Open Access Article
Open Access ArticlePaper-based analytical devices for colorimetric detection of S. aureus and E. coli and their antibiotic resistant strains in milk†
Muhammad
Asif
 abc,
Fazli Rabbi
Awan
abc,
Fazli Rabbi
Awan
 ab,
Qaiser Mahmood
Khan
*ab,
Bongkot
Ngamsom
ab,
Qaiser Mahmood
Khan
*ab,
Bongkot
Ngamsom
 c and
Nicole
Pamme
c and
Nicole
Pamme
 *c
*c
aNational Institute for Biotechnology and Genetic Engineering (NIBGE), Jhang Road, Faisalabad, Punjab 44000, Pakistan
bPakistan Institute of Engineering and Applied Sciences (PIEAS), Islamabad, Pakistan. E-mail: qk_5@yahoo.com
cDepartment of Chemistry and Biochemistry, Faculty of Science and Engineering, University of Hull, Hull, HU6 7RX, UK. E-mail: n.pamme@hull.ac.uk
First published on 10th August 2020
Abstract
Animal derived milk which is an important part of human diet due to its high nutritional value not only supports humans but also presents a growth environment for pathogenic bacteria. Milk may become contaminated with bacteria through udder infections or through contact within the dairy farm environment. Infections are treated with antibiotics, with β-lactams most commonly used in veterinary medicine. However, their frequent use leads to the emergence of β-lactam resistant bacterial strains, which causes difficulties in the treatment of infections in both humans and animals. Detection of pathogens as well as their antibiotic sensitivity is a pre-requisite for successful treatment and this is generally achieved with laboratory-based techniques such as growth inhibition assays, enzyme-linked immunosorbent assays (ELISA) or polymerase chain reactions (PCRs), which are unavailable in resource-limited settings. Here, we investigated paper-based analytical devices (μPADs) for the presumptive detection of Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) and their antibiotic resistant bacterial strains in milk samples. The μPADs were fabricated on filter paper using wax printing, and then impregnated with chromogenic substrates, which reacted with bacterial enzymes to form coloured products. Limits of detection of S. aureus and E. coli and their antibiotic resistant strains in milk samples were found to be 106 cfu mL−1. Enrichment of milk samples in a selective medium for 12 h enabled detection as low as 10 cfu mL−1. The paper devices tested on a set of 640 milk samples collected from dairy animals in Pakistan demonstrated more than 90% sensitivity and 100% selectivity compared to PCR, showing promise to provide inexpensive and portable diagnostic solutions for the detection of pathogenic bacteria in resource-limited settings.
Introduction
Milk is an essential component of the human diet and forms part of the official nutritional recommendations in many countries. It provides nutrients such as amino acids, vitamins and minerals that are difficult to obtain from dairy-free diets.1 In addition to being nutritious for humans and animals, milk is also a favourable growth medium for a range of bacteria. Dairy animals can harbour pathogenic bacteria and continuously excrete them in their secretions. Milk can thus become contaminated with pathogenic bacteria either through udder infections, i.e. mastitis,2 or via direct contact with contaminated materials.3 This contaminated milk is a potential source of transmission of foodborne pathogenic bacteria and its nutrient-rich environment allows bacteria to proliferate rapidly and produce toxins, thus making it an extremely vulnerable commodity for human health.4 In spite of strict quality controls, pasteurization of milk and improved health and well-being of dairy animals in developed countries, outbreaks of milk-borne illness have been reported.5–7 According to the Centers for Disease Control and Prevention (CDC): contaminated milk, and milk products, accounted for the most hospitalizations relating to food-borne illnesses.8 The European Union legislation (Regulation 853/2004) stipulates that only milk from healthy animals can be used for human consumption with a specific limit of <500 cfu mL−1 for S. aureus and <100 cfu mL−1 for E. coli (coliforms) in raw milk for drinking (Council Directive 92/46/EEC).9 The US Food and Drug Administration suggests lower levels of 10–100 organisms of S. aureus and E. coli in milk to be of public health concern.10 Compliance with such legislations can be achieved by constant vigilance over udder health and the quality of milk until it reaches the consumers. This is particularly challenging in resource-limited countries, where preventive measures are not always followed,8 and where reliable and affordable point-of-care diagnostic methods are lacking.Escherichia coli and Staphylococcus aureus are major milk-borne pathogenic bacteria that cause a wide range of diseases including mastitis, food poisoning and gastroenteritis in both humans and animals. Mastitis, the most costly and common disease in dairy animals, is mainly treated with beta-lactam antibiotics such as penicillin and cephalosporin, typically for more than 3 days.11,12 Frequent and prolonged use of antibiotics in feedstock animals is one of the major reasons for the emergence of antibiotic resistant bacterial strains. Beta-lactam resistant strains of both E. coli and S. aureus have been reported, posing a growing threat for the effective treatment of infections related to these pathogens in both humans and animals.13,14
Identification of pathogens and their antibiotic sensitivity is a pre-requisite to tailoring effective prevention and treatment plans. This is usually achieved by cell-culture based methods and growth inhibition assays which are cumbersome, expensive and time-consuming.15,16 Alternatively, PCR16 and ELISA17 techniques have been used for the detection of these pathogens in milk, but they require the availability of a dedicated laboratory facility, highly trained personnel, stable reagents, and multistep sample handling. Management of the logistical considerations associated with sample collection and transport to central laboratories is also required. Due to the lack of basic infrastructure, these techniques are of limited use in resource-poor settings.18 According to the World Health Organization (WHO), diagnostic tests for resource-limited environments should be affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end-users (ASSURED).19 More recently, Land et al. proposed REASSURED with the addition of R (real-time connectivity) and E (ease of specimen collection and environmental friendliness) to the existing ASSURED, for the design of future diagnostic tests to address important priorities such as global health emergencies and antimicrobial resistance (AMR).20
Microfluidic paper-based analytical devices (μPADs) are a relatively new diagnostic platform that may satisfy the ASSURED and REASSURED criteria.20,21 Being inexpensive, and simple to manufacture and operate, μPADs have attracted significant interest as emerging alternatives to conventional tests, showing promise for scalable, low-cost monitoring, and user-operating analytical devices. The paper matrix, most commonly hydrophilic cellulose fibres, allows passive liquid transport through capillary forces.22,23 Hydrophobic barriers embedded into the fibre allow liquid flow to be confined.23 A range of methods has been reported for the fabrication of paper-based devices including inkjet printing,24 photolithography,21 cutting,25 stamping,26 screen printing27 and wax printing.28 Wax printing is probably the simplest and most rapid of these fabrication techniques, requiring only a commercially available office ‘wax’ printer, which operates like an ordinary office printer, but instead of ink, deposits wax on the paper. The printed paper is then heated to allow the wax to penetrate through the paper thickness, thus generating a hydrophobic barrier in the paper to enable control over fluid flow during an assay.
