Open Access Article
Open Access ArticleA computational and experimental study of the fragmentation of L-leucine, L-isoleucine and L-allo-isoleucine under collision-induced dissociation tandem mass spectrometry†
Candy
Jiang
 ,
Christopher J.
Arthur
,
Christopher J.
Arthur
 and
Paul J.
Gates
and
Paul J.
Gates
 *
*
School of Chemistry, University of Bristol, Cantock's Close, Bristol, BS8 1TS, UK. E-mail: paul.gates@bristol.ac.uk
First published on 6th August 2020
Abstract
The isomeric amino acids L-leucine, L-isoleucine and L-allo-isoleucine, are essential to many vital biological processes and are therefore of interest to the fields of metabolomics and proteomics. Their discrimination can be problematic however due to their isomeric natue. This study demonstrates a systematic investigation of the fragmentations of L-leucine, L-isoleucine and L-allo-isoleucine in combination with a thorough theoretical rationalisation. Collision induced dissociation (CID) tandem mass spectra (MS/MS) of all three amino acids were collected under a range of different collision energies to identify spontaneous and sequential fragmentation processes. We demonstrate that the three structural isomers can be distinguished by their CID MS/MS spectra, and additional computational modelling is used to rationalise these differences.
Introduction
The amino acids, the building blocks of proteins, have attracted wide ranging interests in multidisciplinary studies. L-Leucine (Leu) and L-isoleucine (Ile) are essential amino acids to humans. They are involved in vital biological processes such as haemoglobin formation1 and detection of maple syrup urine disease in new-born screening programs.2 One of the popular areas of research involving Leu and Ile is their characterisation in metabolomics and sequencing of peptides by mass spectrometry (MS).3 As they are structural isomers, routine differentiation of Leu and Ile can be problematic.4The first study to address this challenge, using fast atom bombardment (FAB) ionisation and tandem mass spectrometry (MS/MS), reported a difference in relative abundances of the product ions observed.4 Collisionally activated dissociation (CAD) of the ion [M + H-COOH]+ (m/z 86, formed readily through in-source fragmentation of the protonated molecular ion [M + H]+) showed a higher abundance of the [Precursor-C3H6]+ ion (m/z 44), but a lower abundance of the [Precursor-NH3]+ ion (m/z 69) for Leu when compared to Ile. In a sequencing study of deuterated peptides containing both Leu and Ile, CAD spectra demonstrated a formation of two possible ion structures when Ile was involved in the formation of the N-terminus of a wn ion (Roepstorff/Fohlman designation).5 Either the methyl or ethyl group at the side chain β-carbon of Ile can be retained, this allows the differentiation of Ile from Leu.6 Additionally, the differentiation could also be achieved using higher cone voltages (75–100 V) or low energy collision-induced dissociation (CID) MS/MS with electrospray ionisation (ESI).7
Studies using liquid secondary ion mass spectra of N-substituted Leu and Ile, have reported that the relative abundance of [M + H-HCOOH]+ ion is greater in N-substituted Ile compared to that of N-substituted Leu.8 Transition metals such as copper(II) have also been used to form complexes with Leu and Ile, to distinguish and quantify the isomers by MS/MS.9,10 High field asymmetric waveform ion mobility spectrometry (FAIMS) has also been employed to separate the gas-phase molecular anions of Leu and Ile in mixtures. Discrimination was possible when either of the amino-acids had a concentration 625 times more than the other in a mixture.11 Techniques such as hot-electron capture-dissociation have been used to distinguish Ile from Leu in the analysis of intact α- and β-globin chains.12
Recently, the differentiation of Leu and Ile in peptide sequencing was reported using integrated LC-MS strategies in combination with high energy CID-MS (HCD-MSn), multistage electron transfer dissociation (ETD) and higher energy CAD with an Orbitrap Fusion mass spectrometer.13,14 Another recent study of the fragmentation of proteinogenic amino acids stated that under lower collision energy (<30% NCE), product ion m/z 69 was only observed for Ile.15 ETD and HCD have also been used to produce reliable differentiation of the isomeric amino acids in neighbouring positions of short tryptic peptides.16
In this study, a systematic analysis of the CID-MS/MS of L-leucine, L-isoleucine and L-allo-isoleucine obtained from both an Orbitrap Elite and a Synapt G2S instruments is reported. Energy breakdown graphs and full fragmentation pathways are presented and, in combination with insights from quantum mechanical calculations, possible rationalisations for the differences in the fragmentation of these three isomers is presented.
