Multiplex-invasive reaction-assisted qPCR for quantitatively detecting the abundance of EGFR exon 19 deletions in cfDNA†
Abstract
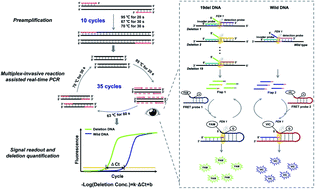
Exon 19 deletions (19-Del) on the epidermal growth factor receptor (EGFR) gene are vital biomarkers for guiding tyrosine kinase inhibitor (TKI) treatment and the diagnosis of non-small cell lung cancer (NSCLC). However, it is difficult for conventional qPCR to quantitatively detect all 19-Del targets of EGFR, especially for cfDNA samples. Herein, a multiplex invasive reaction-assisted qPCR was proposed by employing a multiplex invasive reaction to distinguish 19-Del DNA targets from wild DNA targets and report them with different fluorescence signals in each PCR cycle. As all 19-Del targets have the same amplification efficiency and very similar invasive reaction efficiencies, the 19-Del abundance in a sample could be quantified by using the difference between the Ct values (ΔCt) of the deletion targets and the wild targets without the requirement of a standard calibration curve. Combining the high sensitivity of PCR and the high specificity of the invasive reaction, this method can detect 10 copies of the deletion targets and lower than 0.1% deletion abundance. The results were 100% consistent with ARMS-PCR for the 38 tumor tissues tested and were in good agreement with next-generation sequencing for quantifying the abundance of EGFR 19-Del in 15 cfDNA samples, showing the great potential of the method for liquid biopsies.


 Please wait while we load your content...
Please wait while we load your content...