Recently, there have been reports on μPADs for bacterial diagnostics. Henry's group developed μPADs with colorimetric reactions between bacterial enzymes and chromogenic substrates, i.e. chlorophenol red-β-D-galactopyranoside (CPRG) to detect E. coli O157:H7, 5-bromo-6-chloro-3-indolyl caprylate for Salmonella typhimurium, and 5-bromo-4-chloro-3-indolyl-myo-inositol phosphate for Listeria monocytogenes from ready-to-eat meat29,30 and from agricultural water samples.31 Exploiting the same motif, μPADs were further developed for the detection of β-lactam resistance in E. coli in water samples by measuring the colour developed from the reaction between nitrocefin substrates and the bacterial secreted β-lactamase.30 A similar concept was also reported on μPADs for E. coli BL21 detection from water samples.32
Globally, Pakistan is the fourth largest milk producing country,33 and S. aureus and E. coli as well as their antibiotic resistant strains can cause heavy losses to its dairy industry. Early and rapid screening systems can help minimise such losses and also help reduce the development of antibiotic resistance for mastitis treatment. The emerging Extended Spectrum Beta Lactamases (ESBLs) are not only able to inactivate narrow-spectrum antibiotics such as penicillins, and 1st- and 2nd-generation cephalosporins, but also hydrolyse 3rd, 4th and 5th-generation β-lactam antibiotics such as ceftazidime (CAZ) and cefotaxime (CTX).34,35 Infections caused by ESBL-producing bacteria are very difficult to cure, have limited treatment options, and often result in treatment failure.36,37 Knowledge of antimicrobial susceptibility patterns of local bacterial population will significantly help clinicians with rapid identification of antibiotic resistant bacterial species and prescription of appropriate antibiotics in a timely manner.
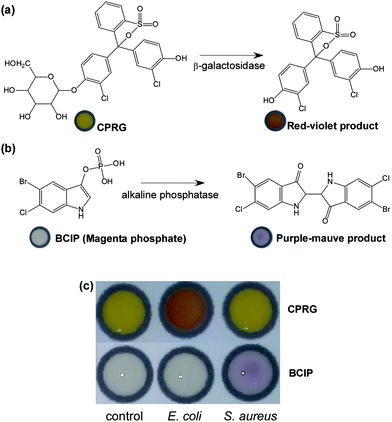
In this study, μPADs were explored for presumptive diagnosis of E. coli and S. aureus and their antibiotic-resistant strains in milk samples employing a range of chromogenic substrates (Table 1). For the first time, 5-bromo-6-chloro-3-indolyl phosphate p-toluidine salt (BCIP, magenta phosphate) and HMRZ-86 were employed on paper-based devices for the detection of S. aureus and ESBL-positive bacteria, respectively. The developed μPADs were tested with 640 milk samples collected from healthy animals from dairy farms in Pakistan. By combining the various colorimetric assays on the μPADs, a simultaneous indication of the presence of particular bacteria and their antibacterial resistance with readout can be achieved by the unaided eye or photographing via a smartphone.
