Open Access Article
Open Access ArticleA platform for multiplexed colorimetric microRNA detection using shape-encoded hydrogel particles†
Nidhi
Juthani
 and
Patrick S.
Doyle
and
Patrick S.
Doyle
 *
*
Department of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA. E-mail: pdoyle@mit.edu; Tel: +1-617-253-4534
First published on 17th June 2020
Abstract
We report a platform utilizing a reporter enzyme, which produces a chromogenic indigo precipitate that preferentially localizes within a hydrogel microparticle. The 3D network of the hydrogel maintains the rapid target binding kinetics found in solution, while multiplexed target detection is achieved through shape-encoding of the particles. Moreover, the precipitate-laden hydrogels can be imaged with a simple phone camera setup. We used this system to detect microRNA (miRNA) down to 0.22 fmol. We then showed the compatibility of this system with real samples by performing multiplexed miRNA measurements from total RNA from matched colon cancer and normal adjacent tissue.
As diagnostics move out of the lab into clinical settings, the use of specialized equipment for signal detection greatly hinders technology translation.1,2 To this end, colorimetric detection systems typically require fewer instrumentation requirements and are more amenable to translation to resource limited settings.3,4 This is most evidently seen by the success of lateral flow assays, which use colloidal nanoparticles for target labelling.5 These surface-based detection systems (i.e. lateral flow assays and contact printing methods) facilitate spatial multiplexing, but one loses the favorable hybridization kinetics offered in solution.6–10 Conversely, solution-based colorimetric detection systems can realize fast hybridization kinetics but are not capable of simultaneous multiplexing.11–16
Hydrogels offer a superior alternative to solution and surface-based biomolecule detection systems due to their biocompatibility, solution-like kinetics, non-fouling nature, and ability to incorporate capture agents inside a 3D network.17,18 Hydrogels have previously been used to detect microRNA, mRNA, proteins and cytokines from a variety of complex samples such as total RNA extract, live and lysed cells, serum, and FFPE tissue.18–26 However, like many other detection platforms, hydrogel-based detection assays typically employ a fluorescent reporter to label the captured target. This could involve direct labelling with a fluorescent dye, or labelling with an enzyme, which in turn produces a fluorescent product. The latter labelling technique necessitates containment of the fluorescent product and hence has found most utility in droplet-based or oil-encapsulated systems, where the soluble fluorescent product can be contained for sufficient signal generation and multiplexing.27–32 Thus, a limited amount of substrate is accessible for the reaction. Moreover, specialized equipment such as fluorescent microscopes, fluorescent plate readers or fluorescent scanners are necessary to detect the fluorescent signal.19,24,29,32 Efforts to develop a colorimetric readout for hydrogels have used gold nanoparticle labelling combined with dark-field microscopy. While the sensitivity of this method was comparable to fluorescent labelling, it still requires the use of a microscope for detection using dark-field imaging.33 To this end, we present a hydrogel-based multiplexed colorimetric detection platform which can be imaged using a cell phone.
In this work, we utilize a reporter enzyme, which catalyzes the production of a chromogenic precipitate. Specifically, the target is labelled with streptavidin–alkaline phosphatase (S-AP), a streptavidin-enzyme conjugate, which acts upon a substrate solution of nitro-blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (NBT-BCIP). This is a precipitating substrate traditionally used with S-AP in blotting and in situ hybridization techniques.34 NBT-BCIP starts as a clear, pale yellow substrate. S-AP cleaves the phosphate group on BCIP, which upon dimerization produces an indigo dye. The dimerization also reduces NBT into an insoluble indigo-purple diformazan in the vicinity of the enzyme.35,36 The enzymatic reaction occurs within the hydrogel particle, which contains the reaction, resulting in the localization of the precipitate.
These hydrogel particles are created using stop-flow lithography (SFL), a technique that can produce 2D-extruded particles in arbitrary shapes dependent on the photomask used.37 This enables a variety of graphical and shape encoding schemes for multiplexing. In this platform, we use shape-encoding, whereby a different particle shape encodes for the target it captures, to enable facile multiplexing, while maintaining the familiar solution assay-like workflow.38,39 As such, this platform is able to combine the benefits of solution-based colorimetric detection systems, with its rapid kinetics, with the multiplexing offered by surface-based colorimetric detection. Moreover, the hydrogel particles can be imaged using a cell-phone camera with no other expensive optics or detection systems necessary.
