang, L. Matia-Merino, R. G. Haverkamp and A. J. Carr, Soft Matter, 2015, 11, 2723–2725 RSC.
- C. G. de Kruif, T. Huppertz, V. S. Urban and A. V. Petukhov, Adv. Colloid Interface Sci., 2012, 171–172, 36–52 CrossRef CAS PubMed.
- A. Jukkola and O. J. Rojas, Adv. Colloid Interface Sci., 2017, 245, 92–101 CrossRef CAS PubMed.
- Q. Chen, S. Rozovsky and W. Chen, Chem. Commun., 2017, 53, 7569–7572 RSC.
- F. A. W. Coumans, A. R. Brisson, E. I. Buzas, F. Dignat-George, E. E. E. Drees, S. El-Andaloussi, C. Emanueli, A. Gasecka, A. Hendrix, A. F. Hill, R. Lacroix, Y. Lee, T. G. van Leeuwen, N. Mackman, I. Mager, J. P. Nolan, E. van der Pol, D. M. Pegtel, S. Sahoo, P. R. M. Siljander, G. Sturk, O. de Wever and R. Nieuwland, Circ. Res., 2017, 120, 1632–1648 CrossRef CAS PubMed.
- S. Busatto, G. Vilanilam, T. Ticer, W. L. Lin, D. W. Dickson, S. Shapiro, P. Bergese and J. Wolfram, Cells, 2018, 7(12), 273 CrossRef CAS PubMed.
- K. W. Witwer, E. I. Buzas, L. T. Bemis, A. Bora, C. Lasser, J. Lotvall, E. N. Nolte- 't Hoen, M. G. Piper, S. Sivaraman, J. Skog, C. Thery, M. H. Wauben and F. Hochberg, J. Extracell. Vesicles, 2013, 2, 20360 CrossRef PubMed.
- C. Y. Yang, J. L. Raya, H. H. Chen, C. H. Chen, Y. Abe, H. J. Pownall, A. A. Taylor and C. V. Smith, Arterioscler. Thromb. Vasc. Biol., 2003, 23, 1083–1090 CrossRef CAS PubMed.
- C. Gardiner, D. Di Vizio, S. Sahoo, C. Thery, K. W. Witwer, M. Wauben and A. F. Hill, J. Extracell. Vesicles, 2016, 5, 32945 CrossRef PubMed.
- M. A. Livshits, E. Khomyakova, E. G. Evtushenko, V. N. Lazarev, N. A. Kulemin, S. E. Semina, E. V. Generozov and V. M. Govorun, Sci. Rep., 2015, 5, 17319 CrossRef PubMed.
- J. Z. Nordin, Y. Lee, P. Vader, I. Mager, H. J. Johansson, W. Heusermann, O. P. Wiklander, M. Hallbrink, Y. Seow, J. J. Bultema, J. Gilthorpe, T. Davies, P. J. Fairchild, S. Gabrielsson, N. C. Meisner-Kober, J. Lehtio, C. I. Smith, M. J. Wood and S. El Andaloussi, Nanomedicine, 2015, 11, 879–883 CrossRef CAS PubMed.
- W. Wang, X. J. Huang, J. D. Cao, P. Lan and W. Wu, Acta Biomater., 2014, 10, 234–243 CrossRef CAS PubMed.
- T. A. Hartjes, S. Mytnyk, G. W. Jenster, V. van Steijn and M. E. van Royen, Bioengineering, 2019, 6(1), 7 CrossRef PubMed.
- I. Furi, F. Momen-Heravi and G. Szabo, Ann. Transl. Med., 2017, 5, 263 CrossRef PubMed.
- A. Salvi, M. Vezzoli, S. Busatto, L. Paolini, T. Faranda, E. Abeni, M. Caracausi, F. Antonaros, A. Piovesan, C. Locatelli, G. Cocchi, G. Alvisi, G. De Petro, D. Ricotta, P. Bergese and A. Radeghieri, Int. J. Mol. Med., 2019, 43, 2303–2318 CAS.
- M. Y. Konoshenko, E. A. Lekchnov, A. V. Vlassov and P. P. Laktionov, BioMed. Res. Int., 2018, 2018, 8545347 Search PubMed.
- L. Paolini, A. Zendrini, G. Di Noto, S. Busatto, E. Lottini, A. Radeghieri, A. Dossi, A. Caneschi, D. Ricotta and P. Bergese, Sci. Rep., 2016, 6, 23550 CrossRef CAS PubMed.
- J. Karttunen, M. Heiskanen, V. Navarro-Ferrandis, S. Das Gupta, A. Lipponen, N. Puhakka, K. Rilla, A. Koistinen and A. Pitkanen, J. Extracell. Vesicles, 2019, 8, 1555410 CrossRef CAS PubMed.
- S. Busatto, A. Giacomini, C. Montis, R. Ronca and P. Bergese, Anal. Chem., 2018, 90, 7855–7861 CrossRef CAS PubMed.
- A. Gamez-Valero, M. Monguio-Tortajada, L. Carreras-Planella, M. Franquesa, K. Beyer and F. E. Borras, Sci. Rep., 2016, 6, 33641 CrossRef CAS PubMed.