| Substrate | Enzyme | Colour change | Indicative | Ref. |
|---|---|---|---|---|
| CPRG | β-Galactosidase | Yellow to red-violet | The presence of E. coli | 29 and 31 |
| BCIP, magenta phosphate | Alkaline phosphatase | Colourless to purple-mauve | The presence of S. aureus | 38 |
| Nitrocefin | β-Lactamase | Yellow to red | Antimicrobial resistance to traditional β-lactams | 30, 39 and 40 |
| HMRZ-86 | ESBL | Yellow to red | Antimicrobial resistance to extended β-lactams | 34, 35, and 41–43 |
Experimental
Materials
Bacterial strains of E. coli (NCTC 12241, 11560, 13351) and S. aureus (NCTC 12981, 12973) were obtained from Pro-Lab Diagnostics (UK). Nitrocefin was obtained from Merck Millipore (UK). Chlorophenol red-β-D-galactopyranoside (CPRG), 5-bromo-6-chloro-3-indolyl phosphate p-toluidine salt (BCIP, magenta phosphate) or magenta phosphate, β-galactosidase, β-lactamase and rAPid alkaline phosphatase were all acquired from Sigma-Aldrich (UK). HMRZ-86 was purchased from Bio-Rad Laboratories (UK). Filter paper (Whatman grade 4) was bought from GE Healthcare Life Sciences (UK). Mueller-Hinton broth and Buffered Peptone Water broth were purchased from Oxoid Ltd, UK. Pasteurised cow's milk was purchased from local stores (Hull, UK).Preparation of bacterial culture
Pure β-lactamase negative (NCTC 12241), β-lactamase positive (NCTC 11560) and ESBL positive (NCTC 13351) E. coli strains were cultured overnight in buffered peptone water (BPW) supplemented with vancomycin hydrochloride (8 mg L−1). Similarly, pure β-lactamase negative (NCTC 12981) and β-lactamase positive (NCTC 12973) S. aureus strains were grown overnight in Mueller-Hinton (MH) broth supplemented with 5% sodium hydroxide (NaCl). Supplementation of the BPW medium with vancomycin hydrochloride renders the medium selective for E. coli, while addition of 5% NaCl in MH broth makes the medium selective for S. aureus.31,44Fabrication of μPADs
Designs of the desired wax features were drawn in AutoCAD software (Student Version 2018). An array of 7 mm diameter circles with a 0.5 mm line thickness was printed on the Whatman paper using a wax printer (Xerox ColorQube). The paper was then wrapped in a single layer of aluminium foil and placed on a hot plate at 200 °C for 2–3 min. The heat melted the wax and allowed it to pass through the paper to create a hydrophobic wax barrier. One side of the paper was sealed with masking tape to prevent leaking of reagents during an experiment.Preparation and deposition of chromogenic substrates in μPADs
Solutions of the chromogenic substrates were prepared in HEPES buffer (0.1 M HEPES, 0.1% BSA, pH 7.5). CPRG for the detection of E. coli was made up as a 3 mM solution according to Bisha et al., 2014.31 BCIP for the detection of S. aureus was prepared at 5.7 mM (the maximum concentration at which the reagent can be dissolved in PBS without precipitation). For the detection of β-lactam resistant strains of E. coli and S. aureus, a solution of nitrocefin (1 mM) was prepared as described by Boehle et al. 2017.30 HMRZ-86 was available in the soluble form in a β-LACTA test kit (Bio-Rad Laboratories) and was used for the detection of ESBL. For a point-of-care device to be used under field conditions, it is desired to have minimal user intervention. To this end, the chromogenic substrates were pre-deposited and dried onto the paper prior to use, in contrast to previous reports where they were added in liquid form.29,31 Prior to substrate deposition, the bottom of the printed paper was sealed with masking tape. Then, each chromogenic substrate solution (6 μL) was deposited on the μPADs and allowed to dry at room temperature for 5–10 min (visually inspected). The μPADs were placed in a Petri dish covered with aluminium foil to prevent light exposure, and stored at 4 °C for 1–2 days prior to use.Optimisation of paper-based assays
The colorimetric reactions for paper-based assays were optimised using enzyme solutions prepared in PBS; alkaline phosphatase (0–80 mU mL−1), β-lactamase (0–0.8 mU mL−1), and β-galactosidase (0–80 mU mL−1). The enzyme solutions were freshly prepared at the start of each experiment.Assays for bacterial culture, spiked milk and real milk samples
Visual detection and data analysis
Colorimetric results were analysed through visual inspection of the μPAD at 3–4 h after sample deposition. The presence of E. coli in the samples was determined by a change in the colour of CPRG embedded in the μPADs from yellow to red-violet.29,31 Similarly, S. aureus was detected by a change in the colour of BCIP embedded in the μPADs from colourless to mauve/purple.38 A change in the colour of nitrocefin24 and HMRZ-8635,41–43,45 embedded in the μPADs from yellow to red are the indicatives of β-lactamase and ESBL positive bacteria, respectively.Semi-quantitative analysis of the colorimetric end results was performed according to the previously reported protocol by Boehle et al.30 Briefly, following the assay, Petri dishes containing the μPADs were placed inside a light box (16 cm × 16 cm × 16 cm) to avoid light interference during image capture. The camera part of the smart phone (Vodafone 890N, operated with flash) was placed between the slit of the light box (2 cm × 5 cm) to obtain images. The images were analysed using ImageJ freeware (National Institute of Health, USA), following the protocol previously described.24 Initially, the images were processed through a Red Green Blue (RGB) stack and the Green channel was selected as it is the complementary colour of red, which is the end point colour of all reactions except the S. aureus experiment which was mauve/purple. Although the complementary colour of mauve/purple is yellow, it gave a relatively high sensitivity with the green image amongst the RGB stack. Therefore, it was also analysed through the green channel. Next, the image was inverted. Circles were drawn to outline the measurement areas within the reaction zones. Utilising the “Measure” tab under “Analyze”, the average colour intensity within the defined areas was obtained and further analysed with Microsoft Excel. The colour intensity of μPADs containing water was also analysed as a blank: the colour intensity of μPADs containing samples was normalized by subtracting the colour intensity of μPADs containing water. Normalised data were processed in Microsoft Excel to obtain mean and standard deviations from three separate repeats (n = 3).
