Engineering at the nano-bio interface: harnessing the protein corona towards nanoparticle design and function
Rebecca L.
Pinals
 a,
Linda
Chio
a,
Linda
Chio
 a,
Francis
Ledesma
a,
Francis
Ledesma
 a and
Markita P.
Landry
a and
Markita P.
Landry
 *abcd
*abcd
aDepartment of Chemical and Biomolecular Engineering, University of California, Berkeley, Berkeley, California 94720, USA. E-mail: landry@berkeley.edu
bCalifornia Institute for Quantitative Biosciences, QB3, University of California, Berkeley, Berkeley, California 94720, USA
cInnovative Genomics Institute (IGI), Berkeley, California 94720, USA
dChan-Zuckerberg Biohub, San Francisco, California 94158, USA
First published on 26th June 2020
Abstract
Unpredictable and uncontrollable protein adsorption on nanoparticles remains a considerable challenge to achieving effective application of nanotechnologies within biological environments. Nevertheless, engineered nanoparticles offer unprecedented functionality and control in probing and altering biological systems. In this review, we highlight recent advances in harnessing the “protein corona” formed on nanoparticles as a handle to tune functional properties of the protein–nanoparticle complex. Towards this end, we first review nanoparticle properties that influence protein adsorption and design strategies to facilitate selective corona formation, with the corresponding characterization techniques. We next focus on literature detailing corona-mediated functionalities, including stealth to avoid recognition and sequestration while in circulation, targeting of predetermined in vivo locations, and controlled activation once localized to the intended biological compartment. We conclude with a discussion of biocompatibility outcomes for these protein–nanoparticle complexes applied in vivo. While formation of the nanoparticle–corona complex may impede our control over its use for the projected nanobiotechnology application, it concurrently presents an opportunity to create improved protein–nanoparticle architectures by exploiting natural or guiding selective protein adsorption to the nanoparticle surface.
1. Introduction
When engineered nanoparticles are introduced into a biological medium, proteins swiftly adsorb to and coat the nanoparticle surfaces. This phenomenon is at present well-established, duly termed formation of the nanoparticle's “protein corona” to provoke imagery of the corona surrounding the sun during a solar eclipse, with tendrils of light (or rather, adsorbed proteins) reaching outwards. As our repertoire of engineered nanoparticles becomes ever-more diverse, these nanoparticles are continually applied for broader functions across vastly differing biological environments. A comprehensive understanding of the protein corona remains one of the greatest challenges in successfully developing and implementing nanobiotechnologies. Moreover, by delving into the fundamental interactions governing protein corona formation, we realize the opportunities to be had in taking advantage of this phenomenon.In this review, we begin by discussing the unpredictable protein corona formed upon exposure of nanoparticles to biological environments, then expand into how recent work has employed this information towards a priori design of corona-mediated functionalities. We highlight certain corona design examples, alongside the relevant development and characterization techniques. Corona design discussion is centered on applications towards corona-mediated nanoparticle stealth, targeting, and activation, with a corresponding discussion of nanoparticle construct biocompatibility to follow.
