 Open Access Article
Open Access ArticleA new genre of fluorescence recovery assay to evaluate polo-like kinase 1 ATP-competitive inhibitors†
Kohei
Tsuji
 ab,
David
Hymel
a and
Terrence R.
Burke
Jr
ab,
David
Hymel
a and
Terrence R.
Burke
Jr
 *a
*a
aChemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, Maryland, 21702 USA. E-mail: burkete@mail.nih.gov
bDepartment of Medicinal Chemistry, Institute of Biomaterials and Bioengineering, Tokyo Medical and Dental University (TMDU), 2-3-10 Kandasurugadai, Chiyoda-ku, Tokyo 101-0062, Japan
First published on 21st August 2020
Abstract
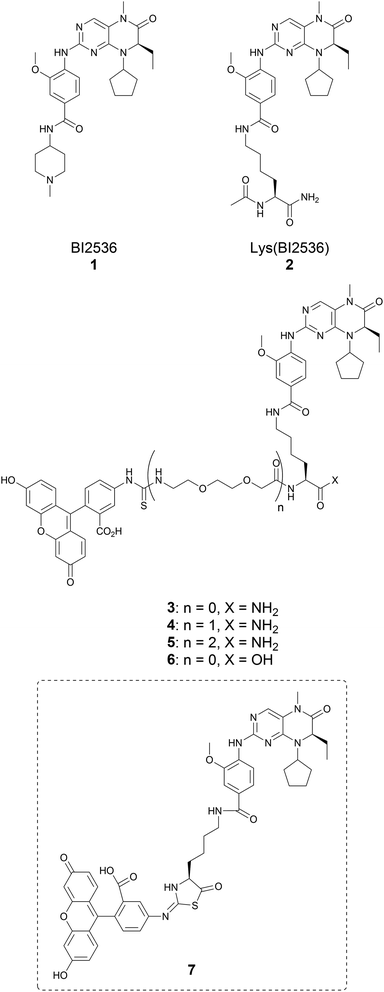
Using a probe consisting of a fluorescein-labeled variant of the potent polo-like kinase 1 (Plk1) inhibitor BI2536 [FITC-PEG-Lys(BI2536) 4], we were able to determine half maximal inhibitory concentration (IC50) of ATP-competitive Type 1 inhibitors of Plk1 by means of a fluorescence recovery assay. This methodology represents a cost-effective and simple alternative to traditional kinase assays for initial screening of potential Plk1 inhibitors.
The serine/threonine-specific polo-like kinase 1 (Plk1) plays crucial roles in mitosis, having both its location and catalytic activity strictly regulated at critical points in the cell cycle.1,2 It is over-expressed in a variety of cancers and associated with aggressiveness and poor prognosis.3 Inhibition of Plk1 can lead to abnormal spindle distribution and apoptotic cell death and it is considered to be an attractive target for anticancer drug development.4 In addition to an N-terminal catalytic kinase domain (KD), Plk1 contains a C-terminal polo-box domain (PBD), which is connected to the KD by an internal domain linker (IDL). A primary function of the PBD is to recognize and bind to phosphoserine/phosphothreonine-containing protein sequences and in so doing, provide spatiotemporal regulation of Plk1 action. The KD and PBD interact in autoinhibitory fashions through mechanisms that are not clearly understood.5,6 Both the KD and PBD of Plk1 are recognized as targets for anticancer therapeutic development, with the discovery of bioavailable PBD-binding inhibitors having proven to be particularly challenging.7,8 Identification of KD-binding agents has been more tractable and a number of agents have entered clinical trials. Yet there are no FDA-approved Plk1 kinase inhibitors and continued efforts in this area are warranted. There are a number of assay methods for evaluating kinase inhibitory activities; however, these often require expensive assay kits, specific instruments or complex assay procedures.9 In contrast, the binding assay that we describe herein serves as a simple and straight forward alternative that can not only identify Plk1 kinase inhibitors, but can do so while distinguishing between difference classes of inhibitors. In addition, to the best of our knowledge, there are no assays yet reported that evaluate ligand binding affinities to the Plk1 KD. The ability to determine binding affinities may have value beyond relevance to kinase inhibition. The PROteolysis Targeting Chimera (PROTAC) approach10 is emerging as an important means of targeting the destruction of proteins that may otherwise be undruggable or difficult to target. This approach requires binding of the PROTAC molecule to target protein and recruiting E3 ligase close to target protein. Therefore, in addition to enzyme inhibitory potency, ligand affinity may also be an important parameter. Accordingly, developing a simple and rapid assay protocol to determine affinity for the Plk1 KD could contribute significantly to the field of Plk1 KD inhibitor exploration.
