Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
RespiDisk: a point-of-care platform for fully automated detection of respiratory tract infection pathogens in clinical samples†
Markus
Rombach
*a,
Sebastian
Hin
 a,
Mara
Specht
a,
Benita
Johannsen
a,
Jan
Lüddecke
a,
Nils
Paust
a,
Mara
Specht
a,
Benita
Johannsen
a,
Jan
Lüddecke
a,
Nils
Paust
 ab,
Roland
Zengerle
ab,
Louis
Roux
c,
Thomas
Sutcliffe
c,
Johannes R.
Peham
d,
Christopher
Herz
d,
Marcus
Panning
e,
Oliver
Donoso Mantke
f and
Konstantinos
Mitsakakis
ab,
Roland
Zengerle
ab,
Louis
Roux
c,
Thomas
Sutcliffe
c,
Johannes R.
Peham
d,
Christopher
Herz
d,
Marcus
Panning
e,
Oliver
Donoso Mantke
f and
Konstantinos
Mitsakakis
 *ab
*ab
aHahn-Schickard, Georges-Koehler-Allee 103, 79110 Freiburg, Germany. E-mail: Markus.Rombach@Hahn-Schickard.de
bLaboratory for MEMS Applications, IMTEK – Department of Microsystems Engineering, University of Freiburg, Georges-Koehler-Allee 103, 79110 Freiburg, Germany. E-mail: Konstantinos.Mitsakakis@Hahn-Schickard.de
cLifeAssay Diagnostics (Pty) Ltd, Westlake Business Park, 7945 Cape Town, South Africa
dAIT Austrian Institute of Technology, Molecular Diagnostics, Center for Health and Bioresources, Giefinggasse 4, 1210 Vienna, Austria
eInstitute of Virology, Medical Center – University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Hermann-Herder-Strasse 11, 79104 Freiburg, Germany
fQuality Control for Molecular Diagnostics (QCMD), Unit 5, Technology Terrace, Todd Campus, West of Scotland Science Park, Glasgow G20 0XA Scotland, UK
First published on 2nd September 2020
Abstract
We present the RespiDisk enabling the fully automated and multiplex point-of-care (POC) detection of (currently) up to 19 respiratory tract infection (RTI) pathogens from a single sample based on reverse transcriptase polymerase chain reaction (RT-PCR). RespiDisk comprises a RTI-specific implementation of the centrifugal microfluidic LabDisk platform and combines new and existing advanced unit operations for liquid control, thereby automating all assay steps only by a spinning frequency and temperature protocol in combination with the use of a permanent magnet for in situ bead handing. The capabilities of the system were demonstrated with 36 tested quality samples mimicking clinical conditions (clinical and/or cultured material suspended in transport medium or synthetic bronchoalveolar lavage (BAL)) from past external quality assessment (EQA) panels covering 13 of the 19 integrated RTI detection assays. In total, 36 samples × 19 assays/sample resulting in 684 assays were performed with the RespiDisk, and its analytical performance was in full agreement with the routine clinical workflow serving as reference. A strong feature of the platform is its universality since its components allow the simultaneous detection of a broad panel of bacteria and viruses in a single run, thereby enabling the differentiation between antibiotic-treatable diseases. Furthermore, the full integration of all necessary biochemical components enables a reduction of the hands-on time from manual to automated sample-to-answer analysis to about 5 min. The study was performed on an air-heated LabDisk Player instrument with a time-to-result of 200 min.
Introduction
Lower respiratory tract infections (RTIs) are a major cause of hospitalizations among adults and children globally, resulting in a broad range of disease severities, from a common cold to death. Severe RTIs can lead to prolonged hospitalization, which is a burden of the healthcare and insurance systems.1 Upper RTIs, such as pharyngitis, rhinitis, sinusitis, otitis media and others are some of the most common reasons for antibiotic prescriptions, frequently unnecessary because of viral causative pathogens. As a consequence, this promotes the rise of antibiotic resistances.2,3In several occasions, such as in Emergency Rooms or in remote clinics, the diagnosis of RTI pathogens can be especially challenging4 as it must be done rapidly and at the patient side instead of following the routine procedure of sample collection and dispatching it to a central laboratory for analysis. Therefore, on-site testing enables immediate patient management at the point of care.5
In addition, RTIs are mostly of viral or bacterial nature and may be caused by one or more candidate pathogens from each of these two pathogen groups6,7 (fungal and parasitic lung infections are less-common8,9). Given the fact that treatment with antibiotics is effective only in case of bacterial infections, it becomes imperative that the diagnostic method is able to differentiate between viruses and bacteria but also to identify the specific pathogen that caused the RTI as this will define the targeted prescription.
However, in several cases, empirical or merely symptomatic diagnosis is still used due to the lack of suitable tools and/or the pressure of time (e.g. circumventing culture-based methods due to time constraints).10 Furthermore, indicators supporting the symptomatic diagnosis such as the medical history, the duration of symptoms, the presence of fever or the auscultation of the lung, lack sufficient specificity to distinguish between a viral or bacterial infection.