This method of precipitation-based colorimetric detection is particularly useful for microRNA (miRNA) detection. miRNA are small, noncoding RNAs that are an emerging class of biomarkers as they have been shown to be dysregulated in many diseases including numerous cancers, cardiac diseases, and Alzheimer's.40–42 However, miRNA only represent a small fraction of total RNA (0.01%) present in tissue and their small size (∼22 nt) make quantification difficult.43 Hydrogel particles are an ideal substrate for miRNA hybridization assay due to the fast solution-like kinetics, non-fouling nature, and ability to perform multiplexed measurements.17,18 Moreover, this platform can be outfitted to any detection assay that can accommodate labelling with an enzyme reporter, such as protein and cytokine detection with appropriate biotinylated antibodies followed by the streptavidin–enzyme conjugate. To demonstrate the ability of our hydrogel particles to contain and localize the chromogenic precipitate, we first co-polymerized hydrogels with a biotin moiety and characterized the precipitation reaction in time. Then we used this colorimetric detection method to detect miRNA in a hydrogel-based hybridization assay. We then showed the ability to perform multiplexed detection of miRNA from total RNA extracted from matched colon tumor and normal adjacent tissue using shape-encoded particles.
Results and discussion
Particle fabrication and imaging
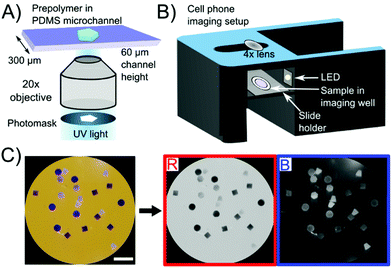
Shape-encoded hydrogel particles were fabricated using SFL in PDMS microfluidic channels using previously published methods.37 The photomask, as shown in Fig. 1A, determined the particle shape. The particles were made from a PEG-based prepolymer solution (20% polyethylene glycol diacrylate 700, 40% polyethylene glycol 600, 5% Darocur 1173 photoinitiator, and 35% 3× Tris-EDTA buffer) and copolymerized with DNA capture probes containing an acrylate group.28 Detailed methods for particle synthesis are contained in the ESI.†Particle imaging was carried out with a Samsung Galaxy S6 with a 4× macro lens attached (easy-macro.com). The phone was placed on a custom-made 3D printed holder (CAD file link is provided in the ESI†), which contained a slide holder for the samples as seen in Fig. 1B. Photos were taken with the stock camera application with orange paper underneath the phone holder. The orange color is complementary to the indigo color of the precipitate and increases the contrast for colorless particles (which would not be visible on a white background). Thus, particles with precipitate were visible in the red channel upon RGB decomposition (where the orange background is nearly white) and particles that were colorless were visible in the blue channel (where the orange background is nearly black) as seen in Fig. 1C. Both red and blue channels were used to detect particles using image thresholding. Of the three channels, as well as a combination of the three (grayscale), the red channel showed the largest signal to noise ratio as seen in Fig. S1.† Hence, the red channel was used for signal quantification, using a custom MATLAB script.