- M. I. Ramirez, M. G. Amorim, C. Gadelha, I. Milic, J. A. Welsh, V. M. Freitas, M. Nawaz, N. Akbar, Y. Couch, L. Makin, F. Cooke, A. L. Vettore, P. X. Batista, R. Freezor, J. A. Pezuk, L. Rosa-Fernandes, A. C. O. Carreira, A. Devitt, L. Jacobs, I. T. Silva, G. Coakley, D. N. Nunes, D. Carter, G. Palmisano and E. Dias-Neto, Nanoscale, 2018, 10, 881–906 RSC.
- J. C. Contreras-Naranjo, H. J. Wu and V. M. Ugaz, Lab Chip, 2017, 17, 3558–3577 RSC.
- P. Cizmar and Y. Yuana, Methods Mol. Biol., 2017, 1660, 221–232 CrossRef CAS PubMed.
- C. Gardiner, M. Shaw, P. Hole, J. Smith, D. Tannetta, C. W. Redman and I. L. Sargent, J. Extracell. Vesicles, 2014, 3, 25361 CrossRef PubMed.
- C. Montis, A. Zendrini, F. Valle, S. Busatto, L. Paolini, A. Radeghieri, A. Salvatore, D. Berti and P. Bergese, Colloids Surf., B, 2017, 158, 331–338 CrossRef CAS PubMed.
- S. L. Maas, J. De Vrij and M. L. Broekman, J. Visualized Exp., 2014, e51623, DOI:10.3791/51623.
- D. Maiolo, L. Paolini, G. Di Noto, A. Zendrini, D. Berti, P. Bergese and D. Ricotta, Anal. Chem., 2015, 87, 4168–4176 CrossRef CAS PubMed.
- A. Gorgens, M. Bremer, R. Ferrer-Tur, F. Murke, T. Tertel, P. A. Horn, S. Thalmann, J. A. Welsh, C. Probst, C. Guerin, C. M. Boulanger, J. C. Jones, H. Hanenberg, U. Erdbrugger, J. Lannigan, F. L. Ricklefs, S. El-Andaloussi and B. Giebel, J. Extracell. Vesicles, 2019, 8, 1587567 CrossRef PubMed.
- C. Campos-Silva, H. Suarez, R. Jara-Acevedo, E. Linares-Espinos, L. Martinez-Pineiro, M. Yanez-Mo and M. Vales-Gomez, Sci. Rep., 2019, 9, 2042 CrossRef PubMed.
- R. Bandu, J. W. Oh and K. P. Kim, Exp. Mol. Med., 2019, 51, 30 CrossRef PubMed.
- H. Shao, H. Im, C. M. Castro, X. Breakefield, R. Weissleder and H. Lee, Chem. Rev., 2018, 118, 1917–1950 CrossRef CAS PubMed.
- B. Hosseinkhani, N. van den Akker, J. D'Haen, M. Gagliardi, T. Struys, I. Lambrichts, J. Waltenberger, I. Nelissen, J. Hooyberghs, D. G. M. Molin and L. Michiels, Nanomedicine, 2017, 13, 1663–1671 CrossRef CAS PubMed.
- J. Mihaly, R. Deak, I. C. Szigyarto, A. Bota, T. Beke-Somfai and Z. Varga, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 459–466 CrossRef CAS PubMed.
- S. Saito, K. Hiemori, K. Kiyoi and H. Tateno, Sci. Rep., 2018, 8, 3997 CrossRef PubMed.
- M. Chalfie, Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, Science, 1994, 263, 802–805 CrossRef CAS PubMed.
- S. Inouye and F. I. Tsuji, FEBS Lett., 1994, 341, 277–280 CrossRef CAS PubMed.
- J. Royes, O. Ilioaia, Q. Lubart, F. Angius, G. V. Dubacheva, M. Bally, B. Miroux and C. Tribet, Angew. Chem., Int. Ed., 2019, 58, 7395–7399 CrossRef CAS PubMed.
- P. Garcia-Manrique, M. Matos, G. Gutierrez, C. Pazos and M. C. Blanco-Lopez, J. Extracell. Vesicles, 2018, 7, 1422676 CrossRef PubMed.
- O. G. de Jong, M. C. Verhaar, Y. Chen, P. Vader, H. Gremmels, G. Posthuma, R. M. Schiffelers, M. Gucek and B. W. van Balkom, J. Extracell. Vesicles, 2012, 1, 18396 CrossRef CAS PubMed.
- D. A. Bricarello, J. T. Smilowitz, A. M. Zivkovic, J. B. German and A. N. Parikh, ACS Nano, 2011, 5, 42–57 CrossRef CAS PubMed.
- J. Chandrapala, G. J. Martin, S. E. Kentish and M. Ashokkumar, Ultrason. Sonochem., 2014, 21, 1658–1665 CrossRef CAS PubMed.
- J. P. Armstrong, M. N. Holme and M. M. Stevens, ACS Nano, 2017, 11, 69–83 CrossRef CAS PubMed.
- E. J. Lee, N. K. Lee and I. S. Kim, Adv. Drug Delivery Rev., 2016, 106, 157–171 CrossRef CAS PubMed.
- C. S. Thaxton, J. S. Rink, P. C. Naha and D. P. Cormode, Adv. Drug Delivery Rev., 2016, 106, 116–131 CrossRef CAS PubMed.