In addition, it can often be the case that RTIs exist simultaneously with underlying co-morbidities, such as tropical diseases in Low- and Middle-Income Countries,11,12 cancer and HIV,12,13 or sudden outbreaks of epidemics/pandemics, especially of respiratory nature like the SARS-CoV-2.14,15 In such occasions, the epidemiological landscape becomes even more complicated, requiring accurate diagnosis of the RTI-causative pathogen for proper management of patients with co-morbidities.16
In this context, nucleic acid amplification testing (NAAT)17–19 is a promising candidate to overcome the aforementioned challenges and offer high sensitivity and specific pathogen identification that can contribute to a reliable downstream patient management. Especially when conducted on an automated platform, NAAT offers the potential for shifting the diagnosis to the patient side. Even though some commercial systems exist for the detection of RTI pathogens, some of them such as the GeneXpert® (Cepheid), the ID NOW™ (Abbott), the cobas® Liat (Roche Diagnostics), the GenePOC™ (GenePOC Inc.) and the Liaison® MDX (DiaSorin Inc.) do not aim at a pathogen panel but instead, at single or very few pathogens. On the other hand, those platforms that aim at a broader panel such as the FilmArray® (bioMérieux), the Verigene® (Luminex Corp.), the VerePLEX™ Biosystem (Veredus Laboratories), the QIAstat-Dx (QIAGEN) and the Unyvero A50 (Curetis) use technically complex cartridges, costly and bulky equipment, which make them more suitable for laboratory settings than for point-of-care use.20
The RespiDisk system we use in the present study is based on previously presented centrifugal microfluidic platform, the LabDisk,21–23 which combines: (i) molecular diagnostics (real-time PCR); (ii) a two layer monolithically and easy-to-manufacture disposable cartridge, aiming at scalable cost-effective manufacturing; (iii) fully integrated reagents for a sample-to-answer analysis, aiming to minimize hands-on intervention; (iv) a point-of-care compatible (compact, lightweight) processing device (the LabDisk Player); (v) a broad pathogen panel currently consisting of 4 bacteria and 15 viruses/subtypes (which can be extended), aiming to demonstrate the system's capability to detect and differentiate main pathogens that lead to RTIs.
The aim of this work was to characterize the analytical performance of the RespiDisk platform by (i) testing several different respiratory tract bacteria- and virus-positive as well as negative samples, and (ii) comparing the results from the RespiDisk with the results from a routine clinical diagnostic method available at the Institute of Virology, University Medical Center Freiburg, Germany. The results constitute a proof-of-principle of the RespiDisk microfluidic and analytical operation using 36 samples from past External Quality Assessment (EQA) panels from QCMD (Glasgow, Scotland, UK).24
Materials & methods
Nucleic acid extraction chemistry (RespiDisk)
A pre-evaluation of three commercially available magnetic bead based extraction kits was conducted with representative RTI viral (Influenza A (H3N2)) and bacterial targets (Bordetella pertussis (Gram-negative) & Streptococcus pneumoniae (Gram-positive)). The kits were rated by overall extraction performance, downscaling of volumes for microfluidic integration and workflow complexity (data not shown).The MagSi-DNA mf kit (cat. no. MD0200010002, magtivio B.V., NL) was selected because it provided the best trade-off between extraction performance and microfluidic integration into the RespiDisk platform. Extraction reagents and volumes as integrated into the RespiDisk are displayed in Table 1, adapted for analyzing a 200 μL patient sample (transport medium).
| Reagent | Volume [μL] | Tolerance | Pre-storage method |
|---|---|---|---|
| Extraction reagents | |||
| Lysis buffer | 150 | ±5% | Stickpack26 |
| Binding buffer | 440 | ±5% | Stickpack26 |
| Washing buffer I | 200 | ±5% | Stickpack26 |
| Washing buffer II | 200 | ±5% | Stickpack26 |
| Elution buffer | 180 | ±5% | Stickpack26 |
| Magnetic beads | 20 | ±2% | Air-dried with trehalose (0.5 M final conc.) |
| Amplification reagents | |||
| TaqMan® Lyo-ready 1-Step qPCR Master Mix (3.5× conc.; Thermo Fisher Scientific, USA) | 46 | ±3% | Lyophilized |
| Oligomixes (per rxn chamber; Pathofinder) | 3 | ±5% | Air-dried with trehalose (0.33 M final conc.) |
Qualitative diagnostic reverse transcriptase (RT)-PCR assays (RespiDisk)
To cover a broad range of organisms for differential diagnostics, qualitative multiplex RT-PCR assay panels were screened, whereas the RealAccurate® Respiratory Quadruplex Assays from Pathofinder, using standardized TaqMan probe technology, were selected for integration due to sufficient coverage of the desired targets.25 The panel consists of six quadruplex RT-PCR assays equipped with four fluorescence labels (FAM, MAX/Yakima yellow, Texas Red and TYE665) allowing for detection and discrimination between 19 organisms and their subtypes, as listed in Table 2. The primers and probes for each quadruplex assay are included in the respective oligomixes.| Quadruplex assay and product code | Pathogens (viruses) | Subtype/strain | Fluor. label | Target genes (regions of interest) |
|---|---|---|---|---|
| Influenza PF0970-R | Influenza virus A | H3N2, H1N1 | FAM | Matrix protein gene |
| Influenza virus B | MAX | Nucleoprotein gene | ||
| Influenza virus A | H1N1-pdm09 | Texas Red | Neuraminidase gene | |
| Corona PF0971-R | Coronavirus | 229E | FAM | Nucleocapsid protein gene |
| Coronavirus | OC43 | MAX | Nucleocapsid protein gene | |
| Coronavirus | HKU1/NL63 | Texas Red | Nucleocapsid protein gene | |