2. Corona-based nanoparticle design
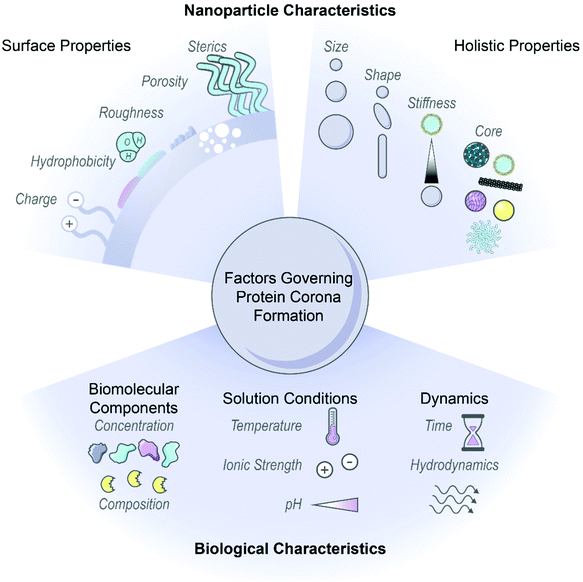
Nanoparticles have emerged as an ideal platform upon which to develop biological sensing, imaging, and delivery tools.1–3 However, pristine nanoparticles undergo significant transformations once injected into biological environments: biomolecules, most notably proteins, rapidly coat the nanoparticle surface in the energetically favorable process of corona formation.4,5 The abruptness of protein adsorption on foreign nanosurfaces causes proteins to interact in unusual modes, contrary to the normal protein–protein interactions governed by precise genetic control, and often produces undesirable outcomes such as protein denaturation.6,7 Further, corona formation unpredictably changes the nanoparticle identity and fate, as the adsorbed proteins mask original surface characteristics and endow new biochemical properties to the nanoparticle.8–10 As a result, how the nanoparticle–corona complex interacts with biological machinery is impacted and in vivo circulation, bioaccumulation, and biocompatibility outcomes are drastically modified.11,12 Consequently, protein corona formation can lead to reduced or abolished nanoparticle efficacy and contradiction of expected in vitro results, whereby the nanoparticle is no longer able to carry out its designated function.13,14 On the contrary, the protein corona can be taken advantage of, where in situ protein adsorption may facilitate stealth targeting and delivery, with improved functionality or therapeutic effect to follow.15,16 Thus, the surface-adsorbed corona may be exploited by avoiding nonselective, deleterious protein adsorption in favor of selective, advantageous protein–surface assembly.17 Regardless, the protein corona displayed on the nanoparticle surface must be appreciated as one of the principle design parameters to ensure successful applications of nanobiotechnologies in vivo.Nanoparticle physicochemical properties and the surrounding bioenvironment are inherent variables affecting protein corona formation. Many studies classify the protein corona around specific nanoparticles in specific biological systems of interest, although findings often lack generality or are contradictory as to which nanoparticle or protein properties drive protein corona composition, dynamics, and subsequent biological outcomes. Additionally, the protein corona is dependent upon a convolution of parameters carrying varying weights, and often these parameters are difficult to decouple without meticulous experimental design. Therefore, while we summarize recent findings and generic design rules as depicted in Fig. 1, we note that these generalities may not always hold depending on the intricacies of the nanoparticle-biosystem under consideration.
2.1. Nanoparticle characteristics
Nanoparticle surface properties are most significant in governing protein corona formation, as compared to the nanoparticle core.8,18,19 These nanoparticle surface features include: (i) electrostatic charge, (ii) hydrophobicity, and (iii) surface structure. These attributes are functions of nanoparticle surface chemistry and ligand functionalization.(i) Electrostatic charge. Nanoparticle charge affects protein corona composition and packing density.20–24 Many studies conclude that proteins possessing opposite charges from the nanoparticles are enriched in the corona. As most proteins are negatively charged at physiological pH, it is often found that cationic nanoparticles adsorb the highest number of proteins.10,20–22,25 However, there are examples in which proteins are able to overcome electrostatically adverse conditions and adsorb by nonelectrostatic driving forces.26–28 It is important to consider that even if nanoparticles are synthesized with appropriate surface chemistries, these nanosurfaces may only retain these engineered features transiently within biosystems.8 Nanoparticle charge further impacts surface packing, where higher magnitude charge leads to more proteins in the corona.18 Other studies conclude that more cationic surfaces increase conformational changes of adsorbed proteins.28 These results are contradictory, in that post-adsorptive protein structural changes generally take place under lower surface packing densities, where proteins have more accessible area to spread out and denature on the surface.6,29 It is unlikely that such conformational changes would be able to occur in a highly crowded corona environment. Beyond the individual nanoparticle, surface charge also directly impacts colloidal stability because neutral surfaces (or surfaces neutralized by protein and ion adsorption) tend to aggregate in the absence of intervening electrostatic repulsions.30 As will be discussed below, the surrounding solution ionic strength determines the importance of electrostatic interactions, as these interactions are screened and play less of a role in high-salt systems. Accordingly, nanoparticles must be designed in such a manner that they are not only colloidally stable as a homogeneous solution, but retain colloidal stability in the presence of proteins in the surrounding bulk and surface-adsorbed state.20
Manipulating nanoparticle charge offers a useful means to tune nanoparticle interactions with cells, with regards to cell internalization and toxicity:31 positively charged nanoparticles have enhanced cell internalization due to interactions with the negatively charged cell membrane,2,32–35 especially enhanced for the case of cancer cells.21 Yet, if positively charged nanoparticles bind too many proteins, this leads to colloidal instability, aggregation, and downstream toxicity.20,36
(ii) Hydrophobicity. Nanoparticle surface chemistry also dictates hydrophobicity, where hydrophobic nanoparticles exhibit increased protein adsorption capacity,9,18 more stable protein adsorption,9,23,30 and cause more protein conformational changes.9,30 Nanoparticles with hydrophobic surfaces are more likely to produce deleterious effects on protein structures, as protein unfolding is driven by the favorable interactions of the protein hydrophobic core with the nanoparticle surface.9 Frequently, such unfolding is irreversible, leading to larger scale aggregates, nonfunctional nanoparticles, and immune activation/clearance.25,30 Again, these conclusions of high packing density and high conformational changes are at odds with each other, for the same reasons as specified for electrostatic charge.