- K. E. Gilligan and R. M. Dwyer, Int. J. Mol. Sci., 2017, 18, 1122 CrossRef PubMed.
- J. O. Jeon, S. Kim, E. Choi, K. Shin, K. Cha, I. S. So, S. J. Kim, E. Jun, D. Kim, H. J. Ahn, B. H. Lee, S. H. Lee and I. S. Kim, ACS Nano, 2013, 7, 7462–7471 CrossRef CAS PubMed.
- M. Liang, C. Gao, Y. Wang, W. Gong, S. Fu, L. Cui, Z. Zhou, X. Chu, Y. Zhang, Q. Liu, X. Zhao, B. Zhao, M. Yang, Z. Li, C. Yang, X. Xie, Y. Yang and C. Gao, Drug Delivery, 2018, 25, 1652–1663 CrossRef CAS PubMed.
- T. Smyth, K. Petrova, N. M. Payton, I. Persaud, J. S. Redzic, M. W. Graner, P. Smith-Jones and T. J. Anchordoquy, Bioconjugate Chem., 2014, 25, 1777–1784 CrossRef CAS PubMed.
- H. Lee, H. Park, G. J. Noh and E. S. Lee, Carbohydr. Polym., 2018, 202, 323–333 CrossRef CAS PubMed.
- J. B. Simonsen, J. Extracell. Vesicles, 2019, 8, 1582237 CrossRef PubMed.
- P. Gangadaran, C. M. Hong and B. C. Ahn, BioMed. Res. Int., 2017, 2017, 9158319 Search PubMed.
- C. P. Lai, B. A. Tannous and X. O. Breakefield, Methods Mol. Biol., 2014, 1098, 249–258 CrossRef CAS PubMed.
- J. Cai, Y. Han, H. Ren, C. Chen, D. He, L. Zhou, G. M. Eisner, L. D. Asico, P. A. Jose and C. Zeng, J. Mol. Cell Biol., 2013, 5, 227–238 CrossRef CAS PubMed.
- K. M. McMahon, R. K. Mutharasan, S. Tripathy, D. Veliceasa, M. Bobeica, D. K. Shumaker, A. J. Luthi, B. T. Helfand, H. Ardehali, C. A. Mirkin, O. Volpert and C. S. Thaxton, Nano Lett., 2011, 11, 1208–1214 CrossRef CAS PubMed.
- N. J. Bitto, R. Chapman, S. Pidot, A. Costin, C. Lo, J. Choi, T. D'Cruze, E. C. Reynolds, S. G. Dashper, L. Turnbull, C. B. Whitchurch, T. P. Stinear, K. J. Stacey and R. L. Ferrero, Sci. Rep., 2017, 7, 7072 CrossRef PubMed.
- K. Isaac-Olive, B. E. Ocampo-Garcia, L. Aranda-Lara, C. L. Santos-Cuevas, N. P. Jimenez-Mancilla, M. A. Luna-Gutierrez, L. A. Medina, B. Nagarajan, N. Sabnis, S. Raut, L. Prokai and A. G. Lacko, Nanoscale, 2019, 11, 541–551 RSC.
- Q. Dai, S. Wilhelm, D. Ding, A. M. Syed, S. Sindhwani, Y. Zhang, Y. Y. Chen, P. MacMillan and W. C. W. Chan, ACS Nano, 2018, 12, 8423–8435 CrossRef CAS PubMed.
- W. C. W. Chan, Acc. Chem. Res., 2017, 50, 627–632 CrossRef CAS PubMed.
- M. T. Larsen, M. Kuhlmann, M. L. Hvam and K. A. Howard, Mol. Cell. Ther., 2016, 4, 3 CrossRef PubMed.
- M. Khoshnejad, H. Parhiz, V. V. Shuvaev, I. J. Dmochowski and V. R. Muzykantov, J. Controlled Release, 2018, 282, 13–24 CrossRef CAS PubMed.
- A. E. Pottash, C. Kuffner, M. Noonan-Shueh and S. M. Jay, J. Biol. Eng., 2019, 13, 19 CrossRef PubMed.
- Y. Zhang, Z. Guo, Z. Cao, W. Zhou, Y. Zhang, Q. Chen, Y. Lu, X. Chen, Q. Guo, C. Li, D. Liang, T. Sun and C. Jiang, Biomaterials, 2018, 183, 243–257 CrossRef CAS PubMed.
- S. Dostalova, H. Polanska, M. Svobodova, J. Balvan, O. Krystofova, Y. Haddad, S. Krizkova, M. Masarik, T. Eckschlager, M. Stiborova, Z. Heger and V. Adam, Sci. Rep., 2018, 8, 8867 CrossRef PubMed.
- J. Li, C. Wu, W. Wang, Y. He, E. Elkayam, L. Joshua-Tor and P. T. Hammond, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E2696–E2705 CrossRef CAS PubMed.
- J. P. K. Armstrong and M. M. Stevens, Adv. Drug Delivery Rev., 2018, 130, 12–16 CrossRef CAS PubMed.
- P. Vader, E. A. Mol, G. Pasterkamp and R. M. Schiffelers, Adv. Drug Delivery Rev., 2016, 106, 148–156 CrossRef CAS PubMed.