| Parainfluenza PF0972-R | Parainfluenza virus | 1 | FAM | Hemagglutinin-neuraminidase gene |
| Parainfluenza virus | 2/4 | MAX | Hemagglutinin-neuraminidase gene/major nucleocapsid protein gene | |
| Parainfluenza virus | 3 | Texas Red | Hemagglutinin-neuraminidase gene | |
| RSV/hMPV PF0973-R | Respiratory syncytical virus (RSV) | A | FAM | Major nucleocapsid protein gene |
| Respiratory syncytical virus | B | MAX | Major nucleocapsid protein gene | |
| Human Metapneumovirus (hMPV) | A/B | Texas Red | Major nucleocapsid protein gene | |
| Adeno/Boca/Rhino/Entero PF0974-R | Adenovirus | A–F and 46 & 49 | FAM | Hexon gene |
| Bocavirus | 1–4 | MAX | Noncapsid gene | |
| Rhinovirus/enterovirus | Texas Red | 5′ untranslated region |
| Quadruplex assay and product code | Pathogens (bacteria) | Fluor. label | Target genes (regions of interest) | |
|---|---|---|---|---|
| Atypical bacteria PF-0966-R | Mycoplasma pneumoniae | FAM | Cytadhesin P1 (P1) gene | |
| Chlamydophila pneumoniae | Yakima yellow | MOMP gene | ||
| Legionella pneumophila | Texas Red | MIP gene | ||
| Bordetella pertussis | TYE665 | Insertion sequence IS481 |
Pre-storage and release on-demand of assay reagents (RespiDisk)
Extraction buffers (Table 1) were pre-stored in stickpacks. The latter were heated to 60 °C during the opening step (ESI, Table S1†) to weaken the peel seam and finally opened centrifugally in parallel at a rotational frequency of fStickpacks = 60 Hz releasing the liquids into the respective, radially outward connected chambers. Magnetic beads were mixed with trehalose to a final concentration of 0.5 M, dispensed into the designated chamber and air-dried at room temperature for 1 h.27 Beads were re-hydrated upon contact with the lysate-binding buffer-mix.RT-PCR reagents (TaqMan® Lyo-ready 1-Step qPCR Master Mix; 3.5×) were lyophilized (Thermo Fisher Scientific, USA) as a single cake (46 μL) for a total reaction volume of 160 μL and manually integrated into the RespiDisk. Re-hydration occurred upon contact with the eluted nucleic acids (NAs). The same RT-PCR reagents were used for both, viral and bacterial assays.
The respective oligomixes were mixed with trehalose to a final concentration of 0.33 M, dispensed into the respective chamber(s) and air-dried at room temperature for 1 h.27
Six reaction chambers were used for the six quadruplex panels shown in Table 2. More target assays as well as controls can be added at a later stage, since the RespiDisk layout comprises 12 reaction chambers. The disk was packaged in an aluminium pouch with nitrogen gas and desiccant and can be stored at room temperature.
The automated RespiDisk platform
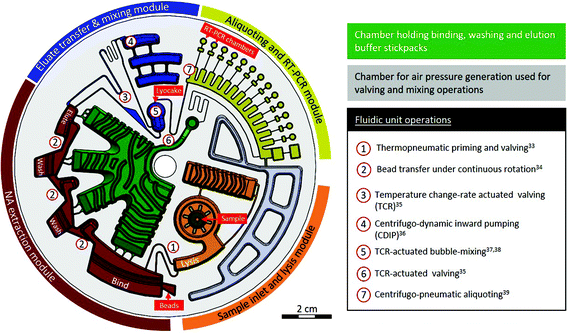
The nucleic acid testing workflow was implemented on the RespiDisk by developing new unit operations and connecting them with the latest ones into process chains28,29 for highly improved performance regarding reduction of inhibitors, increase of the homogeneity in the mixing process of PCR reagents, increase in transfer yields between operations, bubble-free detection of fluorescence signals and an overall increase of robustness. All pneumatic operations for pumping and valving have been designed with the support of network simulations as introduced by Schwarz et al.30 and described in detail by Hess et al.31 An overview of the fluidic network is provided in Fig. 1. Processes for RespiDisk fabrication comprise the thermoforming of the RespiDisk from a polycarbonate (PC) film,32 loading with assay reagents (Table 1), sealing with a pressure-sensitive adhesive film (9795R, 3 M, USA) and packaging in an aluminium pouch filled with nitrogen and desiccant.To start a run, the user needs to place the disk onto the rotor in the LabDisk Player (QIAGEN Lake Constance, Germany), apply the 200 μL sample, close the lid and press the start button. Once the RespiDisk spins, the stickpacks open and release the corresponding reagents into the respective reservoirs, while at the same time the sample is transferred into the lysis reservoir. There, it is mixed with the lysis buffer for 10 min before the lysate is transferred into the binding chamber by using the thermopneumatic pumping principle developed by Abi-Samra et al.33 for siphon priming with a second gas exchange feedback channel above the siphon to ensure complete transfer. The lysate is mixed with rehydrated magnetic silica beads and the binding buffer. A typical bind-wash-elute protocol is applied while transferring the magnetic beads between chambers under controlled rotation,34 with the beads finally ending up in the elution chamber, releasing the collected NAs in the aqueous elution buffer. The eluate is transferred into a mixing chamber by TCR valving35 and centrifugo-dynamic inward pumping.36 The lyophilized RT-PCR reagents (lyocake) are rehydrated by bubble based mixing37,38 with the pneumatic overpressure generated by thermo-pneumatics. Subsequently, the mixture is distributed into the 12 × 10 μL reaction chambers by TCR valving.35 Both TCR valving operations in the RespiDisk are based on a temperature-induced underpressure generation in all connected chambers downstream of the liquid's meniscus position, which is used to pull the liquid over siphon crests and thus enable priming and liquid transfer. After the last valving operation, the liquid is automatically metered and aliquoted39 (13 chambers in total where the 1st chamber is sacrificial to collect pre- and post-flow without holding an oligomix). Upon liquid contact, the oligomixes are re-hydrated in each reaction chamber followed by initiation of the RT-PCR reaction with an initial RT step at 60 °C and subsequent 45 cycles between 60 and 95 °C.