Experimental
The pure amino-acids were purchased from Sigma-Aldrich (Gillingham, UK). They were dissolved in methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (50
water (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) to make up stock solution of 1 mg ml−1. For MS analysis, the stock solution of these three amino acids were diluted to a final concentration 0.1 mg ml−1.
50) to make up stock solution of 1 mg ml−1. For MS analysis, the stock solution of these three amino acids were diluted to a final concentration 0.1 mg ml−1.
Positive ion ESI-MS/MS experiments were performed on an Orbitrap Elite mass spectrometer (ThermoFisher Scientific, Hemel Hempstead, UK). Samples were introduced by autosampler (25 μl min−1) and spectra were recorded in the FTMS mode from 50 and 300 m/z at a resolution of 240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Collision induced dissociation (CID) was performed on the protonated precursor ions with the energy from 1 to 50 eV (in 1 eV steps). The RAW data files were converted to the open mzML format using ProteoWizard msConvert.17 Spectra were then read by KNIME18 and extracted into Excel for later analyses.
000. Collision induced dissociation (CID) was performed on the protonated precursor ions with the energy from 1 to 50 eV (in 1 eV steps). The RAW data files were converted to the open mzML format using ProteoWizard msConvert.17 Spectra were then read by KNIME18 and extracted into Excel for later analyses.
Positive ion Nanospray-MS/MS experiments were performed on a Synapt G2S mass spectrometer (Waters, Manchester, UK) equipped with a Nanomate Triversa chip-based nanospray system (Advion Biosciences, Norwich, UK). The Nanomate was set to aspirate 5 μl of sample solution. The CID collision energy was set in the range from 0 to 25 eV and spectra recorded in the range from 20 to 150 m/z at a resolution of approximately 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.
Density functional theory (DFT) quantum mechanical calculations were performed with Gaussian 0919 using a B3PW91 exchange–correlation functional and a 6-311G++(d,p) basis set. B3PW91 was chosen due to its successful performance in previously published studies, where it out performs B3LYP.23–25 The basis set chosen was 6-311++G (d,p) based on its consistent results obtained within our own research and in a few relevant studies.26–28 All structures were minimised prior to any frequency calculations.
Results
Orbitrap
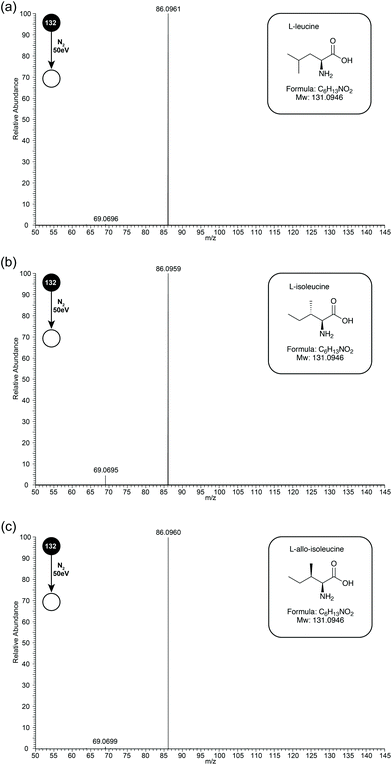