The ability of small molecules to competitive displace KD-binding probes can be an efficient and facile method of screening for kinase inhibitors. Determining KD-occupancy of the probes can be accomplished in a number of fashions. Staurosporine is a non-specific ATP-competitive kinase inhibitor, which binds in the catalytic pockets of a large number of kinases. Intensity of the fluorescence emissions of staurosporine and related analogs can be enhanced when bound within the hydrophobic environment of the protein catalytic pocket. Quantifying decreases in fluorescence intensity of staurosporine as a function of test inhibitor concentrations can provide a means of calculating inhibitor affinities.11 Alternatively, variations in the fluorescence lifetime due to environmental effects incurred by a fluorophore binding to a kinase can be used to determine affinities of competing ligands. Changes in the fluorescence lifetime of an Alexa Fluor 647 – labeled staurosporine probe in response to displacement from the ATP-binding sites of a series of kinases has been used to identify and characterize small molecule inhibitors.12
In our current work, we set out to develop a fluorescence-based assay to determine small molecule binding affinities against the KD of Plk1. For this purpose, we selected the known high-affinity Plk1 inhibitor BI2536 (1), which has been shown to function in an ATP-competitive manner (Fig. 1).13 Fluorescently-labeled derivatives of BI2536 have been used as imaging probes to visualize Plk1 distribution in cells,14,15 and we envisioned that suitably labelled BI2536 constructs could be used in fluorescence polarization (FP)-based competitive binding assays. We found that the methylpiperidinyl moiety of BI2536 could be replaced by a lysine residue (Lys(BI2536), 2, Fig. 1) without loss of Plk1-binding affinity (see ESI, Scheme S1 and Fig. S1†). Accordingly, we designed a series of FITC-tethered BI2536 probes (3–5) (Fig. 1).
The synthesis of 3–5 was approached using NovaSyn® TGR resin and standard Fmoc-based solid-phase peptide synthesis (SPPS) (Scheme S2†).
During the synthesis of 3, we observed M + 2 and M − 17 peaks in the high performance liquid chromatography (HPLC)-purified product. We hypothesized that this might result from an Edman degradation-type reaction during the SPPS (ESI Scheme S3†).16 In order to confirm whether this undesired side reaction was occurring, the C-terminal carboxylic acid derivative 6 was synthesized by solution chemistries (Scheme S4†). We then allowed a solution of 6 to sit at room temperature in HPLC eluent solvent (0.1% TFA containing MeCN/H2O). Monitoring the solution by HPLC over time revealed that 6 slowly converted to the putative cyclized product 7 (Fig. S2†). In light of this instability, further work with probes 3 and 6 was discontinued and efforts were focused on probes 4 and 5.
The probes were intended to be used for FP assays. However, during assay validation we found that signal intensities were extremely low (data not shown). We hypothesized that fluorescence quenching might be arising from interactions between the BI2536 and fluorescein isothiocyanate (FITC) moieties. This did not appear to occur through a Förster resonance energy transfer (FRET) mechanism, because the absorbance of BI2536 moiety does not overlap with FITC excitation at 485 nm (see ESI, Fig. S3 and S4†). We realized that the phenomenon could lead to fluorescence recovery on binding of the probe to Plk1 through disengagement of the BI2536 from the FITC and that this could potentially be used to measure the affinities of small molecule inhibitors by competitive displacement of the probe (Fig. 2). A similar phenomenon has been reported using fluorescein-labelled VX-680, which is an ATP-competitive Aurora-A specific inhibitor.17
We compared the abilities of 4 and 5 to serve as fluorescence recovery probes by examining fluorescence as a function of concentration. We found that both 4 and 5 showed identical Kd values (21 ± 0.37 nM for 4 and 21 ± 0.26 nM for 5) and achieved the same level of maximum fluorescence intensity (Fig. 3 and Table 1). We decided to use 4 for further experiments.