(iii) Surface structure. Features of nanoparticle surface topography that impact protein corona formation include surface roughness, porosity, and sterics. A rough or porous surface creates more available surface area for proteins to coat and so allows proteins to minimize lateral repulsive forces in the adsorbed state. Functionalization with polymers, targeting ligands, or other moieties that extend outward into solution also affects nanoparticle surface topography. The grafting density and conformation of such attached ligands impacts accessibility of proteins to the nanoparticle surface.37,38 Based on the principles of both hydrophobicity and surface structure, corona mitigation techniques often involve surface-grafting of hydrophilic polymers such as polyethylene glycol (PEG) to abrogate protein adsorption and sterically stabilize the nanoparticle.8,39,40 Higher antiadhesive polymer coverage is associated with alleviated protein corona formation, while lower coverage or linear conformations are less effective in deterring protein binding.38,41,42 Combining these concepts of surface roughness and sterically stabilizing polymers, Piloni et al. demonstrated that a patchy polymer-grafted nanoparticle reduced protein adsorption in comparison to a smooth polymer-grafted surface by six-fold (protein content per nanoparticle).11 PEG strategies and efficacies will be discussed further in section 4.1.1.
Beyond surface characteristics, other nanoparticle factors that impact corona formation include: (iv) size, (v) curvature or shape, (vi) stiffness, and, to a much lesser extent, (vii) core material composition.
(iv) Size. Size is found to quantitatively, though not qualitatively, impact corona formation: larger nanoparticles present more high surface free energy, net exposed area and permit higher protein loading per nanoparticle,22,43 although the corona constituents are often no different than those on smaller nanoparticles of identical material properties.30 On the other hand, smaller nanoparticles possess a higher surface area to volume ratio, hence smaller nanoparticles adsorb more proteins on an area normalized basis. Optimizing the metric of protein loading on a nanoparticle number or area basis depends on the desired application, as will be discussed in subsequent sections. Further, increasing nanoparticle size decreases surface curvature (though dependent on the geometry), therefore there exists a threshold above which larger particles do not necessarily adsorb more proteins.44
Another important consideration is the effect of nanoparticle size on targeting and localization efficiencies: biological barrier crossing efficiency and mechanism are both size-dependent, where successful crossing scales inversely with size.34,45 Here, it is critical to consider the hydrodynamic, in situ nanoparticle–corona complex size; an adsorbed protein corona may add up to hundreds of nanometers to in vitro particle size.46 Biological barriers range from vasculature walls to cell membranes, with typical cutoff dimensions including <6 nm for renal clearance24vs. >300 nm for liver or spleen filtration,47 ∼20–200 nm for tumor penetration and retention,3,47,48 <50–100 nm for blood–brain barrier (BBB) crossing and extracellular matrix (ECM) navigation,49,50 and ∼10–100 nm for cell internalization.34 On the cellular level, Shadmani et al. applied a mathematical model based on diffusion of membrane-mobile receptors to examine how protein corona formation impacts internalization of gold nanoparticles by receptor-mediated endocytosis.45 From this model, optimal values for nanoparticle–corona radius (40 nm bare diameter increasing to 60 nm upon corona formation) and targeting ligand density (∼1500 μm−2 on a 100 nm gold nanoparticle) are described to minimize endocytosis time through a balance of membrane tension energy and ligand–receptor interaction density, demonstrating how in silico models are useful towards nanoparticle design.