More details on the fluidic and temperature protocol are given in the ESI, Table S1.† Representative real-time amplification curves are shown in ESI, Fig. S1.† The processing time is currently 50 min for the extraction and 150 min for the RT-PCR reaction.
Routine clinical workflow
Nucleic acids were extracted from respiratory samples (200 μL volume) using the QIAmp MinElute Virus kit on a QIAcube (both QIAGEN, Hilden, Germany) according to the recommendations of the manufacturer. For amplification and detection, the FTD respiratory pathogens 21 kit (Fast Track Diagnostics, Junglingster, Luxemburg) was used for all viral pathogens and complemented with in-house assays for the bacterial pathogens (except for L. longbeachae) by the Institute of Virology Freiburg, Germany (method published by Huzly et al.40). These in-house assays have all been extensively validated and were run in parallel using the same 96 well plate, enzyme chemistry and cycling protocol as the FTD assays. RT-PCRs were carried out on an ABI 7500 machine (Applied Biosystems, Wiesbaden, Germany) as recommended.Design of the validation study with past QCMD EQA programme samples
We selected four different respiratory pathogen panels from past EQA schemes24 provided by QCMD (Quality Control for Molecular Diagnostics, Glasgow, Scotland, UK), a not-for-profit and independent international provider for EQA programmes covering a comprehensive range of infectious diseases. Such past EQA panels can be used to support internal assay evaluations. Our four selected panels consisted of a total number of 37 samples (Table 3):(1) Respiratory I 18S (multiple pathogen panel with various influenza A & B and respiratory syncytial virus strains; 10 samples; QCMD Ref code: RESPI18)
(2) Respiratory II 17S (multiple pathogen panel with focus on human metapneumovirus, respiratory adenoviruses, rhinoviruses, coronaviruses, enterovirus and parainfluenza viruses; 12 samples; QCMD Ref code: RESPII17)
(3) Legionella pneumophila DNA 18S (single pathogen panel; 10 samples; QCMD Ref code: LPDNA18)
(4) Mycoplasma pneumoniae 18S (single pathogen panel; 5 samples; QCMD Ref code: MP18)
| QCMD panel | Sample | Pathogen | Genome | Detected in QCMD programme [%] | C Q (clinical routine) [cycle] | C Q (RespiDisk) [cycle] | ||
|---|---|---|---|---|---|---|---|---|
| a This was a specificity sample included in the panel and was expected to be returned as “negative”. 91.5% of submitted datasets correctly detected this “non-pneumophila species” specificity sample as negative within the LPDNA18S programme. | ||||||||
| RESPI18S | 1 | Influenza virus Type A (H3N2) | RNA | 98.8 | 31 | ✓ | 35 | ✓ |
| 2 | Influenza virus Type B (Victoria) | RNA | 87.7 | 34 | ✓ | 35 | ✓ | |
| 3 | Influenza virus Type A (H1N1 pdm09) | RNA | 98.8 | 32 | ✓ | 35 | ✓ | |
| 4 | Influenza virus Type A (H3N2) | RNA | 98.8 | 33 | ✓ | — | ✗ | |
| 5 | RSV Type A | RNA | 96.4 | 33 | ✓ | 35 | ✓ | |
| 6 | Influenza virus Type B (Yamagata) | RNA | 87.7 | 32 | ✓ | — | ✗ | |
| 7 | RSV Type A | RNA | 95.3 | 33 | ✓ | 37 | ✓ | |
| 8 | Negative | — | 99.4 | — | ✓ | — | ✓ | |
| 9 | RSV Type B | RNA | 97.0 | 31 | ✓ | 32 | ✓ | |
| 10 | RSV Type B | RNA | 98.2 | 29 | ✓ | 27 | ✓ | |
| RESPII17S | 1 | Coronavirus – NL63 | RNA | 100 | 29 | ✓ | 30 | ✓ |
| 2 | Human MPV Type A2 | RNA | 77.6 | — | ✗ | — | ✗ | |
| 3 | Human MPV Type A2 | RNA | 98.4 | 33 | ✓ | 37 | ✓ | |
| 4 | Adenovirus type 1 | RNA | 96.7 | 33 | ✓ | 33 | ✓ | |
| 5 | Negative | — | 97.6 | — | ✓ | — | ✓ | |
| 6 | RV type 5 | RNA | 74.2 | 29 | ✓ | 36 | ✓ | |
| 7 | Coronavirus – NL63 | RNA | 90.1 | 33 | ✓ | 37 | ✓ | |
| 8 | RV type 5 | RNA | 81.7 | 31 | ✓ | 35 | ✓ | |
| 9 | Enterovirus 68 | RNA | 52.2 | 31 | ✓ | 36 | ✓ | |
| 10 | Human MPV Type A1 | RNA | 91.9 | 34 | ✓ | 33 | ✓ | |
| 11 | Coronavirus – OC43 | RNA | 90.1 | 33 | ✓ | 35 | ✓ | |
| 12 | Parainfluenza Type 1 | RNA | 94.3 | 31 | ✓ | 32 | ✓ | |
| LPDNA18S | 1 | L. pneumophila sg6 | DNA | 100.0 | 32 | ✓ | 29 | ✓ |
| 2 | L. pneumophila sg6 | DNA | 99.3 | 30 | ✓ | 27 | ✓ | |