- S. M. Kim and H. S. Kim, Stem Cell Invest., 2017, 4, 74 CrossRef PubMed.
- M. Piffoux, A. Nicolas-Boluda, V. Mulens-Arias, S. Richard, G. Rahmi, F. Gazeau, C. Wilhelm and A. K. A. Silva, Adv. Drug Delivery Rev., 2019, 138, 247–258 CrossRef CAS PubMed.
- D. S. Chulpanova, K. V. Kitaeva, V. James, A. A. Rizvanov and V. V. Solovyeva, Front. Immunol., 2018, 9, 1534 CrossRef PubMed.
- W. M. Usman, T. C. Pham, Y. Y. Kwok, L. T. Vu, V. Ma, B. Peng, Y. S. Chan, L. Wei, S. M. Chin, A. Azad, A. B. He, A. Y. H. Leung, M. Yang, N. Shyh-Chang, W. C. Cho, J. Shi and M. T. N. Le, Nat. Commun., 2018, 9, 2359 CrossRef PubMed.
- A. H. Alhasan, P. C. Patel, C. H. Choi and C. A. Mirkin, Small, 2014, 10, 186–192 CrossRef CAS PubMed.
- M. Garofalo, A. Villa, N. Rizzi, L. Kuryk, B. Rinner, V. Cerullo, M. Yliperttula, V. Mazzaferro and P. Ciana, J. Controlled Release, 2019, 294, 165–175 CrossRef CAS PubMed.
- P. Lv, X. Liu, X. Chen, C. Liu, Y. Zhang, C. Chu, J. Wang, X. Wang, X. Chen and G. Liu, Nano Lett., 2019, 19, 2993–3001 CrossRef CAS PubMed.
- L. Alvarez-Erviti, Y. Seow, H. Yin, C. Betts, S. Lakhal and M. J. Wood, Nat. Biotechnol., 2011, 29, 341–345 CrossRef CAS PubMed.
- E. Cho, G. H. Nam, Y. Hong, Y. K. Kim, D. H. Kim, Y. Yang and I. S. Kim, J. Controlled Release, 2018, 279, 326–335 CrossRef CAS PubMed.
- M. J. Haney, N. L. Klyachko, Y. Zhao, R. Gupta, E. G. Plotnikova, Z. He, T. Patel, A. Piroyan, M. Sokolsky, A. V. Kabanov and E. V. Batrakova, J. Controlled Release, 2015, 207, 18–30 CrossRef CAS PubMed.
- H. Huang, W. Cruz, J. Chen and G. Zheng, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 298–314 CAS.
- A. G. Lacko, N. A. Sabnis, B. Nagarajan and W. J. McConathy, Front. Pharmacol., 2015, 6, 247 Search PubMed.
- Q. L. Zhu, Y. Zhou, M. Guan, X. F. Zhou, S. D. Yang, Y. Liu, W. L. Chen, C. G. Zhang, Z. Q. Yuan, C. Liu, A. J. Zhu and X. N. Zhang, Biomaterials, 2014, 35, 5965–5976 CrossRef CAS PubMed.
- Y. Ding, W. Wang, M. Feng, Y. Wang, J. Zhou, X. Ding, X. Zhou, C. Liu, R. Wang and Q. Zhang, Biomaterials, 2012, 33, 8893–8905 CrossRef CAS PubMed.
- L. Cui, Y. Wang, M. Liang, X. Chu, S. Fu, C. Gao, Q. Liu, W. Gong, M. Yang, Z. Li, L. Yu, C. Yang, Z. Su, X. Xie, Y. Yang and C. Gao, Drug Delivery, 2018, 25, 1865–1876 CrossRef CAS PubMed.
- A. O. Elzoghby, W. S. El-Fotoh and N. A. Elgindy, J. Controlled Release, 2011, 153, 206–216 CrossRef CAS PubMed.
- M. L. C. Picchio, J. C. Cuggino, G. Nagel, S. Wedepohl, R. J. Minari, C. I. Alvarez Igarzabal, L. M. Gugliotta and M. Calderón, Polym. Chem., 2018, 9, 3499–3510 RSC.
- J. Lopez-Sagaseta, E. Malito, R. Rappuoli and M. J. Bottomley, Comput. Struct. Biotechnol. J., 2016, 14, 58–68 CrossRef CAS PubMed.
- G. Zhu, G. M. Lynn, O. Jacobson, K. Chen, Y. Liu, H. Zhang, Y. Ma, F. Zhang, R. Tian, Q. Ni, S. Cheng, Z. Wang, N. Lu, B. C. Yung, Z. Wang, L. Lang, X. Fu, A. Jin, I. D. Weiss, H. Vishwasrao, G. Niu, H. Shroff, D. M. Klinman, R. A. Seder and X. Chen, Nat. Commun., 2017, 8, 1954 CrossRef PubMed.
- M. Kanekiyo, C. J. Wei, H. M. Yassine, P. M. McTamney, J. C. Boyington, J. R. Whittle, S. S. Rao, W. P. Kong, L. Wang and G. J. Nabel, Nature, 2013, 499, 102–106 CrossRef CAS PubMed.