Precipitation reaction characterization within hydrogel particles
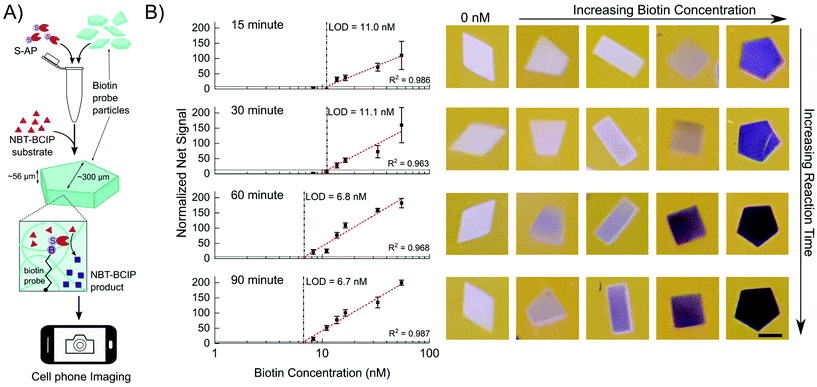
In order to characterize the precipitation reaction independent of target capture and labelling, we fabricated hydrogel particles with different concentrations of a short DNA probe containing a biotin moiety as shown in Fig. 2A. These particles were conjugated to 10 μg ml−1 S-AP over a course of 30 minutes and then mixed with NBT-BCIP substrate for 15, 30, 60 or 90 minutes. After the set reaction time, the particles were deposited inside an imaging well for imaging with a cell phone.The average net red channel signal as a function of biotin probe concentration is shown in Fig. 2B. The limit of detection (LOD) at each time point was determined using the 3σ method as described in the ESI.† The probe concentration is based on the particle volume and it represents how much biotin and consequently enzyme is incorporated into each particle. Hence, the LOD is a measure of the minimum amount of enzyme necessary to produce a detectable signal in a given amount of time. The LOD generally increases with time but there is little improvement after 60 minutes of reaction, where the LOD reaches 6.8 nM. This corresponds to a biotin loading of 20 amol biotin per particle. The net signals start saturating at the highest biotin concentrations tested after 60 minutes as evidenced in the both the plot and photos shown in Fig. 2B. Increasing the substrate concentration up-to 5-fold results in this saturation occurring at lower concentrations as shown in Fig. S2.† This leads to higher net signal for particles with lower biotin concentrations (6–15 nM), accompanied with a slight improvement in the LOD (5.3 nM). Based on these reaction characterization experiments a reaction time of 60 minutes using 5× substrate was used for the colorimetric detection of miRNA.
It should be noted that the particles with different biotin concentrations were all in the same tube during enzyme conjugation and reaction (Fig. 2A), but maintained distinct shades and hence had minimal cross-talk. After 60 minutes, the reaction solution starts turning blue. We hypothesize that at this point in the reaction, the higher concentration biotin particles have reached a saturation limit for how much precipitate they can hold but the enzymes conjugated within keep generating more precipitate, which then seeps out of the hydrogel network and enters the solution phase. Despite this, the control particles stay free of precipitate even after 90 minutes of reaction, when there is free blue reaction product in the solution. This indicates that the precipitate stays considerably localized during the course of reaction and that the precipitate does not simply adsorb onto the particle from solution but must be generated within the particle to stay. Previous reports have shown that the NBT diformazan is more efficiently immobilized in porous 3D structures such as that of hydrogels.3,7 Moreover, there is some evidence that hydrophobic small molecules preferentially partition into the hydrogel matrix.44 Thus a combination of in situ precipitate generation and the relatively hydrophobic environment of the gel both result in precipitate localization and build-up.
miRNA detection
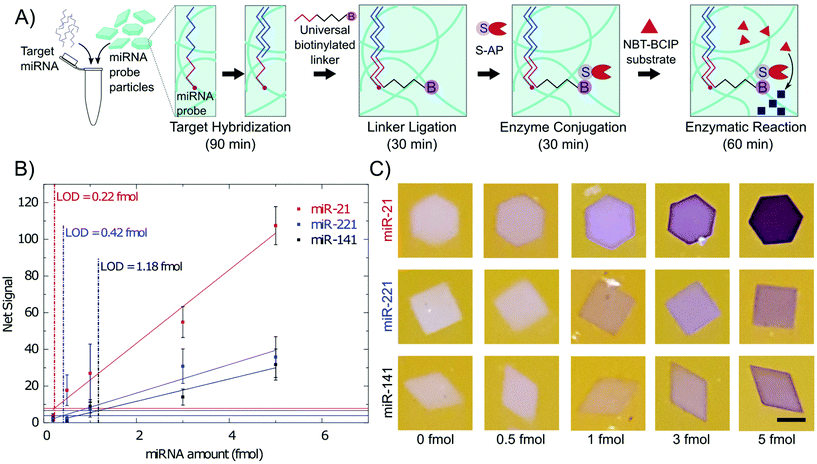
For miRNA detection, particles were co-polymerized with oligonucleotide DNA probes specific to different miRNA.19 The shape of the particle encoded for the miRNA target. Target hybridization was conducted for 90 min at 55 °C followed by 30 min of ligation of a universal biotinylated adapter using T4 DNA ligase as shown in Fig. 3A. Hybridization and ligation times were optimized in a prior study.19 After ligation, the same steps as with a biotinylated probe were followed. The particles were conjugated with 10 μg ml−1 of S-AP enzyme for 30 minutes and then reacted with 5× NBT-BCIP substrate for 60 min, after which the particles were deposited inside an imaging well for imaging with a cell phone. Between each step shown in Fig. 3A, the particles were washed by centrifugation three times to remove unbound target and reagents. For each wash, the particles were centrifuged for 20 s and the supernatant was removed and replaced with fresh buffer as detailed in the ESI.†Calibration curves for 3 different miRNA are shown in Fig. 3B, in which the LODs are 0.22 fmol for miR-21, 0.42 fmol for miR-221, and 1.2 fmol for miR-141. This corresponds to 4.5 pM, 8.4 pM and 23.7 pM respectively based on the 50 μL hybridization volume. LODs were calculated using the 3σ method as was done with the biotin probe particles. Net signal increases linearly with miRNA total amounts. The higher signals with miR-21 could be attributed to the higher probe concentration used for miR-21. Previous work has shown that miR-21 target has a slower binding rate at short times leading to higher probe concentrations being used in the particle.19 Higher probe concentration, resulting in more negative charge in the hydrogel, could also be beneficial to the precipitation reaction.