| 3 | L. pneumophila sg1 (ST62) | DNA | 92.8 | — | ✗ | — | ✗ | |
| 4 | L. pneumophila sg1 (ST62) | DNA | 99.3 | 34 | ✓ | 36 | ✓ | |
| 5 | L. pneumophila sg2–14 | DNA | 98.0 | 32 | ✓ | 30 | ✓ | |
| 6 | L. pneumophila sg2–14 | DNA | 98.7 | 29 | ✓ | 26 | ✓ | |
| 7 | Negative | — | 98.0 | — | ✓ | — | ✓ | |
| 8 | L. pneumophila sg1 (ST47) | DNA | 98.7 | 30 | ✓ | 29 | ✓ | |
| 9 | L. pneumophila sg1 (ST47) | DNA | 96.1 | — | ✗ | 36 | ✓ | |
| 10 | L. longbeachae | DNA | 91.5a | Not tested | Not tested | |||
| MP18S | 1 | M. pneumoniae | DNA | 96.1 | — | ✗ | 36 | ✓ |
| 2 | M. pneumoniae | DNA | 61.8 | — | ✗ | — | ✗ | |
| 3 | M. pneumoniae | DNA | 91.7 | 36 | ✓ | 36 | ✓ | |
| 4 | Negative | — | 97.5 | — | ✓ | — | ✓ | |
| 5 | M. pneumoniae | DNA | 96.1 | 35 | ✓ | 34 | ✓ | |
The samples of panels (1)–(3) contain cultured and/or clinical material suspended in transport medium matrix; panel (4) contains cultured and/or clinical material in transport medium as well as in synthetic bronchoalveolar lavage (BAL). All EQA panels are designed to include the most important and clinically relevant pathogens in line with current clinical practice. Samples were split into 200 μL aliquots and stored at −80 °C until the execution of each study. In case of the routine clinical workflow, the RT-PCR was carried out in triplicates. In case of the RespiDisk, each sample was tested with one disk, in order to retain sufficient material amount to perform both studies.
In total, 36 samples (all except for sample 10 of the LPDNA18S panel, L. longbeachae, because it was not part of the assays) were tested in two different studies:
(a) Routine clinical workflow with standard assays at the Institute of Virology, University Clinic of Freiburg40
(b) Fully automated workflow on the RespiDisk operated on the respective LabDisk Player.
Out of the 36 samples, 32 were expected as QCMD-positive and 4 as QCMD-negative.
Results & discussion
The RespiDisk was evaluated using main RTI-causing pathogens that are reported in literature and are available in commercial, quality assessment samples.7 Since only 1–2 samples per pathogen were tested, and no diverse sample matrices per pathogen were screened (for investigating dependency of sensitivity/specificity on sample matrix),41 the presented results do not aim to provide the full clinical sensitivity and the positive/negative predictive values of the RespiDisk. The scope of our work was to demonstrate the proof-of-principle of the RespiDisk analytical performance in detecting several different viruses and bacteria, analyze and discuss the true/false positive/negative results, and indicate the potential of the platform for future implementation in the field of diagnostics. Previous work by the authors has shown that the LabDisk platform is easily adaptable to different panels by exchanging primers while keeping the non-specific biochemical components and design the same.23 Thus, the RTI panel can be tailored at will, in order to address diverse end-user requirements and settings.The results are summarized in Table 3 showing the RT-PCR CQ values obtained for all samples using the RespiDisk and the routine clinical workflow. The results from the two studies are compared against the QCMD-expected results, but also between each other to gain information about the analytical performance of the RespiDisk platform and its potential for future diagnostic use. The column “Detected in QCMD programme” refers to the percentage of datasets that reported the correct result within the respective programme. For the multiple pathogen programmes (i.e. RESPI18S and RESPII17S), only datasets from laboratories that included the specific pathogens in their molecular workflows were used to calculate the percentage of correct results reported within these EQA programmes (# returned EQA datasets from participants: RESPI18S n = 171; RESPII17S n = 126; LPDNA18S n = 153; MP18S n = 204). In Table 4, we provide a summary of some quantitative performance characteristics of the two methods based on the experimental results.