- Z. Wang, L. Xu, H. Yu, P. Lv, Z. Lei, Y. Zeng, G. Liu and T. Cheng, Biomater. Sci., 2019, 7, 1794–1800 RSC.
- W. Wang, Z. Liu, X. Zhou, Z. Guo, J. Zhang, P. Zhu, S. Yao and M. Zhu, Nanomedicine, 2019, 16, 69–78 CrossRef CAS PubMed.
- K. Tan, R. Li, X. Huang and Q. Liu, Front. Microbiol., 2018, 9, 783 CrossRef PubMed.
- R. Acevedo, S. Fernandez, C. Zayas, A. Acosta, M. Sarmiento, V. Ferro, E. Rosenqvist, C. Campa, D. Cardoso, L. Garcia and J. Perez, Front. Immunol., 2014, 5, 121 Search PubMed.
- G. G. Mekonnen, M. Pearson, A. Loukas and J. Sotillo, Expert Rev. Vaccines, 2018, 17, 197–205 CrossRef CAS PubMed.
- S. Montaner-Tarbes, F. E. Borrás, M. Montoya, L. Fraile and H. A. del Portillo, Vet. Res., 2016, 47, 59 CrossRef PubMed.
- P. Kadiyala, D. Li, F. M. Nuñez, D. Altshuler, R. Doherty, R. Kuai, M. Yu, N. Kamran, M. Edwards, J. J. Moon, P. R. Lowenstein, M. G. Castro and A. Schwendeman, ACS Nano, 2019, 13, 1365–1384 CAS.
- R. Kuai, X. Sun, W. Yuan, L. J. Ochyl, Y. Xu, A. Hassani Najafabadi, L. Scheetz, M.-Z. Yu, I. Balwani, A. Schwendeman and J. J. Moon, J. Controlled Release, 2018, 282, 131–139 CrossRef CAS PubMed.
- D. B. Horváthy, M. Simon, C. M. Schwarz, M. Masteling, G. Vácz, I. Hornyák and Z. Lacza, BioFactors, 2017, 43, 315–330 CrossRef PubMed.
- K. Schandl, D. B. Horvathy, A. Doros, E. Majzik, C. M. Schwarz, L. Csonge, G. Abkarovits, L. Bucsi and Z. Lacza, Int. Orthop., 2016, 40, 2097–2104 CrossRef PubMed.
- D. B. Horvathy, G. Vacz, A. Cselenyak, M. Weszl, L. Kiss and Z. Lacza, Surg. Innov., 2013, 20, 249–255 CrossRef PubMed.
- I. M. Bjorge, S. Y. Kim, J. F. Mano, B. Kalionis and W. Chrzanowski, Biomater. Sci., 2017, 6, 60–78 RSC.
- G. H. Cui, J. Wu, F. F. Mou, W. H. Xie, F. B. Wang, Q. L. Wang, J. Fang, Y. W. Xu, Y. R. Dong, J. R. Liu and H. D. Guo, FASEB J., 2018, 32, 654–668 CrossRef CAS PubMed.
- Y. Ma, L. Dong, D. Zhou, L. Li, W. Zhang, Y. Zhen, T. Wang, J. Su, D. Chen, C. Mao and X. Wang, J. Cell. Mol. Med., 2019, 23, 2822–2835 CrossRef CAS PubMed.
- Y. Wang, L. Zhang, Y. Li, L. Chen, X. Wang, W. Guo, X. Zhang, G. Qin, S. H. He, A. Zimmerman, Y. Liu, I. M. Kim, N. L. Weintraub and Y. Tang, Int. J. Cardiol., 2015, 192, 61–69 CrossRef PubMed.
- J. A. Dougherty, N. Kumar, M. Noor, M. G. Angelos, M. Khan, C. A. Chen and M. Khan, Front. Physiol., 2018, 9, 1794 CrossRef PubMed.
- A. Radeghieri, G. Savio, A. Zendrini, G. Di Noto, A. Salvi, P. Bergese and G. Piovani, Biochem. Biophys. Res. Commun., 2017, 483, 706–711 CrossRef CAS PubMed.
- Y. Wang, D. Yu, Z. Liu, F. Zhou, J. Dai, B. Wu, J. Zhou, B. C. Heng, X. H. Zou, H. Ouyang and H. Liu, Stem Cell Res. Ther., 2017, 8, 189 CrossRef PubMed.
- S. S. Gu, N. Shi and M. P. Wu, Life Sci., 2007, 81, 702–709 CrossRef CAS PubMed.
- B. S. Ding, C. H. Liu, Y. Sun, Y. Chen, S. L. Swendeman, B. Jung, D. Chavez, Z. Cao, C. Christoffersen, L. B. Nielsen, S. R. Schwab, S. Rafii and T. Hla, JCI Insight, 2016, 1, e87058 Search PubMed.
- S. Van Linthout, M. Frias, N. Singh and B. De Geest, Handb. Exp. Pharmacol., 2015, 224, 527–565 CAS.
- X. Wang, J. Zhou and W. Wang, Curr. Pharm. Des., 2015, 21, 1529–1544 CrossRef CAS PubMed.