(v) Curvature/shape. Reiterating the earlier discussion of lateral interactions, now considering nanoparticle shape, a higher curvature surface minimizes adverse lateral protein–protein interactions. Thus, a more curved nanoparticle surface would be expected to adsorb more proteins if unfavorable protein–protein interactions are preeminent (e.g. lateral, repulsive electrostatic interactions). However, if favorable nanoparticle–protein interactions dominate (e.g. attractive dispersion forces), a flatter surface would be advantageous to facilitate more adsorption. This latter case is manifested as higher protein adsorption (per unit surface area) on higher aspect ratio nanoparticles, such as nanorods relative to nanospheres.51 Other studies find that curvature impacts adsorbed protein orientation to result in lower packing.52 Therefore, no generalizable rules can be deduced with regards to the effect of nanoparticle shape on protein adsorption. Once applied in vivo, shape also dictates how nanoparticles behave within convective flow, such as how they interact with bounding walls, and internalization efficiencies and mechanisms, since membrane bending energy is dependent on nanoparticle form factor.34
(vi) Stiffness. As with shape, nanoparticle stiffness has been shown to affect modes of cell internalization and bioaccumulation: less stiff nanoparticles generally exhibit lower cell internalization across many cell types, and correspondingly longer circulation times due to the more difficult uptake and clearance by macrophages.33,53,54 Stiffness here refers to the nanomaterial's ability to resist deformation under applied force, related to the material's Young's Modulus and geometry. Yet, the impact of nanoparticle stiffness on protein corona formation remains relatively under-studied. While proteins are considered soft matter and may be expected to increase the inherent nanoparticle softness, this may not be the dominant factor, as higher protein adsorption often leads to the opposite downstream outcomes as those reported for less stiff nanoparticles. The effect of nanoparticle stiffness on protein adsorption remains an open question.
(vii) Core composition. Finally, while the core material composition does influence corona formation to some extent,22 the core is mostly shielded from direct exposure with the biofluid and thus plays a minor role in determining protein corona formation. However, use of exogenous nanoparticle core materials can lead to immune activation and toxicity during attempted clearance.24
2.2. Biological environment factors
In addition to the influence of innate nanoparticle variables on protein corona formation, the bioenvironment of the intended application must be taken into account. Environmental parameters include: (i) biomolecular components, (ii) solution conditions, and (iii) surrounding dynamics.(i) Biomolecular components. Native biomolecule concentration and composition within a biological environment influences the consequent protein corona formed on nanoparticles. Higher protein concentration in the surrounding fluid frequently leads to more protein adsorption on nanoparticles, as suggested by ideal-solution thermodynamics, and witnessed experimentally.7,55 Nonetheless, relative corona protein concentration does not necessarily correlate with native circulating protein concentration due to preferential protein partitioning into surface vs. bulk solution phases.26,56 More complex mechanisms often govern protein corona formation, giving rise to surprising magnitudes of protein enrichment or depletion on nanoparticles relative to the native biofluid. A frequent example of this phenomenon is the Vroman effect, where highly abundant proteins initially adsorbed to nanoparticles competitively exchange with and are eventually replaced by lower abundance, higher surface-affinity proteins.7,28,57 Cooperative adsorption is another mechanism leading to corona composition unanticipated from circulating concentrations, where initially bound corona proteins provide a scaffold promoting successive protein adsorption.57,58 Regarding native biomolecule composition, the observed corona in the presence of proteases may be a convolution of protease degradation of and exchange with the existing corona.59 These higher order mechanisms offer an explanation as to why corona constituents and kinetics resulting from single