| Routine clinical method | RespiDisk | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| TP | FN | TN | FP | TP | FN | TN | FP on sample level | FP on assay level | |
| Viral QCMD panels | 19/20 (95.0%) | 1/20 (5.0%) | 2/2 (100.0%) | 0/22 (0.0%) | 17/20 (85.0%) | 3/20 (15.0%) | 2/2 (100.0%) | 3/22 (13.6%) | 3/398 (0.8%) |
| Bacterial QCMD panels | 8/12 (66.7%) | 4/12 (33.3%) | 2/2 (100.0%) | 0/14 (0.0%) | 10/12 (83.3%) | 2/12 (16.7%) | 2/2 (100.0%) | 7/14 (50.0%) | 7/254 (2.8%) |
| All panels | 27/32 (84.4%) | 5/32 (15.6%) | 4/4 (100.0%) | 0/36 (0.0%) | 27/32 (84.4%) | 5/32 (15.6%) | 4/4 (100.0%) | 10/36 (27.8%) | 10/652 (1.5%) |
True positive results (TP)
As True Positive (TP) results we consider those that were QCMD-expected positive and the RespiDisk (or the routine clinical workflow) detected indeed positive. Examining the total number of samples, both the routine diagnostic method and the RespiDisk had the same performance as indicated by the 84.4% TP values in Table 4. Interestingly, the routine clinical workflow method performed better than the RespiDisk on the two viral panels (more TP and also lower CQ values except for the RESPI18S-sample #10). However, on the two bacterial panels the RespiDisk workflow exhibited more TP and lower CQ values than the routine clinical workflow in all cases except for the LPDNA18S-sample #4.False negative results (FN)
As False Negative (FN) results we consider those that were QCMD-expected positive but the RespiDisk (or the routine clinical workflow) detected as negative. Regarding the five false negative results on the RespiDisk, we can safely say that these were not due to any microfluidic or assay integration issues, because in confirmatory manual in-tube experiments (in which the exact same protocol and components were used, but just manually instead of the disk; data not shown) the same samples yielded also FN results (apart from the MP18S-sample #2 that was detected in tube but not on RespiDisk). Furthermore, in 3 out of 5 FN RespiDisk results (1 viral and 2 bacterial), the routine clinical workflow also yielded FN results.The observed FN results could be attributed to the EQA samples being very close to, or below the detection limit of the integrated components into this specific RespiDisk configuration and/or the routine clinical workflow:
• RESPI18S-sample #4 (FN in RespiDisk, TP in clinical method) is a 10× dilution of sample RESPI18S-01, the latter already being at high CQ values in RespiDisk method.
• RESPI18S-sample #6 (FN in RespiDisk, TP in clinical method) is a medium range sample.
• RESPII17S-sample #2 (FN in RespiDisk and clinical method) is a 100× dilution of sample #3, the latter already being at high CQ values in RespiDisk and clinical method. Notably, the sample #2 was found positive only by 77.6% of datasets reported from participants who included the specific pathogen in their molecular workflow.
• LPDNA18S-sample #3 (FN in RespiDisk and clinical method) is a 100× dilution of sample #4, the latter already being at high CQ values in RespiDisk and clinical method.
• MP18S-sample #2 (FN in RespiDisk and clinical method) is a 10× dilution of MP18S-sample #5, the latter already being at high CQ values in RespiDisk and clinical method.
• Other samples that were found FN with the clinical method but TP with RespiDisk method are LPDNA18S-sample #9 (which is a 100× dilution of sample #8) and MP18S-sample #1. These FN results could be attributed to the samples being close to the limit of detection of the assay or to some primer-probe mismatches rendering the assays refractory to amplification.
• Regarding the QCMD samples that “scored low” (<70%) by the participants of the QCMD collection studies: RESPII17S-sample #9 was detected by both methods; MP18S-sample #2 was missed by both the clinical method and the RespiDisk.
True negative and false positive results (TN and FP)
As True Negative (TN) results we consider those that were QCMD-expected negative and the RespiDisk (or the routine clinical workflow) detected indeed negative. Four samples were QCMD-expected negative and correctly detected in both the routine clinical workflow and the RespiDisk. As False Positive (FP) results we consider those that were QCMD-expected negative, but the RespiDisk (or the routine clinical workflow) detected as positive. Among the four aforementioned QCMD-expected negative samples, neither the RespiDisk nor the routine clinical workflow detected any FP (Table 4). However, due to the fact that each disk integrates 19 assays (Table 2), each of the 32 samples with one QCMD-expected positive result is expected to give also 18 negative assay results (and equivalently for the routine clinical workflow, where multiplex RT-PCR assays were used). Therefore, broadening our aforementioned definition, a result is considered as FP when at least one of the expected-negative assays is detected as positive (this is why we use the total number of samples of each panel as denominator in the columns “FP” in Table 4). We observed 10 such cases in the RespiDisk, namely samples 3, 5 & 7 of the RESPI18S panel and samples 1, 2, 4–6, 8 & 9 of the LPDNA18S panel (these samples also count as TP since we detected the QCMD-expected positive target as well). The first case of the RESPI18S panel can be attributed to contamination of the sample during handling. All 9 other cases can be attributed to fluorescence cross-talk from the green into the yellow channel (RESPI18S) and from the orange into the red channel (LPDNA18S).This bleed-over from one channel into the next one is a known issue on commercial PCR instruments when running multiplex assays,42 which is addressed by running specific color compensation kits. Unfortunately, this is not or hardly possible on our current R&D instrument. However, this issue will be addressed in the next generation instrument (LabDisk Player 2nd generation). Consequently, on a total assay rather than sample-level, 36 (samples) × 19 (assays/sample) = 684 assays were performed with the RespiDisk workflow, out of which the 32 were QCMD-expected positive and the 652 were QCMD-expected negative. Among the latter, 10/652 (1.5%) assays were FP (3/398 = 0.8% in the viral panel and 7/254 = 2.8% in the bacterial panel). On the other hand, the routine clinical workflow exhibited no FP results.