- F. Spillmann, C. Trimpert, J. Peng, L. G. Eckerle, A. Staudt, K. Warstat, S. B. Felix, B. Pieske, C. Tschöpe and S. Van Linthout, Biochem. Biophys. Res. Commun., 2015, 466, 272–277 CrossRef CAS PubMed.
- C. M. Gibson, M. Kerneis, M. K. Yee, Y. Daaboul, S. Korjian, A. P. Mehr, P. Tricoci, J. H. Alexander, J. J. P. Kastelein, R. Mehran, C. Bode, B. S. Lewis, R. Mehta, D. Duffy, J. Feaster, M. Halabi, D. J. Angiolillo, D. Duerschmied, T. O. Ophuis and B. Merkely, Am. Heart J., 2019, 208, 81–90 CrossRef CAS PubMed.
- C. Gebhard, E. Rheaume, C. Berry, G. Brand, A. E. Kernaleguen, G. Theberge-Julien, M. A. Alam, C. Y. Lee, L. Boileau, M. Chabot-Blanchet, M. C. Guertin, M. A. Lavoie, J. Gregoire, R. Ibrahim, P. L'Allier and J. C. Tardif, PLoS One, 2017, 12, e0168448 CrossRef PubMed.
- M. Mishra, I. Muthuramu, J. P. Aboumsallem, H. Kempen and B. De Geest, Int. J. Mol. Sci., 2018, 19, 3399 CrossRef PubMed.
- T. Tsatralis, A. Ridiandries, S. Robertson, L. Z. Vanags, Y. T. Lam, J. T. M. Tan, M. K. C. Ng and C. A. Bursill, Lipids Health Dis., 2016, 15, 150–150 CrossRef PubMed.
- R. Gao, M. Watson, K. E. Callon, D. Tuari, M. Dray, D. Naot, S. Amirapu, J. T. Munro, J. Cornish and D. S. Musson, J. Tissue Eng. Regener. Med., 2018, 12, e620–e626 CrossRef CAS PubMed.
- T. Fuji, Y. Umeda, A. Nyuya, F. Taniguchi, T. Kawai, K. Yasui, T. Toshima, K. Yoshida, T. Fujiwara, A. Goel and T. Nagasaka, Int. J. Cancer, 2019, 144, 2169–2180 CrossRef CAS PubMed.
- E. Vila-Navarro, S. Duran-Sanchon, M. Vila-Casadesus, L. Moreira, A. Gines, M. Cuatrecasas, J. J. Lozano, L. Bujanda, A. Castells and M. Gironella, Clin. Transl. Gastroenterol., 2019, 10, e00029 CrossRef PubMed.
- N. Asano, J. Matsuzaki, M. Ichikawa, J. Kawauchi, S. Takizawa, Y. Aoki, H. Sakamoto, A. Yoshida, E. Kobayashi, Y. Tanzawa, R. Nakayama, H. Morioka, M. Matsumoto, M. Nakamura, T. Kondo, K. Kato, N. Tsuchiya, A. Kawai and T. Ochiya, Nat. Commun., 2019, 10, 1299 CrossRef PubMed.
- Q. Jiang, X. Lu, P. Huang, C. Gao, X. Zhao, T. Xing, G. Li, S. Bao and H. Zheng, BioMed Res. Int., 2018, 2018, 10 Search PubMed.
- H. Honghai, Y. Haihui and X. Yong, Curr. Pharm. Biotechnol., 2018, 19, 79–86 Search PubMed.
- H. Nakashima, R. Yoshida, A. Hirosue, K. Kawahara, J. Sakata, H. Arita, T. Yamamoto, R. Toya, R. Murakami, A. Hiraki, M. Shinohara, T. Ito, Y. Kuwahara and H. Nakayama, Tumor Biol., 2019, 41, 1–10 Search PubMed.
- D. Turpin, M. E. Truchetet, B. Faustin, J. F. Augusto, C. Contin-Bordes, A. Brisson, P. Blanco and P. Duffau, Autoimmun. Rev., 2016, 15, 174–183 CrossRef CAS PubMed.
- S. Wang, J. Gao and Z. Wang, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2019, 11, e1523 Search PubMed.
- L. A. Aqrawi, H. K. Galtung, B. Vestad, R. Ovstebo, B. Thiede, S. Rusthen, A. Young, E. M. Guerreiro, T. P. Utheim, X. Chen, O. A. Utheim, O. Palm and J. L. Jensen, Arthritis Res. Ther., 2017, 19, 14 CrossRef PubMed.
- W. Zhang, X. Zhou, H. Zhang, Q. Yao, Y. Liu and Z. Dong, Am. J. Physiol. Renal Physiol., 2016, 311, F844–F851 CrossRef CAS PubMed.
- G. Lou, Z. Chen, M. Zheng and Y. Liu, Exp. Mol. Med., 2017, 49, e346 CrossRef PubMed.
- X. Jin, Y. Chen, H. Chen, S. Fei, D. Chen, X. Cai, L. Liu, B. Lin, H. Su, L. Zhao, M. Su, H. Pan, L. Shen, D. Xie and C. Xie, Clin. Cancer Res., 2017, 23, 5311–5319 CrossRef CAS PubMed.