protein adsorption experiments are often not representative of whole biofluid experiments.26 These findings also stress the importance of testing nanoparticles within physiologically relevant biological fluids. A prominent example is the proliferous use of blood serum (absent of blood coagulation proteins) instead of blood plasma (which contains blood coagulation proteins) to test nanoparticles designed for intravenous administration, where coronas formed from plasma proteins have been shown to be different from those of serum and more strongly adhered.22,48,57,60 An additional consideration in terms of biomolecule composition arises in that nanoparticles may be subject to harsh conditions such as enzymatic degradation in the gastrointestinal tract61 and cancer cells,59 or immobilized, tenacious biomolecules in the mucus layer61 and brain extracellular matrix.62 Presence of these biomolecules introduces physical obstacles to penetration and routes to irreversible corona formation with subsequent toxicity. It is of further consequence that disease states alter endogenous protein concentrations and compositions, which leads to deleterious effects if the same such protein is pre-conjugated on nanoparticles for targeting purposes.63
(ii) Solution conditions. Corona formation is a function of surrounding conditions, such as temperature, ionic strength, and pH. Increasing temperature increases the weighting of the entropic term within the net Gibbs free energy change of adsorption (ΔG = ΔH − TΔS). This results in proteins that are entropically favorable to adsorb (+ΔS) becoming more favorable at higher temperature (−ΔG), and vice versa.26,43 Within this analysis, it is key to note that each term is the net system, therefore a function of the protein, nanoparticle, and solution initial and final states during binding. For solution ionic strength, electrostatic forces scale inversely with the square-root of salt ionic strength in solution. These electrostatic forces originate from interactions between electric double layers surrounding the charged colloidal nanoparticles and proteins. This underscores how nanoparticle surface charge is inherently coupled with solution ionic strength, and the two parameters must be co-designed appropriately. In high ionic strength conditions (high salt concentration), nanoparticles and proteins do not “see” each other in solution until they are in closer proximity. Closer approach between entities bearing the same electrostatic charge results in more protein adsorption. In addition, once adsorbed, charge shielding ensures less unfavorable lateral repulsions between adsorbed proteins (again, most bearing the same negative charge at physiological pH). Both phenomena imply more protein adsorption with more salt present. However, when ionic strength becomes too elevated, charge screening leads to undesirable protein–nanoparticle complex aggregation.30,48 Accordingly, ionic strength and even ionic composition are important considerations in protein adsorption and potential downstream toxicity due to aggregation, such as high free calcium ion concentrations in the brain microenvironment leading to nanoparticle aggregation.50 pH is another relevant solution condition in that it governs the protonation state of surface chemistries on the nanoparticle, again influencing aggregation tendency,50 and impacts adsorbed protein extent and stability. As the solution pH approaches the protein isoelectric point, proteins become less stable in solution and tend to self-aggregate or adsorb to available nanoparticle surfaces.64 In terms of corona stability, the pH range that the nanoparticle will encounter is crucial to consider as a design parameter, as biological compartments cover a significant pH range at the organ and cell levels, and can differ as a function of disease state, such as the acidic pH of tumors.7,65 Many protein or peptide drugs externally loaded on nanoparticles may not survive severe conditions,61 or their expected release profile can be negatively impacted.49