Leu, Ile, and L-allo-Ile are structural isomers, separation of which can be a challenge by MS (see Fig. 1 inserts for structures). Early studies have suggested that differences occurred in the relative abundances of fragment ions m/z 69 and 44. During this study, the three isomeric amino-acids were studied by ESI-MS/MS on an Orbitrap Elite mass spectrometer. Unfortunately, due to instrument limitations, ions below m/z 50 cannot be observed on this instrument. Due to this drawback, experiments were also performed on a Synapt instrument, and later discussion will be based on data obtained from both instruments.In initial experiments, CID MS/MS spectra of Leu, Ile and L-allo-Ile were obtained at collision energy 50 eV (see Fig. 1 for CID-MS/MS spectra of the three compounds at 50 eV). The differences in relative abundances of product ion m/z 69, formed through loss of NH3 from product ion m/z 86, can be readily observed (see Table 1). Ile has the most abundant m/z 69, followed by L-allo-Ile, and finally Leu. This agrees with the previous studies even though they were performed by a range of techniques.4–8,15
| Compound | Relative intensity m/z 86 | Relative intensity m/z 69 |
|---|---|---|
| L-Leucine | 0.976 | 0.002 |
| L-Isoleucine | 0.941 | 0.040 |
| L-Allo-Isoleucine | 0.923 | 0.012 |
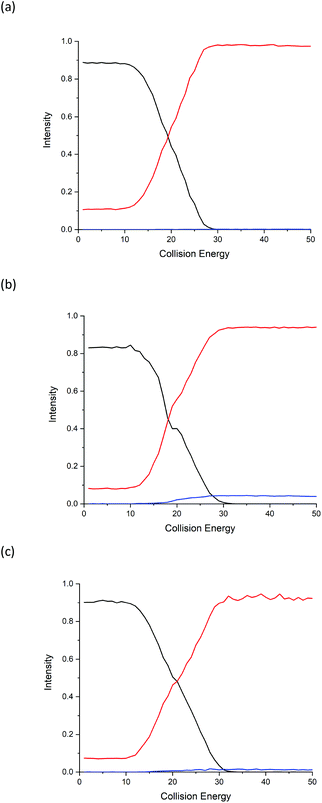
Energy breakdown graphs are 2D plots of collision energy against m/z and represent the behaviour of a select product ion (m/z) at a range of different CID energies of the precursor ion.20,21 Energy breakdown graphs were plotted to further represent the differences (see Fig. 2). The red line is used to represent the product ion m/z 86 and the blue line for m/z 69. All three isomeric amino acids have characteristic increases in the loss of HCOOH (m/z 86). The increase in relative intensity of product ion m/z 69 for Ile is easily seen as the collision energy rises, L-allo-Ile shows a much smaller increase, whereas for Leu m/z 69 is hardly observed, even at the highest collision energies. The summary of the trend in relative abundances of m/z 69 for all three amino acids is: I(69)Ile > I(69)L-allo-Ile > I(69)Leu where I(69) represents the peak intensity of product ion m/z 69.
Synapt G2S
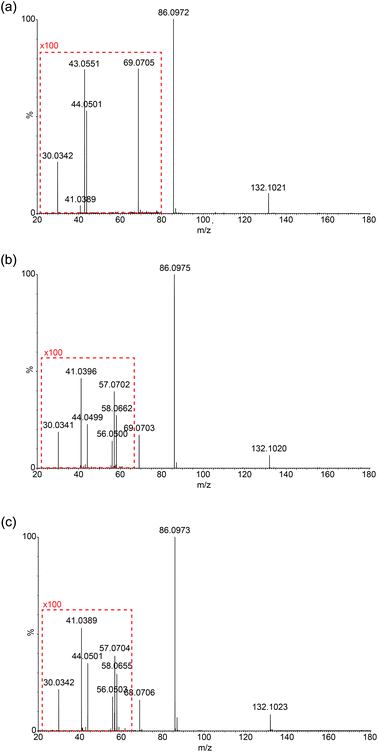
Due to the limitation in the Orbitrap's mass range, the same experiment was repeated on a Synapt G2S mass spectrometer using a chip-based nanospray ionisation. Low mass range scans and energy breakdown graphs (0 to 25 eV) were acquired to try to improve differentiation of the three structural isomers (see Fig. 3 and ESI†). From Fig. 3, the peak intensity of product ion m/z 69 is the highest for Ile but lowest for Leu, which corresponds to the results previously obtained from the CID-MS/MS on Orbitrap. The same trend follows I(69)Ile > I(69)L-allo-Ile > I(69)Leu.Low-mass product ions can be observed for all three structural isomers in the Synapt recorded MS/MS data. The capability to scan down to m/z 20 dramatically improves the discrimination of the isomers. Leu has product ions at m/z 43, 44 and 30; whereas both Ile and L-allo-Ile have product ions at m/z 56, 57, 58, 44, 41 and 30. There is a slight variation in the peak intensities of these product ions for Ile and L-allo-Ile.