| Reported Plk1 IC50 values (nM) | IC50 values determined by a kinase assay* (nM) | IC50 values determined in this work (nM) | |
|---|---|---|---|
| BI2536 (1) | 0.83 (ref. 13) | 19 ± 1.0 | 25 ± 0.83 |
| 2 | — | 17 ± 2.2 | 12 ± 0.34 |
| BI6727 | 0.87 (ref. 20) | — | 31 ± 0.67 |
| Wortmannin | 5.8 (ref. 21) | — | 21 ± 1.1 |
| SBE13 | 0.2 (ref. 22) | — | n.d. |
| ON-01910 | 9–10 (ref. 23) | — | n.d. |
| Poloxin | n.a. | — | n.d. |
| Poloppin II | n.a. | — | n.d. |
In fluorescence-based competitive binding assays, it is important to employ the minimum effective concentration of probe.18,19 In order to determine this value, we performed experiments that measured fluorescence intensity as a function of Plk1 protein with various concentrations of 4. We found that Kd values (32 ± 0.78 nM, 33 ± 0.98 nM, 32 ± 1.1 nM, and 38 ± 1.8 nM for 10, 20, 30, and 40 nM of 4, respectively) were similar over the range of probe concentrations examined (Fig. S6a†). The 20 nM and 30 nM probe concentrations provided similar fluorescence intensities and curve shapes and we selected 20 nM as the minimum acceptable probe concentration. In further assays, we determined that almost identical results were obtained using several incubation times (42 ± 1.5 nM, 34 ± 1.1 nM, and 32 ± 1.5 nM for 15, 30, and 60 minutes, respectively) and we selected as our final assay conditions, 30 minute incubations using 20 nM of probe 4 (Fig. S6b†).
Having established assay conditions, we determined the IC50 values of a variety of Plk1 inhibitors (Fig. 4). These included Type 1 inhibitors, which target the ATP-binding pocket in the active form of the enzyme (BI2536, BI6727, and Wortmannin);13,20,21 Type 2 inhibitors, which target both the ATP-binding and allosteric pockets in the inactive form of the enzyme (SBE13);22 a non-ATP-competitive inhibitor, which targets the substrate binding-pocket (ON01910)23 and PBD-binding inhibitors (Poloxin and Poloppin II).24,25 We found that the Type 1 inhibitors could selectively inhibit probe binding to the Plk1 KD. The Type 2 inhibitors, non-ATP-competitive inhibitors, and PBD-binding inhibitors did not influence probe binding (Fig. 4). Results for the Type 1 inhibitors are consistent with reported IC50 values obtained from kinase assays (Table 1).
In our current work, we designed a series of probes based on the active pharmacophore of BI2536 tethered to a FITC moiety. Our original intent was to use these in competitive FP-based binding assays to determine binding affinities of kinase inhibitors. This methodology would be much more straightforward than is possible with enzymatic kinase assays. We found that the probes did provide a fluorescence-based measure of binding affinity. However, binding of the probes to the protein was evidenced by intramolecular fluorescence quenching, rather than fluorescence polarization. We found that the assay is able to provide IC50 values of Type 1 kinase inhibitors that are in accordance with values obtained from enzymatic assays. Because the assay is insensitive to other classes of kinase inhibitors, it could potentially be used in conjunction with enzymatic kinase assays to distinguish Type 1 inhibitors from these other classes. Importantly, we believe that the assay may afford a facile means for initial screening of Type 1 ATP-competitive Plk1 inhibitors that offers distinct advantages over kinase assays in terms of cost, speed and ease of handling.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the NIH Intramural Program, Center for Cancer Research, National Cancer Institute (ZIA BC 006198) and by a JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at NIH (KT).Notes and references
- V. Archambault, G. Lepine and D. Kachaner, Oncogene, 2015, 34, 4799 CrossRef CAS.
- R. F. Lera, G. K. Potts, A. Suzuki, J. M. Johnson, E. D. Salmon, J. J. Coon and M. E. Burkard, Nat. Chem. Biol., 2016, 12, 411 CrossRef CAS.
- B. D. Cholewa, X. Liu and N. Ahmad, Cancer Res., 2013, 73, 6848 CrossRef CAS.
- K. S. Lee, T. R. Burke, Jr, J.-E. Park, J. K. Bang and E. Lee, Trends Pharmacol. Sci., 2015, 36, 858 CrossRef CAS.