(iii) Dynamics. Finally, temporal dynamics and hydrodynamics should be considered for protein corona formation. Protein adsorption occurs within seconds of contact with biofluids,57 and may either display a dynamic nature, with fast and reversible protein association/dissociation events on the nanoparticle surface, or enter an irreversibly aggregated state.6,30,66 Proteins adsorbed directly to the nanoparticle surface are termed the “hard”, inner corona, characterized by a longer (if not indefinite) residence time in the corona phase and often more prominent conformational changes.7,28,29,57 Proteins interacting predominantly with other adsorbed proteins, instead of directly with the nanoparticle surface, constitute the “soft”, outer corona, and frequently maintain their native conformation as they undergo continuous exchange with proteins in the surrounding media.7,46 Protein corona composition is impacted by the contact time and history of nanoparticles in biofluids: the former, reiterating the likes of the Vroman effect, and the latter, in that nanoparticles evolve to carry a “fingerprint” of adsorbed proteins as they progress from one biological compartment to the next.10,67
Nanoparticles must endure flow conditions during transit, navigating channels or regions of characteristic tortuosity, permeability, and hydrodynamics. A classic example of nanoparticles maneuvering through a highly tortuous path is within the porous extracellular matrix (ECM) of the brain, relevant for neurosensors or brain-targeted therapeutics.49,62 The ECM is a mesh-like structural and biochemical scaffold for brain cells, with channels of widths ∼40–200 nm, that acts as an adhesive and steric barrier for nanoparticles attempting to pass.49,50 Work within the Nance lab has pioneered brain-penetrating nanoparticles, with design principles to ensure that the nanoparticles exhibit minimized electrostatic, hydrophobic, or hydrogen bonding interactions with the ECM.62 Towards nanoparticle permeability, crossing of biological barriers is of paramount importance. Again considering the brain, the blood–brain barrier (BBB) functions as a selective barrier to protectively isolate the brain from an influx of potentially harmful entities within blood circulation, as will be discussed in section 5.3. It must also be taken into consideration that the brain's tortuous ECM and selective BBB vary with pathology and developmental age.13,62 Finally, regarding hydrodynamics, dynamic flow can result in a more rapidly formed and compositionally diverse corona.57,68 From a design perspective, shear stresses imposed on nanoparticle-loaded cargoes must be considered a priori: the required stability of the corona–nanoparticle complex will depend on whether the dominant transport mechanism will be passive diffusion or active convection.57
In sum, a host of intrinsic nanoparticle-based and extrinsic bioenvironmental factors affect protein corona formation (Fig. 1). These relevant factors should be considered beforehand to aid appropriate experimental design and implementation towards rational protein–nanoparticle complex construction. There are frequent discrepancies between in vitro and in vivo corona characterizations that arise from negligence of these factors, such as flow dynamics present in circulation that are absent for in vitro tests.10,17,69 Validation in vivo is preferable to gauge nanoparticle functionality or efficacy, and accordingly, we focus mainly on in vivo studies for the remainder of this review.
3. Corona-based nanoparticle development and characterization
With these design considerations in mind, we now discuss how recent work has leveraged these interactions to develop engineered protein–nanoparticle constructs. Next, we detail characterization methods, both conventional and new, to properly assess protein–nanoparticle complex formation and function.3.1. Nanoparticle-corona complexation considerations and techniques
Development of functional protein–nanoparticle constructs requires special attention to the packing, conformation, and orientation of proteins on the nanoparticle surface. As detailed earlier, less tightly packed proteins may experience damaging post-adsorptive transitions including spreading, reorientation, and denaturation.6,29,70 This control over interfacial protein conformation is critical, in that denatured proteins are generally not functional and increase the risk of triggering nanoparticle aggregation or immune system recognition and clearance.7,9 In turn, packing densities and patterns of biomolecules on nanoparticles can significantly affect targeting abilities23 and clearance pathways.58 Protein orientation on the surface also directly impacts protein function, in that particular