- J. Castillo, V. Bernard, F. A. San Lucas, K. Allenson, M. Capello, D. U. Kim, P. Gascoyne, F. C. Mulu, B. M. Stephens, J. Huang, H. Wang, A. A. Momin, R. O. Jacamo, M. Katz, R. Wolff, M. Javle, G. Varadhachary, I. I. Wistuba, S. Hanash, A. Maitra and H. Alvarez, Ann. Oncol., 2018, 29, 223–229 CrossRef CAS PubMed.
- J. Fredsøe, A. K. Rasmussen, P. Mouritzen, M. Borre, T. Ørntoft and K. D. Sørensen, Int. J. Cancer, 2019, 145, 2558–2567 CrossRef PubMed.
- P. Li, X. Yu, W. Han, Y. Kong, W. Bao, J. Zhang, W. Zhang and Y. Gu, ACS Sens., 2019, 4, 1433–1441 CrossRef CAS PubMed.
- E. Reátegui, K. E. van der Vos, C. P. Lai, M. Zeinali, N. A. Atai, B. Aldikacti, F. P. Floyd Jr., A. H. Khankhel, V. Thapar, F. H. Hochberg, L. V. Sequist, B. V. Nahed, B. S. Carter, M. Toner, L. Balaj, D. T. Ting, X. O. Breakefield and S. L. Stott, Nat. Commun., 2018, 9, 175 CrossRef PubMed.
- H. Shao, J. Chung, K. Lee, L. Balaj, C. Min, B. S. Carter, F. H. Hochberg, X. O. Breakefield, H. Lee and R. Weissleder, Nat. Commun., 2015, 6, 6999 CrossRef CAS PubMed.
- Y. Bei, P. Yu, D. Cretoiu, S. M. Cretoiu and J. Xiao, Adv. Exp. Med. Biol., 2017, 998, 71–88 CrossRef CAS PubMed.
- A. E. Berezin, A. A. Kremzer, Y. V. Martovitskaya, T. A. Samura and T. A. Berezina, BBA Clin., 2015, 3, 18–24 CrossRef PubMed.
- S. Evans and D. L. Mann, Circ. Res., 2013, 113, 242–244 CrossRef CAS PubMed.
- G. Chiva-Blanch, R. Suades, J. Crespo, G. Vilahur, G. Arderiu, T. Padro, D. Corella, J. Salas-Salvado, F. Aros, M. A. Martinez-Gonzalez, E. Ros, M. Fito, R. Estruch and L. Badimon, Int. J. Cardiol., 2016, 208, 147–149 CrossRef PubMed.
- Q. Ji, Y. Ji, J. Peng, X. Zhou, X. Chen, H. Zhao, T. Xu, L. Chen and Y. Xu, PLoS One, 2016, 11, e0163645 CrossRef PubMed.
- G. Di Noto, A. Bugatti, A. Zendrini, E. L. Mazzoldi, A. Montanelli, L. Caimi, M. Rusnati, D. Ricotta and P. Bergese, Biosens. Bioelectron., 2016, 77, 518–524 CrossRef CAS PubMed.
- L.-G. Liang, M.-Q. Kong, S. Zhou, Y.-F. Sheng, P. Wang, T. Yu, F. Inci, W. P. Kuo, L.-J. Li, U. Demirci and S. Wang, Sci. Rep., 2017, 7, 46224 CrossRef PubMed.
- K. Liang, F. Liu, J. Fan, D. Sun, C. Liu, C. J. Lyon, D. W. Bernard, Y. Li, K. Yokoi, M. H. Katz, E. J. Koay, Z. Zhao and Y. Hu, Nat. Biomed. Eng., 2017, 1, 0021 CrossRef CAS PubMed.
- C. Liu, X. Xu, B. Li, B. Situ, W. Pan, Y. Hu, T. An, S. Yao and L. Zheng, Nano Lett., 2018, 18, 4226–4232 CrossRef CAS PubMed.
- S. M. Santana, M. A. Antonyak, R. A. Cerione and B. J. Kirby, Phys. Biol., 2014, 11, 065001 CrossRef PubMed.
- J. S. Yang, J. C. Lee, S. K. Byeon, K. H. Rha and M. H. Moon, Anal. Chem., 2017, 89, 2488–2496 CrossRef CAS PubMed.
- M. Tong, Q. Chen, J. L. James, P. R. Stone and L. W. Chamley, Front. Endocrinol., 2017, 8, 174 CrossRef PubMed.
- B. Whitehead, L. Wu, M. L. Hvam, H. Aslan, M. Dong, L. Dyrskjøt, M. S. Ostenfeld, S. M. Moghimi and K. A. Howard, J. Extracell. Vesicles, 2015, 4, 29685 CrossRef PubMed.
- A. Rivas-Urbina, S. Benitez, A. Perez and J. L. Sanchez-Quesada, Front. Biosci., Landmark Ed., 2018, 23, 1220–1240 CrossRef CAS PubMed.
- A. Trpkovic, I. Resanovic, J. Stanimirovic, D. Radak, S. A. Mousa, D. Cenic-Milosevic, D. Jevremovic and E. R. Isenovic, Crit. Rev. Clin. Lab. Sci., 2015, 52, 70–85 CrossRef CAS PubMed.