UV-vis spectrophotometric detection of colorimetric reactions
UV-vis spectrophotometric analysis of colorimetric reactions employed for μPADs was performed on the same reactions using microtitre plates and a UV-vis plate reader (NanoDrop™ Lite Spectrophotometer, Thermofisher Scientific). For this purpose, 100 μL of ten-fold serial dilutions of S. aureus and E. coli and their β-lactam resistant bacterial strains were reacted with 100 μL of their respective chromogenic substrates whose concentrations were similar to those used on μPADs. The microtiter plate was then incubated at 37 °C for 2 h. Following this, absorbance at various wavelengths was measured; 595 nm for the CPRG reaction, 405 nm for the BCIP reaction, and 490 nm for the nitrocefin and HMRZ-86 reaction.Polymerase chain reactions (PCR)
DNA was extracted from enrichment broth using a GeneJET Genomic DNA Purification Kit according to the manufacturer's instructions. A Nano-drop system (ND-2000C, Thermofisher Scientific) was employed to quantify the concentration and purity of the extracted DNA. The primers used during PCR are summarised in Table S2, ESI.† All the reactions were carried out to a final volume of 50 μL under the following conditions. Initial denaturation at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing (at the temperature summarised in Table S2, ESI†) for 1 min, and extension at 72 °C for 2 min. Final extension was performed at 72 °C for 10 min to complete the reaction. Amplified PCR products were detected by electrophoresis on 1.5% agarose gel and visualised using a gel documentation system (Gel Doc™ EZ Imager, Bio-RAD) and photographed. A GeneRuler 1kb DNA ladder (Thermo Scientific) was included in each run.Results and discussion
Paper-based assays employing colorimetric reactions between bacterial enzymes and their corresponding chromogenic substrates
The development of the paper-based assays initially involved the use of enzyme solutions, rather than enzymes derived from the bacteria, to confirm the feasibility of enzyme–substrate reactions on the paper matrix. For optimisation of the reaction conditions, different dilutions of pure bacterial enzymes were freshly prepared at the start of each experiment. The lowest β-galactosidase concentration to produce a distinguishable colour change from yellow to red-violet was 0.02 U mL−1 (Fig. 1S(a), ESI†), comparable to 0.03 U mL−1 reported for β-galactosidase.46 The lowest enzyme concentration to show a distinctive colour change for β-lactamase assay was 0.1 mU mL−1 (Fig. 1S(b), ESI†). The discrepancy with the value of 10 mU mL−1 reported by Boehle et al.30 may plausibly be due to different sources of beta-lactamase employed for the reaction.For S. aureus detection, the reaction between solutions of BCIP and alkaline phosphatase solution was first attempted on paper. Utilising 3 mM BCIP, a characteristic colour change from colourless to mauve/purple was successfully observed at ≥0.006 U mL−1 of alkaline phosphatase (Fig. 1S(c), ESI†). This demonstrates a novel paper-based assay for S. aureus detection via bacterial alkaline phosphatase and its corresponding chromogenic substrate BCIP (magenta phosphate).
Testing of the developed μPADs with bacterial culture
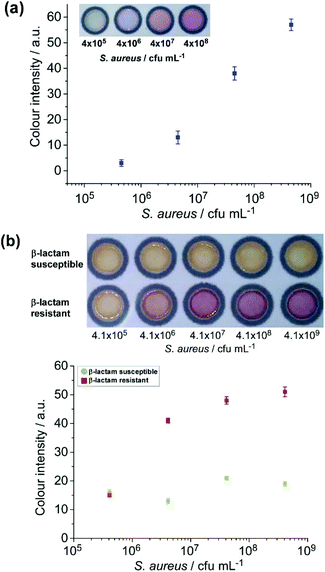
Having successfully developed μPADs for the detection of bacterial enzymes from enzyme solutions, the devices were next tested with live bacteria populations obtained from cell cultures in their respective growth media. Ten-fold serial dilutions of different strains of E. coli and S. aureus were prepared in their respective selective broths and reacted with their relevant chromogenic substrate on the μPADs.In order to determine the presence of β-galactosidase, E. coli was lysed prior to testing on the developed paper device as β-galactosidase is produced inside E. coli cells and is not secreted into the surrounding medium. A distinctive colour change from yellow to red-violet when reacting with CPRG was observed at an E. coli concentration of ≥3.6 × 106 cfu mL−1 (Fig. S2(a), ESI†), in good agreement with the 3.8 × 106 cfu mL−1 value detected on μPADs developed by Boehle et al.30
In contrast to β-galactosidase, alkaline phosphatase is excreted from S. aureus cells into the surrounding medium. The reaction between S. aureus secreted alkaline phosphatase and BCIP was therefore carried out without cell lysis (Fig. S2(b), ESI†) employing the same device shown in Fig. S1(c), ESI.† A distinguishable colour change from colourless to mauve/purple was observed at a concentration exceeding 3.6 × 106 cfu mL−1. To the best of our knowledge, this is the first time the detection of S. aureus was demonstrated on a paper device based on the reaction between bacteria secreted enzyme alkaline phosphatase and 5-bromo-6-chloro-3-indolyl phosphate p-toluidine salt.
Next, the efficiency of the developed μPADs on detecting the β-lactam resistant strain of E. coli was examined using the colorimetric reaction between nitrocefin and beta-lactamase to produce a characteristic colour change from yellow to red. No cell lysis was performed prior to the reaction, even though a marginal 5% increase in colour intensity on μPADs tested with lysed E. coli was observed when compared to that of intact cells.30 As expected, no colour change occurred for the β-lactam susceptible strain of E. coli, where no β-lactamase was produced by the bacterial enzyme (Fig. S3(a), ESI†). For the β-lactam resistant strains, a colour change occurred at concentrations ≥106 cfu mL−1, in good agreement with ≥3.8 × 106 cfu mL−1 bacteria reported from μPADs tested with different β-lactam resistant bacteria in wastewater and sewage.24 The same device tested with a β-lactam resistant strain of S. aureus detected the colour change at ≥1.3 × 106 cfu mL−1 (Fig. S3(b), ESI†).