protein domains must be outwardly displayed in solution, such as enzymatic active sites and targeting moieties for receptor or molecular recognition.52,64,70Protein adsorption on nanoparticle surfaces is accomplished by either noncovalent or covalent means. Within the subset of noncovalent corona formation techniques, we describe both post factum and ab initio routes of protein corona formation. With post factum corona formation, the in vivo corona formed on nanoparticles is characterized, and this knowledge is applied to the advantage of the construct towards specific means.31 For example, spontaneous apolipoprotein adsorption onto peptide-functionalized liposomes assists in brain targeting of drug-loaded liposomes.15 The ab initio protein corona is achieved by initially pre-coating nanoparticles with the desired protein, resulting in noncovalent attachment of the protein on the nanoparticle. Examples of passive incubation to pre-coat nanoparticles with functional protein coronas include: albumin for extended circulation or biobarrier crossing;61,71 antibodies for targeted cell uptake;71 apolipoprotein E for extended circulation or brain targeting;9,72 and transferrin for cancer cell targeting.73 Proteins may also be actively adsorbed, i.e. requiring energy input, such as high-intensity sonication to disperse hydrophobic carbon nanotubes with various plasma protein coatings.74,75 Another aim of passive ab initio corona formation is to passivate exposed nanoparticle surface for biocompatibility,76 or retain the folded protein conformation or orientation of the functional corona components.6
Noncovalent methodologies are simple and rapid, yet inherently less stable than a covalent attachment and thus prone to exchange with other proteins in solution.77 When Zhang et al. probed pre-coated protein stability on spherical nucleic acids, they discovered that ∼45% of the initial corona is removed upon exposure to blood serum.71 Additionally, passive incubation techniques generally lack control over the resulting spatial distribution and functional orientation of proteins on nanoparticle surfaces.3,17 It is difficult to control monolayer vs. multilayer protein corona formation on nanoparticles, where monolayers may be desired, but multilayers often result from a combination of the hard and soft coronas.7 To overcome unpredictable corona organization, Mout et al. present a rational design strategy taking advantage of directed electrostatic assembly to form hierarchical protein–nanoparticle superstructures via coengineering recombinant proteins with ligand-tagged nanoparticles.78 Noncovalent assembly is also ideal in some cases for preserving the intrinsic nanoparticle properties.75,79 A clever bridge between retaining nanoparticle properties and enabling controlled protein attachment has been done by Mann et al., where DNA is noncovalently adsorbed on the surface of single-walled carbon nanotubes, then nanobodies are covalently attached to the DNA.79 This strategy preserves the intrinsic, near-infrared fluorescence of the underlying nanotube by avoiding protein-nanotube covalent attachment chemistries, and simultaneously confers more controlled protein orientation and packing that in turn enables successful nanobody targeting.
Covalent conjugation of proteins to nanoparticles offers another attachment route. While covalent functionalization is more stable and controlled than noncovalent adsorption, the former requires introducing new covalent bonds on both the nanoparticle surface and the protein. Examples of covalent corona attachment methods include maleimide–thiol chemistry,33,40,49 photochemical cross-linking,24N-hydroxysuccinimide (NHS) active ester reactions,21,64 and thiol–ene click chemistry.3 These chemistries can be applied sequentially, such as gold nanoparticle-thiol surface chemistry followed by NHS ester chemistry with a linker to tether the protein.63 New conjugation protocols such as those from Lee et al. offer promising, facile chemistries for more direct gold nanoparticle-PEG linker-peptide attachments.33 New covalent chemistries have also enabled protein attachment to carbon nanotubes, with re-aromatization of the graphitic sidewalls to retain the desired near-infrared fluorescence for nanosensor readout.1
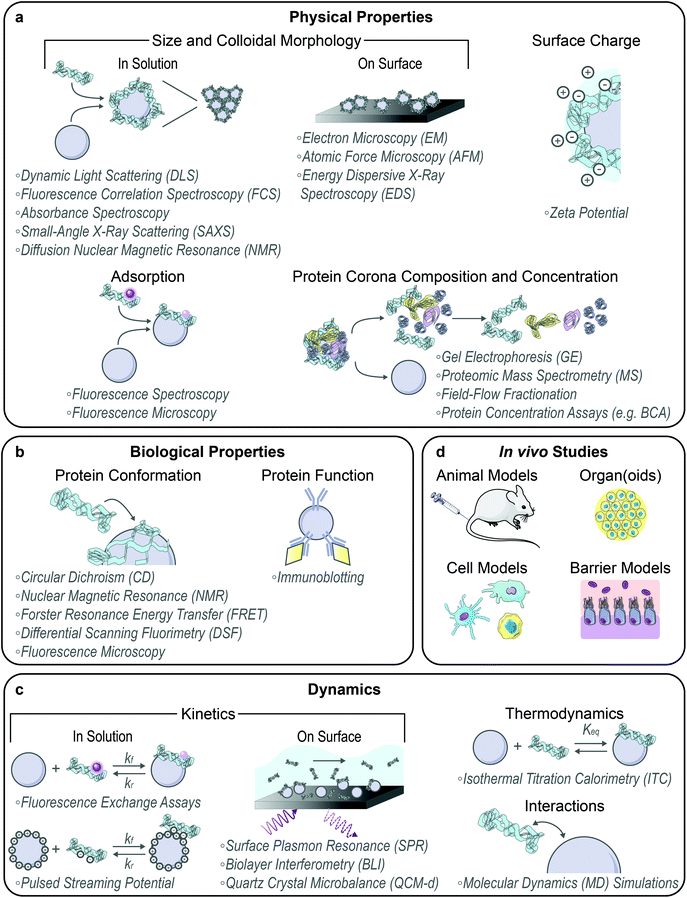
3.2. Nanoparticle-corona characterization methods