Conclusions
The proof-of-principle study successfully demonstrated the capacity of the developed RespiDisk to provide differentiation between viral and bacterial pathogens causing RTIs in a single test run. This will support the treatment decision process for the end-users in point-of-care settings such as Emergency Rooms and remote clinics at high or low/middle income countries. The RespiDisk microfluidic and analytical performance was verified using cultured and/or clinical material from EQA panels, which were also tested with a routine clinical diagnostic method. The results revealed that, compared to the QCMD-expected results, in 84.4% of the cases (27/32) the RespiDisk provided true positive results, exactly as many as the routine diagnostic method. This indicates a high potential of the RespiDisk for future implementation in the diagnostic practice, upon further prospective clinical assessment studies using real-time recruited patients, a broader spectrum of pathogens, and different sample matrices for sensitivity assessment. As a technology outlook, some key next steps are in progress: (i) reproducibility and production robustness assessment; (ii) determination and improvement of the analytical sensitivity levels, by incorporating a pre-amplification step followed by nested PCR; (iii) improvement of detectors to eliminate cross-talk; (iv) reduction of the time-to-result (currently 200 min) by performing in situ crude-sample preparation (instead of bind-wash-elute chemistry) and by implementing fast solid state heating in a next generation LabDisk Player, which achieves heating and cooling rates of ≥3 K s−1. This is expected to reduce the overall processing time to <1 h.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Research reported in this publication was supported by the Strategic Health Innovation Partnerships (SHIP) Unit of the South African Medical Research Council with funds received from Anglo American Platinum Limited and the South African Department of Science and Innovation. The article processing charge was funded by the Baden-Württemberg Ministry of Economics, Labour and Housing under the project CoviDisk-BW. The authors would like to acknowledge Verena Schilling from the Institute of Virology at the University of Freiburg as well as the Hahn-Schickard Lab-on-a-Chip Foundry Service for LabDisk production.References
- B. G. Williams, E. Gouws, C. Boschi-Pinto, J. Bryce and C. Dye, Lancet Infect. Dis., 2002, 2, 25–32 CrossRef.
- K. Mitsakakis, W. E. Kaman, G. Elshout, M. Specht and J. P. Hays, Future Microbiol., 2018, 13, 1157–1164 CrossRef CAS.
- A. van Belkum, T. T. Bachmann, G. Lüdke, J. G. Lisby, G. Kahlmeter, A. Mohess, K. Becker, J. P. Hays, N. Woodford, K. Mitsakakis, J. Moran-Gilad, J. Vila, H. Peter, J. H. Rex, Wm. M. Dunne Jr. and the JPIAMR AMR-RDT Working Group, Nat. Rev. Microbiol., 2019, 17, 51–62 CrossRef CAS.
- W. E. Kaman, G. Elshout, P. J. Bindels, K. Mitsakakis and J. P. Hays, Future Microbiol., 2016, 11, 607–610 CrossRef CAS.
- J. P. Hays, K. Mitsakakis, S. Luz, A. van Belkum, K. Becker, A. van den Bruel, S. Harbarth, J. H. Rex, G. S. Simonsen, G. Werner, V. Di Gregori, G. Lüdke, T. van Staa, J. Moran-Gilad, T. T. Bachmann and the JPIAMR AMR-RDT consortium, Eur. J. Clin. Microbiol. Infect. Dis., 2019, 38, 1015–1022 CrossRef.
- D. M. Musher and A. R. Thorner, N. Engl. J. Med., 2014, 371, 1619–1628 CrossRef CAS.
- M. Ieven, S. Coenen, K. Loens, C. Lammens, F. Coenjaerts, A. Vanderstraeten, B. Henriques-Normark, D. Crook, K. Huygen, C. C. Butler, T. J. M. Verheij, P. Little, K. Zlateva, A. van Loon, E. C. J. Claas, H. Goossens and the GRACE consortium, Clin. Microbiol. Infect., 2018, 24, 1158–1163 CrossRef CAS.
- A. Cheepsattayakorn and R. Cheepsattayakorn, BioMed Res. Int., 2014, 2014, 874021 Search PubMed.
- E. D. Lease and B. D. Alexander, Semin. Respir. Crit. Care Med., 2011, 32, 663–672 CrossRef.
- H. J. Zar, P. Jeena, A. Argent, R. Gie, S. A. Madhi and the Working Groups of the Paediatric Assembly of the South African Thoracic Society, South Afr. J. Epidemiol. Infect., 2009, 24, 25–36 CrossRef.
- V. D'Acremont, M. Kilowoko, E. Kyungu, S. Philipina, W. Sangu, J. Kahama-Maro, C. Lengeler, P. Cherpillod, L. Kaiser and B. Genton, N. Engl. J. Med., 2014, 370, 809–817 CrossRef.
- S. A. Madhi, C. L. Cutland, S. Downs, S. Jones, N. van Niekerk, E. A. F. Simoes and M. C. Nunes, Clin. Infect. Dis., 2018, 66, 1658–1665 CrossRef.
- M. L. McMorrow, S. Tempia, S. Walaza, F. K. Treurnicht, J. Moyes, A. L. Cohen, M. Pretorius, O. Hellferscee, N. Wolter, A. von Gottberg, A. Nguweneza, J. M. McAnerney, F. Naby, O. Mekgoe, M. Venter, S. A. Madhi and C. Cohen, Clin. Infect. Dis., 2019, 68, 773–780 CrossRef.
- X. Zhu, Y. Ge, T. Wu, K. Zhao, Y. Chen, B. Wu, F. Zhu, B. Zhu and L. Cui, Virus Res., 2020, 285, 198005 CrossRef CAS.
- J. A. Bengoechea and C. G. G. Bamford, EMBO Mol. Med., 2020, 12, e12560 CAS.
- F. Muro, R. Reyburn and H. Reyburn, Pneumonia, 2015, 6, 6–17 CrossRef.
- K. Loens, A. M. van Loon, F. Coenjaerts, Y. van Aarle, H. Goossens, P. Wallace, E. J. C. Claas, M. Ievena and on behalf of the GRACE Study Group, J. Clin. Microbiol., 2012, 50, 977–987 CrossRef CAS.