The experiments confirmed the feasibility of μPADs for E. coli and the β-lactam resistant strain of E. coli, as well as feasibility for the detection of S. aureus employing the reaction between their secreted alkaline phosphatase and the chromogenic BCIP substrate. In addition, a β-lactam resistant strain of S. aureus was effectively detected using the nitrocefin-embedded μPAD. Distinctive colour changes were observed in all cases at bacterial levels of ≥106 cfu mL−1.
Cross reactivity study of μPADs for S. aureus and E. coli
The colorimetric assays employed for the developed paper devices utilised enzymes which could also be produced by other bacteria, and hence a compromised specificity might be expected when performed under field conditions. In laboratory settings, specific supplements can be added into the enrichment media in order to promote the growth of target bacteria, whilst simultaneously inhibiting the growth of other competing bacteria. Herein, vancomycin hydrochloride was utilised to support the growth of E. coli, and to suppress the growth of S. aureus. Similarly, the high salt content present in MH broth only permits S. aureus to grow. A cross-reactivity study between the two assays was carried out after incubating the bacteria in selective enrichment broths. The colour change was only observed when the correct pair of bacterial enzyme and specific substrate was present, with no false positive results found from the two bacterial species (Fig. 1). Although cross reactivity can be eliminated in laboratory analysis using a growth inhibitor to suppress competing bacteria, this can be problematic in field tests and false positive results are inevitable. Nevertheless, the method can still be suitable as an initial screening test providing impetus for further testing and confirmation to be conducted. Further investigations will include protocols to minimise other related contaminating bacteria in the sample using selective enrichment media.μPADs for the differentiation of β-lactam resistant and ESBL resistant bacteria
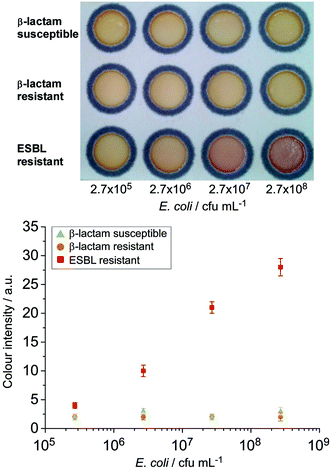
Next, we aimed to develop paper-based assays for the detection of ESBL bacteria which not only show resistance to penicillins, such as cephalosporins and aztreonam, but also to other classes of antibiotics, such as aminoglycosides, cotrimoxazole, tetracycline and fuoroquinolones.Nitrocefin was reported as a suitable substrate for the detection of ESBL bacteria. A colour change from yellow to red could be observed on the developed μPAD when pathogen concentrations exceeded 106 cfu mL−1 for both β-lactam resistant E. coli and ESBL resistant E. coli (Fig. S4(a), ESI†) as nitrocefin cannot differentiate between these two groups.36 As such, a specific chromogenic cephalosporin, HMRZ-86, was herein exploited as a selective chromogenic substrate embedded on the μPAD to detect ESBL-resistant bacteria. A carboxypropyl-oxyimino group that binds to the side chain at position 7 in compound protects the β-lactam ring from a range of β-lactamases, but not from extended-spectrum β-lactamases (ESBLs).45 In contrast to nitrocefin embedded μPADs, a colour change from yellow to red only occurred from the presence of ESBL-resistant E. coli at concentrations exceeding 106 cfu mL−1 (Fig. S4(b), ESI†). This development can be applied to real samples where differentiation of antibiotic resistant E. coli strains can prove to be crucial.
The sensitivity of the paper-based reactions performed on the developed μPADs was validated against the UV-visible spectrophotometry results obtained from the same liquid-phase reactions conducted in conventional microtitre plates. A comparable level of 106 cfu mL−1 bacteria detection observed from the absorbance values of the spectrophotometric assay confirming the viability of the developed μPADs (Fig. S5–S7, ESI†), and demonstrating a cost-effective approach of μPADs coupled with a smart phone for the detection of bacteria in samples without the need for sophisticated laboratory equipment and trained staff.
μPADs for the analysis of bacterial spiked-milk samples
Having demonstrated the viability of the paper devices for the detection of E. coli and S. aureus and their antibiotic resistant strains from bacterial culture media, the μPADs were next tested with milk samples spiked with bacteria in a range of ten-fold serial dilutions.First, the μPAD embedded with BCIP was investigated with milk samples spiked with S. aureus. A colour change from the reaction between S. aureus secreted alkaline phosphatase and BCIP was visually detectable at a concentration exceeding 4.5 × 106 cfu mL−1 (Fig. 2a), similar to the result previously obtained from S. aureus in culture media. The nitrocefin reaction also worked well distinguishing between β-lactam susceptible and β-lactam resistant strains of S. aureus spiked into milk samples (Fig. 2b). The resistant strain of S. aureus revealed a red colour at concentrations exceeding 4.5 × 106 cfu mL−1. These results showed the successful paper-based detection of S. aureus and its β-lactam resistant strain from spiked milk samples, which can potentially be utilised for detecting such pathogenic bacteria from real milk samples and other complex matrices.