Table 2 summarises the data, from Fig. 3 and demonstrates the differences in the product ions observed in the lower mass range (<m/z 60). More peaks are produced for Ile and L-allo-Ile compared to Leu. Molecular formulae are proposed for these peaks and possible mechanisms for the generation of a selection of them are proposed in the discussion.
| Ion formula | Theoretical mass | Observed masses | ||
|---|---|---|---|---|
| L-Leucine | L-Isoleucine | L-Allo-isoleucine | ||
| C6H14NO2+ | 132.1025 | 132.1021 | 132.1020 | 132.1023 |
| C5H12N+ | 86.0970 | 86.0972 | 86.0975 | 86.0973 |
| C5H9+ | 69.0704 | 69.0705 | 69.0703 | 69.0706 |
| C3H8N+ | 58.0657 | — | 58.0662 | 58.0655 |
| C4H9+ | 57.0704 | — | 57.0702 | 57.0704 |
| C3H6N+ | 56.0500 | — | 56.0500 | 56.0503 |
| C2H6N+ | 44.0500 | 44.0501 | 44.0499 | 44.0501 |
| C3H7+ | 43.0548 | 43.0551 | — | — |
| C3H5+ | 41.0391 | 41.0389 | 41.0396 | 41.0389 |
| CH4N+ | 30.0344 | 30.0342 | 30.0341 | 30.0342 |
Discussion
Based on observations in their MS/MS spectra and energy breakdown graphs mechanisms for the generation of the product ions of Leu and Ile are proposed in Scheme 1. L-Allo-Ile shares the same route as for Ile and will therefore not be discussed further. Gaussian calculations performed during this study demonstrate that the most energetically favourable protonation site is on the nitrogen atom, which agrees with previous studies.22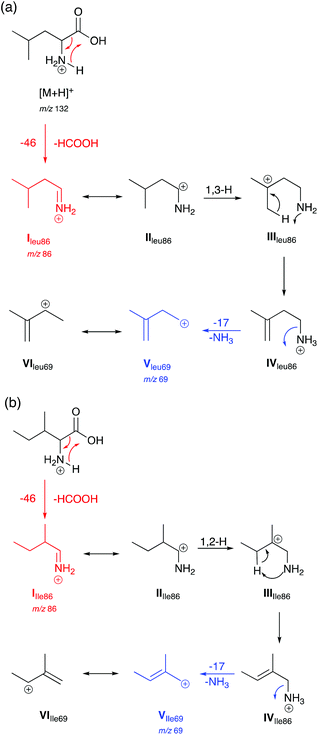 | ||
| Scheme 1 Proposed mechanisms for the generation of selected product ions in the CID-MS/MS of (a) Leu and (b) Ile in positive ion mode. Product ions are m/z 86 and 69. | ||
Both Leu and Ile have an initial loss of HCOOH, which has been found to be a non-discriminating fragmentation for almost all the 20 proteinogenic α-amino acids (data not shown). The mechanism for Leu fragmentation involves a 1,3-hydride shift from structure II to III. This results in the positive charge moving from the secondary carbon onto a much more favoured tertiary carbon that can stabilise the structure. This is followed by a proton transfer to the nitrogen. Intermediate IV then undergoes an entropically favoured neutral loss of ammonia to form intermediate V (m/z 69) which is stabilised through delocalisation with the adjacent alkene (structure VI).
In the case of Ile and L-allo-Ile (not shown), as the methyl group is now on the β-carbon, a 1,2-hydride shift from intermediate II to III forms a tertiary carbocation in both cases, which can better stabilise the positive charge than the secondary carbocation in the case of Leu. This is then accompanied by proton transfer and a loss of NH3 as seen for Leu.
Mechanisms for the formation of product ions m/z 44, 43 and 30 for Leu and m/z 58, 57, 56 and 41 for Ile are proposed in Scheme 2. To obtain ions m/z 58, 44 and 30, a McLafferty-type rearrangement occurs.29 Ion m/z 58 then undergoes dehydrogenation to form ion m/z 56. Ions m/z 58, 57 and 56 are unique for Ile due to the position of the methyl group on the β-carbon sidechain. This can be used as a characteristic factor to differentiate between Leu and Ile.