We hypothesize that the key source of variability in this detection scheme comes from the ability to ensure the particles are being well mixed in solution. The large size of these particles make them prone to settling and sticking to the reaction tube even with continuous mixing. Improvements to the mixing protocol should result in reduced variability. As was demonstrated with the biotin probe experiments, there is a saturation limit on how much NBT precipitate can be contained within the hydrogels, limiting the dynamic range of this bioassay. However, we believe that this saturation limit should not be a concern due to the focus on analytes of low concentration. Moreover, for multiplexed experiments, the capture probe concentrations can be adjusted to account for large variances in analyte concentrations. While there exist solution-based colorimetric assays with higher sensitivity (by using nucleic acid amplification), they cannot perform multiplexing.11–14,45 Moreover, it is possible to increase the sensitivity of this platform by combining it with upstream nucleic acid amplification, which is compatible with hydrogels, such as rolling circle amplification or hybridization chain reaction.46,47 These amplification schemes result in more biotin labels per target bound, resulting in more enzyme reporter labelling within the hydrogel particle.
Based on previous work with fluorescent reporters, the acquired signal increases with decreasing overall hydrogel volume and with increasing ratio of surface area to 2D projected imaging area.18,23,48 Thus, it should be possible to increase signal similarly in the colorimetric assay by manipulating these variables. For example, decreasing overall particle size (whilst staying within the imaging resolution of a cell phone), or hollowing out the particles to create a frame-like structure should concentrate the target into a smaller volume and increase the flux of target into the hydrogels.18 Also, it is interesting to note that a boundary layer (dark edge) is visible on the particles after reaction as shown in the images in Fig. 3C. In fluorescence-based assays, a boundary layer is often visible and is indicative of a high Damköhler number (Da ≫ 1), where the reaction (hybridization in this case) is much faster than diffusion of target into the hydrogel.18,23 Fig. S3† shows that the profile of this boundary layer is similar to that visible by direct fluorescent labelling, highlighting how well the NBT precipitate stays localized even within the particle.
To assess whether precipitation-based colorimetric detection could measure miRNA from complex samples containing a variety of other nucleic acids and other contaminants, miR-21 was detected from 500 ng total RNA extracted from 3 different types of healthy tissue: prostate, breast, and colon. As a negative control, particles with a probe for cel-miR-54, a miRNA only found in C. elegans, were also added for net signal calculations. All three tissues had similar miR-21 signal as shown in Fig. S4.† miR-21 is a known oncogenic miRNA upregulated in cancers.41,49,50 Since the tissue samples were all healthy tissue, it follows that only baseline levels of miR-21 were present, and hence similar levels were found in all three tissues.