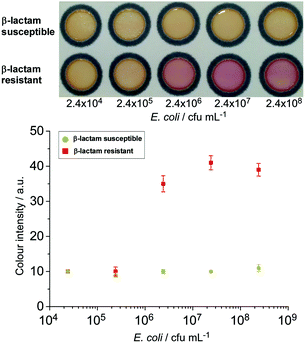
The nitrocefin-embedded μPAD tested with E. coli spiked into milk samples showed a colour change to red for the resistant strain at a concentration ≥2.4 × 106 cfu mL−1 (Fig. 3), similar to the observation from the culture broth. Boehle et al. also detected a colour change in their nitrocefin embedded μPADs tested with wastewater and sewage with ≥3.8 × 106 cfu mL−1 bacteria, but not with lower concentrations.30 This shows the versatility of the paper-based β-lactamase-nitrocefin assays for the detection of β-lactam resistant E. coli at ca. 106 cfu mL−1, regardless of the complexity of the matrices.
The assessment of the HMRZ-86 impregnated μPAD with spiked milk samples also demonstrated the specificity to the EMBL-resistant strain of E. coli at a similar visual detection level of 2.7 × 106 cfu mL−1 as found in pure culture samples (Fig. 4).
However, the reaction between CPRG and β-galactosidase from lysed E. coli was not successfully tested with spiked milk samples (Fig. S8, ESI†). It is postulated that β-galactosidase released by the bacteria might have hydrolysed a large amount of lactose contained in the milk samples, and therefore only a small quantity of β-galactosidase was available to react with embedded CPRG.
Following on, pre-enrichment of spiked milk samples was attempted in order to supply nutrients and to encourage bacteria growth over a period of a few hours prior to testing. The supplements contained within the enrichment media selectively promote the growth of target bacteria, whilst simultaneously inhibiting the growth of other competing bacteria. Thus, the desired bacteria can be brought to a concentration level that is detectable through the colorimetric reaction on the μPADs.
Fig. 5 depicts the colorimetric results obtained from milk samples spiked with E. coli and S. aureus and their antibiotic resistant strains after pre-enrichment. Levels of bacteria as low as 10 cfu per 1 mL of the spiked milk sample could be detected following 12 or 16 h of pre-enrichment in their respective selective broths. This suggests that the sensitivity of the paper-based assays can be vastly improved by including a bacterial pre-enrichment step. Pre-enrichment amplifies the analyte target, and when combined with selective growth media, it reduces the growth of unwanted bacteria in samples that could contribute to false positive results as also reported previously.24,31 This step only required an incubator, which is commonly used in microbiology laboratories. With further investigations, a suitable pre-concentration step can be included in the platform for field tests. The final detection performed on μPADs can offer a simple, cost-effective, and easy-to-interpret solution for the screening of major milk borne pathogens. The entire process, overnight pre-enrichment and colorimetric detection, can be performed within one day, and it allows the detection of bacterial contamination as low as 10 cfu mL−1 from the initial sample.
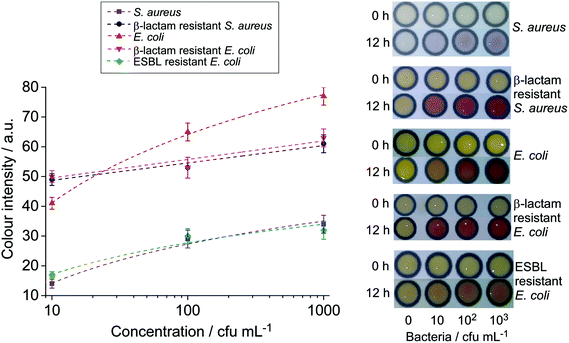 | ||
| Fig. 5 Photographs and colour intensity analysis of milk samples spiked with various bacteria after 12 h enrichment (n = 3). | ||
μPADs for the analysis of bacteria in raw milk samples
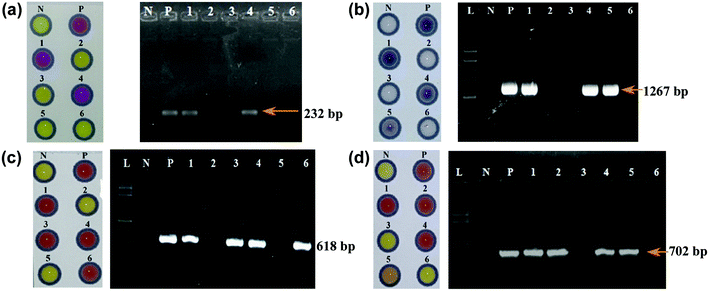
The ultimate goal of this present study was to develop a simple, cost-effective and user-friendly device for the detection of pathogenic bacteria causing mastitis suitable for resource-poor areas in the developing countries. The viability of the developed paper devices was therefore examined for the detection of S. aureus and E. coli with raw milk collected from a group of cattle and buffaloes in Pakistan. The samples were collected from healthy animals from the field. Due to very low level of bacteria from the samples, pre-enrichment with selective media was required to increase the number of bacteria to meet with the detectable level of the paper devices (ca. 106 cfu mL−1) previously observed from bacterial spiked milk samples. In order to achieve this, a milk sample (10 v%) was mixed with selective media (90 v%) and incubated at 37 °C for 12 h. This step not only increased the bacterial level, but also diluted the lactose concentration in the samples, thereby diminishing the consequent competing lactose hydrolysis which was problematic in the β-galactosidase-CPRG reaction previously observed in E. coli spiked milk samples. Subsequently, the pre-enriched samples were analysed on μPADs and PCR gene analysis was conducted for result confirmation (Fig. 6a and b).Out of 640 milk samples, μPADs detected 77 samples with E. coli positive (84 samples confirmed by PCR), and 129 samples with S. aureus positive (143 confirmed by PCR) as summarised in Table 2. These results suggested that the sensitivity of our developed μPADs was higher than 90%. The positive results obtained on μPADs also showed positive from PCR, indicating that the μPADs were also 100% specific.