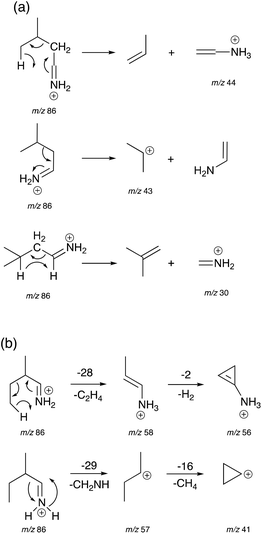 | ||
| Scheme 2 Proposed mechanisms for the generation of product ions m/z 44, 43, and 30 for Leu (a), and product ions m/z 58, 57, 56 and 41 for Ile (b). | ||
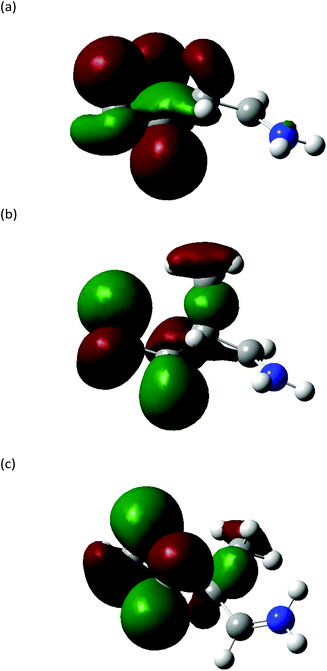
The Highest Occupied Molecular Orbitals (HOMO, molecular orbital = 24) of structure I (product ion m/z 86) of Leu, Ile and L-allo-Ile were determined using DFT calculations. These HOMOs showed distinctive differences (Fig. 4). With two methyl groups at the end of Leu, a delocalisation of orbitals could be observed, leaving CH2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH2+ (m/z 30) to be a more accessible product on the end of the structure. This agrees with previous studies of Leu4,22 and results obtained from the Synapt mass spectra in this study, where Leu has a higher peak intensity of product ion m/z 30 compared to m/z 69. The HOMO of Ile reveals a diffusion of orbitals from the second to the fourth carbon, encouraging charge transfer from structure IIle86 to IIIIle86. This leads to the fragmentation of this ion shown in Scheme 2 and leads to the higher abundance of product ion m/z 69 in the fragmentation spectrum of Ile and its corresponding energy breakdown graph (Fig. 2). L-Allo-Ile shows a HOMO in between of Leu and Ile. This corresponds to its MS/MS spectrum, where the peak intensity of m/z 69 lies between that of Leu and Ile.
NH2+ (m/z 30) to be a more accessible product on the end of the structure. This agrees with previous studies of Leu4,22 and results obtained from the Synapt mass spectra in this study, where Leu has a higher peak intensity of product ion m/z 30 compared to m/z 69. The HOMO of Ile reveals a diffusion of orbitals from the second to the fourth carbon, encouraging charge transfer from structure IIle86 to IIIIle86. This leads to the fragmentation of this ion shown in Scheme 2 and leads to the higher abundance of product ion m/z 69 in the fragmentation spectrum of Ile and its corresponding energy breakdown graph (Fig. 2). L-Allo-Ile shows a HOMO in between of Leu and Ile. This corresponds to its MS/MS spectrum, where the peak intensity of m/z 69 lies between that of Leu and Ile.
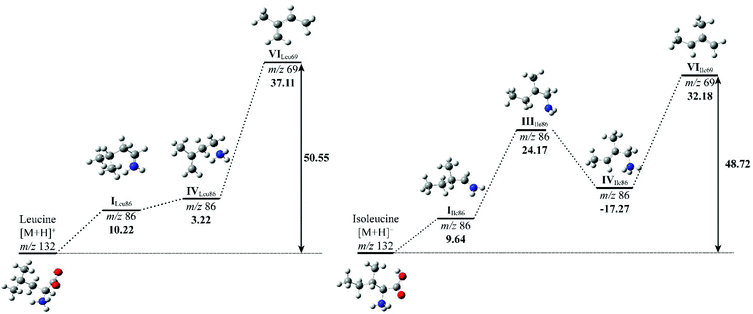
To compare the energy costs for each fragmentation pathway to form product ion m/z 69, potential energy diagrams were constructed for Leu and Ile (Fig. 5). Each pathway followed their proposed mechanisms respectively and are labelled with corresponding structures. In comparison, Leu requires more energy to initiate the loss of formic acid in the first step. Although the energy difference between structure I and IV is only 3.22 kcal mol−1, the overall energy costs to form structure VI are higher than that of Ile. Ile has more stabilised structures in its suggested fragmentation pathway. From structures I to III of Ile, there is a noticeable increase in energy, however, this is offset by a decrease of 17.27 kcal mol−1 from structure III to IV. The total energy costs to form product ion m/z 69 for Ile are lower when compared to Leu, suggesting a more energetically favourable fragmentation route.