- D. Andrews, Y. Chetty, B. S. Cooper, M. Virk, S. K. Glass, A. Letters, P. A. Kelly, M. Sudhanva and D. Jeyaratnam, BMC Infect. Dis., 2017, 17, 671 CrossRef.
- M. Raymaekers, B. de Rijke, I. Pauli, A.-M. Van den Abeele and R. Cartuyvels, J. Clin. Virol., 2011, 52, 314–316 CrossRef CAS.
- K. Mitsakakis, V. D'Acremont, S. Hin, F. von Stetten and R. Zengerle, Microelectron. Eng., 2018, 201, 26–59 CrossRef CAS.
- G. Czilwik, T. Messinger, O. Strohmeier, S. Wadle, F. von Stetten, N. Paust, G. Roth, R. Zengerle, P. Saarinen, J. Niittymäki, K. McAllister, O. Sheils, J. O'Leary and D. Mark, Lab Chip, 2015, 15, 3749–3759 RSC.
- F. Stumpf, F. Schwemmer, T. Hutzenlaub, D. Baumann, O. Strohmeier, G. Dingemanns, G. Simons, C. Sager, L. Plobner, F. von Stetten, R. Zengerle and D. Mark, Lab Chip, 2016, 16, 199–207 RSC.
- S. Hin, B. Lopez-Jimena, M. Bakheit, V. Klein, S. Stack, C. Fall, A. Sall, K. Enan, S. Frischmann, L. Gillies, M. Weidmann, S. Goethel, V. Rusu, O. Strohmeier, N. Paust, R. Zengerle and K. Mitsakakis, Proceedings of the 21st International Conference on Miniaturized Systems for Chemistry and Life Sciences, μTAS 2017, USA; (full paper under revision in PLoS Negl. Trop. Dis., 2020).
- Quality Control for Molecular Diagnostics (QCMD), http://www.qcmd.org, (accessed July 2020).
- Pathofinder RealAccurate® Quadruplex real-time PCR, https://www.pathofinder.com/technology/realaccurate (accessed July 2020).
- T. van Oordt, Y. Barb, J. Smetana, R. Zengerle and F. von Stetten, Lab Chip, 2013, 13, 2888–2892 RSC.
- M. Rombach, D. Kosse, B. Faltin, S. Wadle, G. Roth, R. Zengerle and F. von Stetten, BioTechniques, 2014, 57, 151–155 CrossRef CAS.
- R. Gorkin, J. Park, J. Siegrist, M. Amasia, B. S. Lee, J.-M. Park, J. Kim, H. Kim, M. Madou and Y.-K. Cho, Lab Chip, 2010, 10, 1758–1773 RSC.
- O. Strohmeier, M. Keller, F. Schwemmer, S. Zehnle, D. Mark, F. von Stetten, R. Zengerle and N. Paust, Chem. Soc. Rev., 2015, 44, 6187–6229 RSC.
- I. Schwarz, S. Zehnle, T. Hutzenlaub, R. Zengerle and N. Paust, Lab Chip, 2016, 16, 1873–1885 RSC.
- J. F. Hess, S. Zehnle, P. Juelg, T. Hutzenlaub, R. Zengerle and N. Paust, Lab Chip, 2019, 22, 3745–3770 RSC.
- M. Focke, D. Kosse, D. Al-Bamerni, S. Lutz, C. Müller, H. Reinecke, R. Zengerle and F. von Stetten, J. Micromech. Microeng., 2011, 21, 115002 CrossRef.
- K. Abi-Samra, L. Clime, L. Kong, R. Gorkin III, T.-H. Kim, Y.-K. Cho and M. Madou, Microfluid. Nanofluid., 2011, 11, 643–652 CrossRef CAS.
- S. Hin, N. Paust, M. Rombach, J. Lüddecke, M. Specht, R. Zengerle and K. Mitsakakis, Proceedings of the 20th International Conference on Solid-State Sensors, Actuators and Microsystems & Eurosensors XXXIII, 2019, DOI:10.1109/TRANSDUCERS.2019.8808542.
- M. Keller, G. Czilwik, J. Schott, I. Schwarz, K. Dormanns, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2017, 17, 864–875 RSC.
- S. Zehnle, F. Schwemmer, G. Roth, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2012, 12, 5142–5145 RSC.
- S. Burger, M. Schulz, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2016, 16, 261–268 RSC.
- S. Hin, N. Paust, M. Keller, M. Rombach, O. Strohmeier, R. Zengerle and K. Mitsakakis, Lab Chip, 2018, 18, 362–370 RSC.
- D. Mark, P. Weber, S. Lutz, M. Focke, R. Zengerle and F. von Stetten, Microfluid. Nanofluid., 2011, 10, 1279–1288 CrossRef CAS.
- D. Huzly, S. Kurz, W. Ebner, M. Dettenkofer and M. Panning, J. Clin. Virol., 2015, 73, 47–51 CrossRef.
- J. C. Holter, F. Müller, O. Bjørang, H. H. Samdal, J. B. Marthinsen, P. A. Jenum, T. Ueland, S. S. Frøland, P. Aukrust, E. Husebye and L. Heggelund, BMC Infect. Dis., 2015, 15, 64 CrossRef.
- C. T. Wittwer, M. G. Herrmann, C. N. Gundry and K. S. Elenitoba-Johnson, Methods, 2001, 25, 430–442 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0an01226b |
| This journal is © The Royal Society of Chemistry 2020 |