| Bacteria | Positive samples | |
|---|---|---|
| μPAD | PCR | |
| E. coli | 77 (12%) | 84 (13.1%) |
| S. aureus | 129 (20.2%) | 143 (22.3%) |
Beta-lactam is the most widely used class of antibiotics in veterinary medicine. As a result, β-lactam resistant pathogens are very common in livestock. For the detection of β-lactam resistance in the E. coli- and S. aureus-positive milk samples, enrichment cultures were sub-cultured in BPW supplemented with cefotaxime (2 μg mL−1) and tested on nitrocefin embedded μPADs. Beta-lactam resistance in the same samples was also confirmed by PCR through amplification of the blaTEM gene (Fig. 6(c and d)). The μPAD detected β-lactam resistant pathogens in 67% and 62.7% of E. coli and S. aureus isolates, respectively, without false positive results confirmed by PCR (Table 3). Therefore, the μPAD accurately detected β-lactam resistant isolates.
| Bacteria | Positive samples | |
|---|---|---|
| μPAD | PCR | |
| β-Lactam resistant E. coli | 52/77 (67.5%) | 52/77 (67.5%) |
| β-Lactam resistant S. aureus | 81/129 (62.7%) | 81/129 (62.7%) |
We have shown a successful implementation of paper-based colorimetric assays for the detection of E. coli and S. aureus and β-lactam resistant strains in milk samples that could be achieved within a day. The present setup required no sophisticated equipment, and yet yielded similar sensitivity and selectivity to the molecular diagnostic technique of PCR for results obtained from 640 raw milk samples. In developing countries like Pakistan, dairy farms are located in remote and resource limited areas and local veterinary diagnostic laboratories are not equipped with sophisticated equipment like micro-plate readers, thermal cyclers, etc., and they tend to have only bacterial culture facilities such as incubators and simple microscopes.1,47 In these laboratories, routine identification of bacteria and their antibiotic sensitivity is usually carried out by their culture characteristics (less specific) and growth inhibition assays (i.e. analysis of bacterial growth in the presence of antimicrobial agents), which give results in 2–4 days.
Conclusions
In the present study, paper-based assays have been devised for the detection of S. aureus and E. coli and their antibiotic resistant strains in milk samples, with similar sensitivity to the sophisticated laboratory-based technique, UV-vis spectrophotometry. Employing specific chromogenic substrates, colour-change reactions on μPADs have successfully distinguished specific bacteria without cross-reactivity between their β-lactam susceptible and β-lactam resistant strains. They also showed, for the first time, a successful differentiation between ESBL resistant and β-lactam resistant strains. The paper devices showed ≥90% sensitivity and 100% selectivity, compared to the advanced and sophisticated PCR method, demonstrating a cost-effective platform for the presumptive diagnosis of bacteria in milk, especially in resource limited areas.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the Commonwealth Scholarship Commission, United Kingdom, for providing funding to this project. Dr Fazli Rabbi Awan acknowledges funding (Project No. CRP/PAK14-02; Contract No. CRP/14/012) from the International Centre for Genetic Engineering and Biotechnology (ICGEB), Italy for partial support for research in this study.References
- L. Parsons, A. Somoskovi, C. Gutierrez, E. Lee, C. Paramasivan, A. Abimiku, S. Spector, G. Roscigno and J. Nkengasong, Clin. Microbiol. Rev., 2011, 24, 314–350 CrossRef.
- S. Oliver, B. Jayarao and R. Almeida, Foodborne Pathog. Dis., 2005, 2, 115–129 CrossRef CAS.
- M. Pal, S. Mulu, M. Tekle, S. V. Pintoo and J. Prajapati, Beverage Food World, 2016, 43, 1–3 Search PubMed.
- E. Fox, Y. Jiang and K. Gobius, Int. Dairy J., 2018, 84, 28–35 CrossRef.
- D. Morgan, C. Newman, D. Hutchinson, A. Walker, B. Rowe and F. Majid, Epidemiol. Infect., 1993, 111, 181–187 CrossRef CAS.
- M. De Buyser, B. Dufour, M. Maire and V. Lafarge, Int. J. Food Microbiol., 2001, 67, 1–17 CrossRef CAS.
- D. Schmid, R. Fretz, P. Winter, M. Mann, G. Hoger, A. Stoger, W. Ruppitsch, J. Ladstatter, N. Mayer, A. de Martin and F. Allerberger, Wien. Klin. Wochenschr., 2009, 121, 125–131 CrossRef CAS.
- J. Painter, R. Hoekstra, T. Ayers, R. Tauxe, C. Braden, F. Angulo and P. Griffin, Emerging Infect. Dis., 2013, 19, 407–415 CrossRef.
- J. E. Hillerton and E. A. Berry, NMC Annual Meeting Proceedings, 2004 Search PubMed.
- US Food and Drug Administration, Compliance Policy Guide Sec. 527.300, 2012.
- N. Fejzic, M. Begagic, S. Serie-Haracic and M. Smajlovic, Bosnian J. Basic Med. Sci., 2014, 14, 155–159 CrossRef.
- S. Pyorala, Ir. Vet. J., 2009, 62, 40–44 CrossRef.
- T. Ali, S. Rahman, L. Zhang, M. Shahid, S. Zhang, G. Liu, J. Gao and B. Han, Front. Microbiol., 2016, 7, 1931 Search PubMed.
- B. Robles, D. Nobrega, F. Guimaraes, G. Wanderley and H. Langoni, Pesqui. Vet. Brasil, 2014, 34, 325–328 CrossRef.