The relative energy of formation of product ion m/z 69 has been summarised, alongside with their peak intensity at collision energy 50 eV of all three isomeric amino acids (Table 3). It confirms the trend that Leu requires the highest amount of energy to produce ion m/z 69, hence the observation of the lowest peak intensity in its MS/MS spectrum. A calculation of the relative formation energy of product ion m/z 30 (Scheme 2) of Leu revealed that it costs relatively less in energy (34.17 kcal mol−1) than to form product ion m/z 69. Hence the observation of the higher peak intensity of m/z 30 to m/z 69 in mass spectrum of Leu (Fig. 3).
| Compound | Relative intensity m/z 69 | ΔH69 (kcal mol−1) | ΔHTotal (kcal mol−1) |
|---|---|---|---|
| L-Leucine | 0.002 | 37.11 | 50.55 |
| L-Isoleucine | 0.040 | 32.18 | 48.72 |
| L-Allo-Isoleucine | 0.012 | 34.58 | 49.93 |
Conclusions
In conclusion, during this study differences have been observed in the CID MS/MS mass spectra of Leu, Ile and L-allo-Ile on both Orbitrap Elite and Synapt G2S instruments. It is possible to distinguish the three isomeric amino acids by the energy breakdown graphs generated for the Orbitrap data and additionally by thorough examination of the lower mass range MS/MS spectra obtained from the Synapt instrument. Using high-performance instrumentation with wide dynamic ranges allows for the generation of spectra which are highly reliable with very little variation in peak intensities. Computational modelling of relative formation energies and molecular orbital visualisations are necessary to provide theoretical explanations for any discrimination.Distinguishable product ions for all three isomeric α-amino-acids have been identified and the results from their MS/MS spectra revealed a noticeable difference in peak intensities of key product ions. Computational data has been utilised to explain the differences observed in the peak intensities of product ion m/z 69 for Leu, Ile and L-allo-Ile. Constructions of HOMOs of Leu, Ile and L-allo-Ile showed that Leu has a preference to generate product ion m/z 30 instead of m/z 69. Potential energy diagrams of Leu and Ile also demonstrated that Ile has a more energetically favourable fragmentation pathway to produce ion m/z 69.
Previous studies have experimented with various methods to distinguish Leu and Ile, however, none of those have had any detailed investigation to rationalise these differences. This study provides sufficient evidence to allow full differentiation of Lue and Ile using results obtained from their MS/MS spectra. Additionally, computational modelling has been employed to rationalise these differences in terms of the relative formation energies and molecular orbital constructions of the product ions. For future work, analysis performed in this study can have further applications in metabolomics and proteomics. We believe the techniques used here provide a solid foundation for the structural elucidation of more complex analytes and isomeric species.
Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
The authors would like to thank to Dr Natalie Fey for helpful discussions with the computational modelling contained in this study. CJ also thanks the mass spectrometry facility in the University of Bristol for providing access to all of the facilities.Notes and references
- R. Schweet, H. Lamfrom and E. Allen, Proc. Natl. Acad. Sci. U. S. A., 1958, 44, 1029 CrossRef CAS PubMed.
- K. A. Strauss, E. G. Puffenberger and D. H. Morton, in Maple Syrup Urine Disease, ed. M. P. Adam, University of Washington, Seattle, GeneReviews®, Seattle (WA), 1993, 1993–2020, PMID: 20301495 Search PubMed.
- K. Biemann, Biol. Mass Spectrom., 1988, 16, 99 Search PubMed.
- I. L. Aubagnac and B. Elamrani, Org. Mass Spectrom., 1985, 20, 428 CAS.
- P. Roepstorff and J. Fohlman, Biomed. Mass Spectrom., 1984, 11, 601 CrossRef CAS PubMed.
- J.T. Stults and J.T. Watson, Biol. Mass Spectrom., 1987, 14, 583 Search PubMed.
- A. G. Hulst and C. E. Kientz, J. Mass Spectrom., 1996, 31, 1188 CrossRef CAS.
- P. Krishna, S. Prabhakar and M. Vairamani, Rapid Commun. Mass Spectrom., 1998, 12, 1429 CrossRef CAS.
- J. L. Seymour and F. Turecek, J. Mass Spectrom., 2000, 35, 566 CrossRef CAS.
- W. A. Tao, L. M. Wu and R. G. Cooks, J. Am. Soc. Mass Spectrom., 2001, 12, 490 CrossRef CAS PubMed.