Multiplexed miRNA detection from total RNA
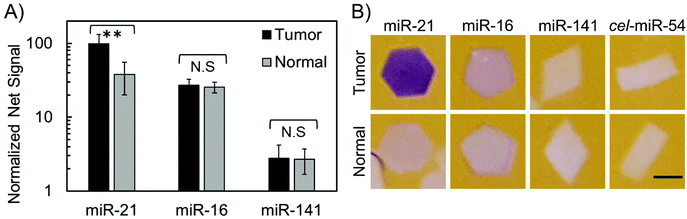
When considering miRNA dysregulation patterns, a small panel of miRNA has better diagnostic value than individual miRNA, and hence multiplexed measurements from a single sample are especially important for miRNA detection.2,27,40 Each particle shape encodes for a different miRNA probe, allowing for multiplexed measurements from the same sample. All the particles used for multiplexed miRNA detection have a similar surface area to volume ratio (coefficient of variation of 5.2%) for comparable target and enzyme diffusion.In order to assess whether miRNA dysregulation patterns could be determined with this colorimetric platform, a multiplexed miRNA detection assay was performed with total RNA extracted from colon tumor tissue and normal adjacent tissue. Again, particles specific to cel-miR-54 were added to each sample as a negative control. As seen in Fig. 4A, high miR-21 concentrations were found in tumor total RNA. miR-21 is an oncogenic miRNA found to be upregulated in multiple different cancers and diseases including colon cancer tissue, and this is reflected in the data from colorimetric measurements (Fig. 4A) as wells as the clearly discernable purple hexagon particle in Fig. 4B.41,49,51,52 In contrast, similar amounts of miR-16 were found in both tumor and normal total RNA (Fig. 4A and B). Previous reports have shown that in colon cancer specifically, miR-16 has fairly stable, high expression levels and hence has been used as an endogenous standard in miRNA analysis of colon cancer.41,52 Upregulation of miR-141 has shown to be associated with poor prognosis in advanced colon cancer.53 While miR-141 levels in this specific patient's tissue were found to be similar in both tumor and normal tissue, their low abundance, close to the limit of detection, prevents a true comparison.
Conclusions
In conclusion, we developed and characterized a precipitation-based colorimetric detection system that can be measured using a simple cell phone camera. This system combines the superior advantages offered by non-fouling, biocompatible hydrogel particles as a substrate for biomolecule detection assays with a facile and robust enzymatic, colorimetric precipitation reaction. The hydrogels act as both the reactor vessel and concentrator, leading to sensitive target detection. The indigo-purple NBT precipitate stays localized exceptionally well within the particles, even outlining the formation of a boundary layer at high target concentrations. The shape-encoded particles were fabricated using stop-flow lithography, enabling multiplexing even in complex samples, while preserving the superior kinetics of solution assays. As a result, multiplexed, colorimetric detection of miRNA was possible down to sub-femtomole amounts using a simple phone camera setup. Finally, this detection system is compatible with upstream nucleic acid amplification to improve sensitivity and is applicable to many other biomolecules of interest such as protein and cytokine detection. The precipitation reaction within the hydrogel is independent of the type of target and only requires the ability to conjugate the enzyme alkaline phosphatase to the location of the target. We hope that the low-cost and ease of this cell-phone compatible, multiplexed, colorimetric detection system helps in translation of this platform to low resource settings where multiplexed measurements are ideal but fluorescence measurements are not possible.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the NIH-NIBIB Grant 5R21EB024101-02. N. J. was supported in part by a fellowship from the Natural Sciences and Engineering Research Council (NSERC) of Canada. We thank Dr Maxwell Nagarajan, Dr Dana Al-Sulaiman, and Dr Sarah Shapiro for insightful discussions.References
- D. Kim, Q. Wei, D. H. Kim, D. Tseng, J. Zhang, E. Pan, O. Garner, A. Ozcan and D. Di Carlo, Anal. Chem., 2018, 90, 690–695 CrossRef CAS PubMed.
- Q. Feng, Y. Qi, D. Chen and C. Liu, Sens. Actuators, B, 2019, 301, 127119 CrossRef CAS.
- G. C. Le Goff, B. P. Corgier, C. A. Mandon, G. De Crozals, C. Chaix, L. J. Blum and C. A. Marquette, Biosens. Bioelectron., 2012, 35, 94–100 CrossRef CAS PubMed.
- I. Hernández-Neuta, F. Neumann, J. Brightmeyer, T. Ba Tis, N. Madaboosi, Q. Wei, A. Ozcan and M. Nilsson, J. Intern. Med., 2019, 285, 19–39 CrossRef PubMed.
- L. Huang, S. Tian, W. Zhao, K. Liu, X. Ma and J. Guo, Analyst, 2020, 145, 2828–2840 RSC.
- S. Lathwal and H. D. Sikes, Lab Chip, 2016, 16, 1374–1382 RSC.
- G. C. Le Goff, L. J. Blum and C. A. Marquette, Macromol. Biosci., 2013, 13, 227–233 CrossRef CAS PubMed.