- J. Jorgensen and M. Ferraro, Clin. Infect. Dis., 2009, 49, 1749–1755 CrossRef CAS.
- A. Ashraf, M. Imran, T. Yaqub, M. Tayyab, W. Shehzad and P. Thomson, Mol. Cell. Probes, 2017, 33, 57–64 CrossRef CAS.
- R. Bu, J. Wang, C. DebRoy, J. Wu, L. Xi, Y. Liu and Z. Shen, Iran J. Vet. Res., 2015, 16, 283–287 CAS.
- C. Rodrigues, N. Desai and H. Fernandes, Clin. Microbiol. Newsl., 2016, 38, 51–56 CrossRef.
- G. Wu and M. Zaman, Bull. W. H. O., 2012, 90, 914–920 CrossRef.
- K. Land, D. Boeras, X. Chen, A. Ramsay and R. Peeling, Nat. Microbiol., 2019, 4, 46–54 CrossRef CAS.
- A. Martinez, S. Phillips, G. Whitesides and E. Carrilho, Anal. Chem., 2010, 82, 3–10 CrossRef CAS.
- D. Liana, B. Raguse, J. Gooding and E. Chow, Sensors, 2012, 12, 11505–11526 CrossRef CAS.
- T. Satarpai, J. Shiowatana and A. Siripinyanond, Talanta, 2016, 154, 504–510 CrossRef CAS.
- K. Abe, K. Suzuki and D. Citterio, Anal. Chem., 2008, 80, 6928–6934 CrossRef CAS.
- E. Fenton, M. Mascarenas, G. Lopez and S. Sibbett, ACS Appl. Mater. Interfaces, 2009, 1, 124–129 CrossRef CAS.
- C. Cheng, A. Mazzeo, J. Gong, A. Martinez, S. Phillips, N. Jain and G. Whitesides, Lab Chip, 2010, 10, 3201–3205 RSC.
- Z. Nie, C. Nijhuis, J. Gong, X. Chen, A. Kumachev, A. Martinez, M. Narovlyansky and G. Whitesides, Lab Chip, 2010, 10, 477–483 RSC.
- Y. Lu, W. Shi, L. Jiang, J. Qin and B. Lin, Electrophoresis, 2009, 30, 1497–1500 CrossRef CAS.
- J. Jokerst, J. Adkins, B. Bisha, M. Mentele, L. Goodridge and C. Henry, Anal. Chem., 2012, 84, 2900–2907 CrossRef CAS.
- K. Boehle, J. Gilliand, C. Wheeldon, A. Holder, J. Adkins, B. Geiss, E. Ryan and C. Henry, Angew. Chem., Int. Ed., 2017, 56, 6886–6890 CrossRef CAS.
- B. Bisha, J. Adkins, J. Jokerst, J. Chandler, A. Perez-Mendez, S. Coleman, A. Sbodio, T. Suslow, M. Danyluk, C. Henry and L. Goodridge, J. Vis. Exp., 2014, 51414 Search PubMed.
- D. Lin, B. Li, J. Qi, X. Ji, S. Yang, W. Wang and L. Chen, Sens. Actuators, B, 2020, 303, e51414 Search PubMed.
- M. M. Ajmal, C. X. Li and W. Aslam, J. Econ. Sustain. Dev., 2015, 6, 19–28 Search PubMed.
- H. Hanaki, Y. Koide, H. Yamazaki, R. Kubo, T. Nakano, K. Atsuda and K. Sunakawa, J. Infect. Chemother., 2007, 13, 390–395 CrossRef CAS.
- M. El-Jade, M. Parcina, R. Schmithausen, C. Stein, A. Meilaender, A. Hoerauf, E. Molitor and I. Bekeredjian-Ding, PLoS One, 2016, 11, 127213 CrossRef.
- D. Rawat and D. Nair, J. Global Infect. Dis., 2010, 2, 263–274 CrossRef.
- R. Colodner, Am. J. Infect. Control, 2005, 33, 104–107 CrossRef.
- A. Rambach, US Patent, 6548268B1, 2003 Search PubMed.
- S. Bidya and R. S. Suman, Open J. Clin. Diagn., 2014, 4, 47–52 CrossRef.
- S. Sharma, P. Ramnani and J. Virdi, J. Antimicrob. Chemother, 2004, 54, 401–405 CrossRef CAS.
- H. Hanaki, R. Kubo, T. Nakano, M. Kurihara and K. Sunagawa, J. Antimicrob. Chemother, 2004, 53, 888–889 CrossRef CAS.
- S. Jain, J. Andrews, A. Fraise and N. Brenwald, J. Antimicrob. Chemother, 2007, 60, 652–654 CrossRef CAS.
- D. Livermore, M. Warner and S. Mushtaq, J. Antimicrob. Chemother, 2007, 60, 1375–1379 CrossRef CAS.
- M. Bruins, P. Juffer, M. Wolfhagen and G. Ruijs, J. Clin. Microbiol., 2007, 45, 682–683 CrossRef.
- H. Hanaki, Y. Koide, H. Yamazaki, R. Kubo, T. Nakano, K. Atsuda and K. Sunakawa, J. Antimicrob. Chemother, 2007, 13, 390–395 CAS.
- A. Lokur, J. Pharm. Innov., 2018, 7, 103–106 Search PubMed.
- J. Jacobs, L. Hardy, M. Semret, O. Lunguya, T. Phe, D. Affolabi, C. Yansouni and O. Vandenberg, Front. Med., 2019, 6, 205 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0an01075h |
| This journal is © The Royal Society of Chemistry 2020 |