- D. A. Barnett, B. Ells, R. Guevremont and R. W. Purves, J. Am. Soc. Mass Spectrom., 1999, 10, 1279 CrossRef CAS.
- J. P. Williams, A. J. Creese, D. R. Roper, B. N. Green and H. J. Cooper, J. Am. Soc. Mass Spectrom., 2009, 20, 1707 CrossRef CAS PubMed.
- A.T. Lebedev, E. Damoc, A. A. Makarov and T. Y. Samgina, Anal. Chem., 2014, 86, 7017 CrossRef CAS PubMed.
- Y. Xiao, M. M. Vecchi and D. Wen, Anal. Chem., 2016, 88, 10757 CrossRef CAS PubMed.
- P. Zhang, W. Chan, I. L. Ang, R. Wei, M. M. T. Lam, K. M. K. Lei and T. C. W. Poon, Sci. Rep., 2019, 9, 6453 CrossRef PubMed.
- S. V. Kovalyov, S. S. Zhokhov, L. V. Onoprienko, B. V. Vaskovsky and A.T. Lebedev, Eur. J. Mass Spectrom., 2017, 23, 376 CrossRef CAS PubMed.
- M. C. Chambers, B. Maclean, R. Burke, D. Amodei, D. L. Ruderman, S. Neumann, L. Gatto, B. Fischer, B. Pratt, J. Egertson, K. Hoff, D. Kessner, N. Tasman, N. Shulman, B. Frewen, T. A. Baker, M. -Y. Brusniak, C. Paulse, D. Creasy, L. Flashner, K. Kani, C. Moulding, S. L. Seymour, L. M. Nuwaysir, B. Lefebvre, F. Kuhlmann, J. Roark, P. Rainer, S. Detlev, T. Hemenway, A. Huhmer, J. Langridge, B. Connolly, T. Chadick, K. Holly, J. Eckels, E. W. Deutsch, R. L Moritz, J. E. Katz, D. B. Agus, M. MacCoss, D. L. Tabb and P. Mallick, Nat. Biotechnol., 2012, 30, 918 CrossRef CAS PubMed.
- M. R. Berthold, N. Cebron, F. Dill, G. Di Fatta, T. R. Gabriel, F. Georg, T. Meinl, P. Ohl, C. Sieb and B. Wiswedel, Studies in Classification, Data Analysis and Kowledge Organization, Springer-Verlag Berlin Heidelberg, Germany, 2007 Search PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- C. P. G. Butcher, P. J. Dyson, B. F. G. Johnson, P. R. R. Langridge-Smith, J. S. McIndoe and C. Whyte, Rapid Commun. Mass Spectrom., 2002, 16, 1595 CrossRef CAS.
- A. G. A. Harrison, Rapid Commun. Mass Spectrom., 1999, 13, 1663 CrossRef CAS PubMed.
- S. Bouchonnet and Y. Hoppilliard, Org. Mass Spectrom., 1992, 27, 71 CrossRef CAS.
- J. B. Foresman and A. Frisch, Exploring Chemistry with Electronic Structure Methods, Gaussian, Inc, Wallingford, CT, 2015, ISBN: 0963676938.
- N. Chéron, D. Jacquemin and P. Fleurat-Lessard, Phys. Chem. Chem. Phys., 2012, 14, 7170 RSC.
- J. N. Dahanayake, C. Kasireddy, J. M. Ellis, D. Hildebrandt, O. A. Hull, J. P. Karnes, D. Morlan and K. R. Mitchell-Koch, J. Comput. Chem., 2017, 38, 2605 CrossRef CAS PubMed.
- T. Shoeib, A. Cunje, A.C. Hopkinson and K. W. M. Siu, J. Am. Soc. Mass Spectrom., 2002, 13, 408 CrossRef CAS PubMed.
- W. Yu, L. Liang, Z. Lin, S. Ling, M. Haranczyk and M. Gutowski, J. Comput. Chem., 2009, 30, 589 CrossRef CAS PubMed.
- N. Kerru, L. Gummidi, S. V. H. S. Bhaskaruni, S. N. Maddila, P. Singh and S. B. Jonnalagadda, Sci. Rep., 2019, 9, 19280 CrossRef CAS PubMed.
- K. Levsen and F. W. McLafferty, J. Am. Chem. Soc., 1974, 96, 139 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0an00778a |
| This journal is © The Royal Society of Chemistry 2020 |