- E. H. Yee and H. D. Sikes, ACS Sens., 2020, 5, 308–312 CrossRef CAS PubMed.
- X. Gao, L. P. Xu, T. Wu, Y. Wen, X. Ma and X. Zhang, Talanta, 2016, 146, 648–654 CrossRef CAS PubMed.
- A. E. Urusov, A. V. Zherdev and B. B. Dzantiev, Biosensors, 2019, 9, 89 CrossRef PubMed.
- S. Wei, G. Chen, X. Jia, X. Mao, T. Chen, D. Mao, W. Zhang and W. Xiong, Anal. Chim. Acta, 2020, 1095, 179–184 CrossRef CAS PubMed.
- J. Jiang, B. Zhang, C. Zhang and Y. Guan, Int. J. Mol. Sci., 2018, 19, 3374 CrossRef PubMed.
- J. Dong, G. Chen, W. Wang, X. Huang, H. Peng, Q. Pu, F. Du, X. Cui, Y. Deng and Z. Tang, Anal. Chem., 2018, 90, 7107–7111 CrossRef CAS PubMed.
- S. Persano, M. L. Guevara, J. Wolfram, E. Blanco, H. Shen, M. Ferrari and P. P. Pompa, ACS Omega, 2016, 1, 448–455 CrossRef CAS PubMed.
- W. Shen, H. Deng, Y. Ren and Z. Gao, Chem. Commun., 2013, 49, 4959–4961 RSC.
- Q. Wang, R. D. Li, B. C. Yin and B. C. Ye, Analyst, 2015, 140, 6306–6312 RSC.
- G. C. Le Goff, R. L. Srinivas, W. A. Hill and P. S. Doyle, Eur. Polym. J., 2015, 72, 386–412 CrossRef CAS PubMed.
- S. J. Shapiro, D. Dendukuri and P. S. Doyle, Anal. Chem., 2018, 90, 13572–13579 CrossRef CAS PubMed.
- S. C. Chapin, D. C. Appleyard, D. C. Pregibon and P. S. Doyle, Angew. Chem., Int. Ed., 2011, 50, 2289–2293 CrossRef CAS PubMed.
- N. W. Choi, J. Kim, S. C. Chapin, T. Duong, E. Donohue, P. Pandey, W. Broom, W. A. Hill and P. S. Doyle, Anal. Chem., 2012, 84, 9370–9378 CrossRef CAS PubMed.
- Y. H. Roh, H. J. Lee, H. J. Moon, S. M. Kim and K. W. Bong, Anal. Chim. Acta, 2019, 1076, 110–117 CrossRef CAS PubMed.
- H. J. Kim, W. Choi, J. Kim, J. Choi, N. Choi and K. S. Hwang, Sens. Actuators, B, 2020, 302, 127190 CrossRef CAS.
- A. M. Tentori, M. B. Nagarajan, J. J. Kim, W. C. Zhang, F. J. Slack and P. S. Doyle, Lab Chip, 2018, 18, 2410–2424 RSC.
- M. B. Nagarajan, A. M. Tentori, W. C. Zhang, F. J. Slack and P. S. Doyle, Anal. Chem., 2018, 90, 10279–10285 CrossRef CAS PubMed.
- S. C. Chapin, PhD Thesis, Massachusetts Institute of Technology, 2012.
- H. J. Lee, Y. H. Roh, H. U. Kim, S. M. Kim and K. W. Bong, Lab Chip, 2019, 19, 111–119 RSC.
- H. Lee, R. L. Srinivas, A. Gupta and P. S. Doyle, Angew. Chem., Int. Ed., 2015, 54, 2477–2481 CrossRef CAS PubMed.
- J. J. Kim, L. Chen and P. S. Doyle, Lab Chip, 2017, 17, 3120–3128 RSC.
- L. Cohen, M. R. Hartman, A. Amardey-Wellington and D. R. Walt, Nucleic Acids Res., 2017, 45, e137 CrossRef CAS PubMed.
- C. Liu, X. Xu, B. Li, B. Situ, W. Pan, Y. Hu, T. An, S. Yao and L. Zheng, Nano Lett., 2018, 18, 4226–4232 CrossRef CAS PubMed.
- M. N. Hatori, S. C. Kim and A. R. Abate, Anal. Chem., 2018, 90, 9813–9820 CrossRef CAS.