- M. P. Hermans and P. Valensi, Curr. Opin. Endocrinol. Diabetes Obes., 2018, 25, 118–129 CrossRef CAS PubMed.
- N. Clouet-Foraison, F. Gaie-Levrel, P. Gillery and V. Delatour, Journal, 2017, 55, 1453 CAS.
- I. Rosado-Sánchez, E. Rodríguez-Gallego, J. Peraire, C. Viladés, P. Herrero, F. Fanjul, F. Gutiérrez, E. Bernal, R. Pelazas, M. Leal, S. Veloso, M. López-Dupla, J. Blanco, F. Vidal, Y. M. Pacheco and A. Rull, Clin. Sci., 2019, 133, 997 CrossRef PubMed.
- S. De Bruyne, T. Monteyne, M. Speeckaert and J. Delanghe, Journal, 2017, 55, 876 CAS.
- J. Wagner, M. Riwanto, C. Besler, A. Knau, S. Fichtlscherer, T. Röxe, A. M. Zeiher, U. Landmesser and S. Dimmeler, Arterioscler., Thromb., Vasc. Biol., 2013, 33, 1392–1400 CrossRef CAS PubMed.
- L. S. Niculescu, N. Simionescu, G. M. Sanda, M. G. Carnuta, C. S. Stancu, A. C. Popescu, M. R. Popescu, A. Vlad, D. R. Dimulescu, M. Simionescu and A. V. Sima, PLoS One, 2015, 10, e0140958 CrossRef PubMed.
- K. C. Vickers, B. T. Palmisano, B. M. Shoucri, R. D. Shamburek and A. T. Remaley, Nat. Cell Biol., 2011, 13, 423 CrossRef CAS PubMed.
- D. Liu, M. Zhang, W. Xie, G. Lan, H.-P. Cheng, D. Gong, C. Huang, Y.-C. Lv, F. Yao, Y.-L. Tan, L. Li, X.-L. Zheng and C.-K. Tang, Biochem. Biophys. Res. Commun., 2016, 472, 418–424 CrossRef CAS PubMed.
- K. C. Vickers, S. R. Landstreet, M. G. Levin, B. M. Shoucri, C. L. Toth, R. C. Taylor, B. T. Palmisano, F. Tabet, H. L. Cui, K.-A. Rye, P. Sethupathy and A. T. Remaley, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 14518 CrossRef CAS PubMed.
- C. Paganini, U. C. Palmiero, G. Pocsfalvi, N. Touzet, A. Bongiovanni and P. Arosio, Biotechnol. J., 2019, e1800528, DOI:10.1002/biot.201800528.
- A. Goes and G. Fuhrmann, ACS Infect. Dis., 2018, 4, 881–892 CrossRef CAS PubMed.
- M. Faria, M. Bjornmalm, K. J. Thurecht, S. J. Kent, R. G. Parton, M. Kavallaris, A. P. R. Johnston, J. J. Gooding, S. R. Corrie, B. J. Boyd, P. Thordarson, A. K. Whittaker, M. M. Stevens, C. A. Prestidge, C. J. H. Porter, W. J. Parak, T. P. Davis, E. J. Crampin and F. Caruso, Nat. Nanotechnol., 2018, 13, 777–785 CrossRef CAS PubMed.
- X. Li, M. Soler, C. Szydzik, K. Khoshmanesh, J. Schmidt, G. Coukos, A. Mitchell and H. Altug, Small, 2018, 14, e1800698 CrossRef PubMed.
- K. Powell, Nature, 2018, 563, 172–175 CrossRef CAS PubMed.
- R. Desplantes, C. Leveque, B. Muller, M. Lotierzo, G. Ferracci, M. Popoff, M. Seagar, R. Mamoun and O. El Far, Sci. Rep., 2017, 7, 1032 CrossRef PubMed.
- C. Montis, S. Busatto, F. Valle, A. Zendrini, A. Salvatore, Y. Gerelli, D. Berti and P. Bergese, Adv. Biosyst., 2018, 2, 1700200 CrossRef.
- J. Tang, D. Shen, T. G. Caranasos, Z. Wang, A. C. Vandergriff, T. A. Allen, M. T. Hensley, P. U. Dinh, J. Cores, T. S. Li, J. Zhang, Q. Kan and K. Cheng, Nat. Commun., 2017, 8, 13724 CrossRef CAS PubMed.
- A. K. Silva, R. Di Corato, T. Pellegrino, S. Chat, G. Pugliese, N. Luciani, F. Gazeau and C. Wilhelm, Nanoscale, 2013, 5, 11374–11384 RSC.
- M. Weiss, J. P. Frohnmayer, L. T. Benk, B. Haller, J. W. Janiesch, T. Heitkamp, M. Borsch, R. B. Lira, R. Dimova, R. Lipowsky, E. Bodenschatz, J. C. Baret, T. Vidakovic-Koch, K. Sundmacher, I. Platzman and J. P. Spatz, Nat. Mater., 2018, 17, 89–96 CrossRef CAS PubMed.
- F. Wu, S. A. Wuensch, M. Azadniv, M. R. Ebrahimkhani and I. N. Crispe, et al. , Mol. Pharm, 2009, 6(5), 1506–1517 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9bm01007f |
| This journal is © The Royal Society of Chemistry 2020 |