- A. E. Elia, P. Rellos, L. F. Haire, J. W. Chao, F. J. Ivins, K. Hoepker, D. Mohammad, L. C. Cantley, S. J. Smerdon and M. B. Yaffe, Cell, 2003, 115, 83 CrossRef CAS.
- J. Xu, C. Shen, T. Wang and J. Quan, Nat. Struct. Mol. Biol., 2013, 20, 1047 CrossRef CAS.
- A. Berg and T. Berg, ChemBioChem, 2016, 17, 650 CrossRef CAS.
- J. E. Park, D. Hymel, T. R. Burke, Jr and K. S. Lee, F1000Research, 2017, 6, 1024, DOI:10.12688/f1000research.11398.1.
- H. Ma, S. Deacon and K. Horiuchi, Expert Opin. Drug Discovery, 2008, 3, 607 CrossRef CAS.
- (a) K. M. Sakamoto, K. B. Kim, A. Kumagai, F. Mercurio, C. M. Crews and R. J. Deshaies, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 8554 CrossRef CAS; (b) H. Gao, X. Sun and Y. Rao, ACS Med. Chem. Lett., 2020, 11, 237 CrossRef.
- G. H. Iyer, P. Taslimi and S. Pazhanisamy, Anal. Biochem., 2008, 373, 197 CrossRef CAS.
- C. S. Lebakken, H. C. Kang and K. W. Vogel, J. Biomol. Screening, 2007, 12, 828 CrossRef CAS.
- M. Steegmaier, M. Hoffmann, A. Baum, P. Lenart, M. Petronczki, M. Krssak, U. Guertler, P. Garin-Chesa, S. Lieb, J. Quant, M. Grauert, G. R. Adolf, N. Kraut, J.-M. Peters and W. J. Rettig, Curr. Biol., 2007, 17, 316 CrossRef CAS.
- G. Budin, K. S. Yang, T. Reiner and R. Weissleder, Angew. Chem., Int. Ed. Engl., 2011, 50, 9378 CrossRef CAS.
- Z. Zhang, N. Kwiatkowski, H. Zeng, S. M. Lim, N. S. Gray, W. Zhang and P. L. Yang, Mol. BioSyst., 2012, 8, 2523 RSC.
- P. Edman, Acta Chem. Scand., 1950, 4, 283 CrossRef CAS.
- A. F. Slatter, S. Campbell and R. M. Angell, J. Biomol. Screening, 2013, 18, 219 CrossRef CAS.
- N. J. Moerke, Curr. Protoc. Chem. Biol., 2009, 1, 1 CrossRef.
- F. Ansideri, A. Lange, A. El-Gokha, F. M. Boeckler and P. Koch, Anal. Biochem., 2016, 503, 28 CrossRef CAS.
- D. Rudolph, M. Steegmaier, M. Hoffmann, M. Grauert, A. Baum, J. Quant, C. Haslinger, P. Garin-Chesa and G. R. Adolf, Clin. Cancer Res., 2009, 15, 3094 CrossRef CAS.
- Y. Liu, K. R. Shreder, W. Gai, S. Corral, D. K. Ferris and J. S. Rosenblum, Chem. Biol., 2005, 12, 99 CrossRef CAS.
- S. Keppner, E. Proschak, G. Schneider and B. Spaenkuch, ChemMedChem, 2009, 4, 1806 CrossRef CAS.
- K. Gumireddy, M. V. R. Reddy, S. C. Cosenza, R. B. Nathan, S. J. Baker, N. Papathi, J. Jiang, J. Holland and E. P. Reddy, Cancer Cell, 2005, 7, 275 CrossRef CAS.
- W. G. Reindl, J. P. Yuan, A. Kramer, K. Strebhardt and T. Berg, Chem. Biol., 2008, 15, 459 CrossRef CAS.
- A. J. Narvaez, S. Ber, A. Crooks, A. Emery, B. Hardwick, E. Guarino Almeida, D. J. Huggins, D. Perera, M. Roberts-Thomson, R. Azzarelli, F. E. Hood, I. A. Prior, D. W. Walker, R. Boyce, R. G. Boyle, S. P. Barker, C. J. Torrance, G. J. McKenzie and A. R. Venkitaraman, Cell Chem. Biol., 2017, 24, 1017 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ay01223h |
| This journal is © The Royal Society of Chemistry 2020 |



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M) and the
M) and the 