- G. Destgeer, M. Ouyang, C.-Y. Wu and D. Di Carlo, bioRxiv, 2020, Preprint, DOI: DOI:10.1101/2020.03.15.992321.
- H. Lee, J. Lee, S.-G. Lee and P. S. Doyle, Anal. Chem., 2020, 92, 5750–5755 CrossRef CAS.
- J. A. Schumacher, E. J. Zhao, M. J. Kofron and S. Sumanas, BioTechniques, 2014, 57, 254–256 CrossRef CAS PubMed.
- Sigma-Aldrich, Colorimetric Alkaline Phosphatase and Peroxidase Substrate Detection Systems, 2008, vol. 3 Search PubMed.
- R. C. Murdock, L. Shen, D. K. Griffin, N. Kelley-Loughnane, I. Papautsky and J. A. Hagen, Anal. Chem., 2013, 85, 11634–11642 CrossRef CAS PubMed.
- D. Dendukuri, S. S. Gu, D. C. Pregibon, T. A. Hatton and P. S. Doyle, Lab Chip, 2007, 7, 818 RSC.
- E. Kang, S. Jung, J. H. Abel, A. Pine and H. Yi, Langmuir, 2016, 32, 5394–5402 CrossRef CAS PubMed.
- C. L. Lewis, C.-H. Choi, Y. Lin, C.-S. Lee and H. Yi, Anal. Chem., 2010, 82, 5851–5858 CrossRef CAS PubMed.
- S. Müller, F. Janke, S. Dietz and H. Sültmann, in Tumor Liquid Biopsies. Recent results in cancer research., NLM (Medline), 2019, vol. 215, pp. 299–318 Search PubMed.
- S. Anfossi, A. Babayan, K. Pantel and G. A. Calin, Nat. Rev. Clin. Oncol., 2018, 15, 541–563 CrossRef PubMed.
- P. S. Mitchell, R. K. Parkin, E. M. Kroh, B. R. Fritz, S. K. Wyman, E. L. Pogosova-Agadjanyan, A. Peterson, J. Noteboom, K. C. O'Briant, A. Allen, D. W. Lin, N. Urban, C. W. Drescher, B. S. Knudsen, D. L. Stirewalt, R. Gentleman, R. L. Vessella, P. S. Nelson, D. B. Martin and M. Tewari, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 10513–10518 CrossRef CAS PubMed.
- C. C. Pritchard, H. H. Cheng and M. Tewari, Nat. Rev. Genet., 2012, 13, 358–369 CrossRef CAS PubMed.
- R. L. Srinivas, PhD Thesis, Massachusetts Institute of Technology, 2015.
- J. Miao, J. Wang, J. Guo, H. Gao, K. Han, C. Jiang and P. Miao, Sci. Rep., 2016, 6, 1–7 CrossRef PubMed.
- S. C. Chapin and P. S. Doyle, Anal. Chem., 2011, 83, 7179–7185 CrossRef CAS.
- L. Wu, Y. Wang, R. He, Y. Zhang, Y. He, C. Wang, Z. Lu, Y. Liu and H. Ju, Anal. Chim. Acta, 2019, 1080, 206–214 CrossRef CAS.
- D. C. Pregibon and P. S. Doyle, Anal. Chem., 2009, 81, 4873–4881 CrossRef CAS.
- G. A. Calin and C. M. Croce, Nat. Rev. Cancer, 2006, 6, 857–866 CrossRef CAS.
- M. Mahdiannasser and Z. Karami, Biosens. Bioelectron., 2018, 107, 123–144 CrossRef CAS.
- C. You, L. Jin, Q. I. Xu, B. O. Shen, X. Jiao and X. Huang, Oncol. Lett., 2019, 17, 2271–2277 CAS.
- J. Yang, D. Ma, A. Fesler, H. Zhai, A. Leamniramit, W. Li, S. Wu and J. Ju, Oncotarget, 2017, 8, 52403–52412 CrossRef PubMed.
- H. Cheng, L. Zhang, D. E. Cogdell, H. Zheng, A. J. Schetter, M. Nykter, C. C. Harris, K. Chen, S. R. Hamilton and W. Zhang, PLoS One, 2011, 6(3), e17745 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed materials and methods; supplementary data referenced throughout the text; CAD file for phone imaging setup; sample MATLAB script for image analysis. See DOI: 10.1039/d0an00938e |
| This journal is © The Royal Society of Chemistry 2020 |