With the formation of these protein–nanoparticle complexes, their physical, biological, and dynamic properties must be characterized, alongside testing in suitable ex vivo or in vivo systems (Fig. 2). Many requisite bioanalytical methods are well-established for this purpose and can be directly applied or adapted to study corona formation and outcomes.80 We also highlight novel methodologies being developed towards this.Techniques commonly applied to assess in-solution physical properties of protein corona formation include: zeta potential to assess surface charge;22,44,49 dynamic light scattering (DLS)22,44,49 or fluorescence correlation spectroscopy (FCS)30,81 for hydrodynamic size; absorbance spectroscopy for colloidal morphology and concentration;8,22,76 fluorescence quenching to track adsorption,56 with Stern–Volmer analysis of the mechanism;52,63 and fluorescence microscopy to confirm adsorption via colocalization.14,61,64 Although zeta potential is not rigorously equivalent to the electric surface potential nor the Stern potential and there are implicit geometry assumptions in the calculation, zeta potential still provides a proxy for colloidal charge and stability, where often the zeta potential tends to zero in the presence of destabilizing protein adsorbates. Small-angle X-ray scattering (SAXS) has recently been applied to acquire in-solution colloidal morphology of nanoparticle–corona systems, including protein–nanoparticle complexation to verify binding and higher order aggregate formation to examine potential routes of in vivo toxicity.10,26,78 Diffusion nuclear magnetic resonance (NMR) has also been employed to infer protein adsorption on nanoparticles via increasing hydrodynamic radius, offering the advantage of in situ characterization in turbid bioenvironments by virtue of not being an optics-based measurement.82 Regarding the aforementioned techniques used to measure protein–nanoparticle size (DLS, FCS, etc.), the readout must be carefully interpreted. Large increases in hydrodynamic size may indicate aggregation of the nanoparticles in the presence of proteins via polymer bridging or other noncovalent interactions, rather than formation of protein multilayers on individual nanoparticles.30,68,81 Surface techniques are also applied to assess dried-state physical properties of protein corona formation, including: electron microscopy (EM),49,70,74 atomic force microscopy (AFM),9,75 and energy dispersive X-ray spectroscopy (EDS mapping)83 for size and morphology. However, these methods all require drying samples on a substrate for observation, which results in conclusions not representative of the solubilized system. Recent work has also implemented cryogenic transmission electron microscopy (cryo-TEM) to enable visualization of protein–nanoparticle morphology in a closer-to-native state.17,84
Beyond the whole-complex attributes, the composition of the protein corona is of paramount importance to take advantage of post factum corona formation. To study corona constituents, the protein–nanoparticle complexes are first isolated from non-binding entities, typically accomplished by some variation of a pull-down assay.14 After corona proteins are unbound from the nanoparticle, characterization methods to identify the protein constituents include gel electrophoresis (GE)61,63,71 and proteomic mass spectrometry (MS).8,29,46 Separation techniques to isolate the soft, more loosely bound corona from the hard corona are currently in development, such as asymmetric field-flow fractionation by Weber et al.85 The Sutherland lab has also developed an in situ click-chemistry reaction to separately characterize the soft and hard coronas formed on model nanoparticles.29 To gauge whether corona loading or mitigation strategies are successful, net protein adsorption can be measured by protein assays such as the bicinchoninic assay (BCA) for protein loading,8,49,72 gel electrophoresis again, immunoblotting (e.g. Western blots),59 and enzyme-linked immunosorbent assays (ELISAs).59 The accuracy of colorimetric protein assays such as BCA in the presence of nanoparticles must be critically assessed prior to experiments, as nanoparticles often interfere by adsorbing the reporter molecule or absorbing the output light used to quantify protein concentration. Moreover, the specific chemistry of the assay will determine whether proteins in solution, in the adsorbed state, or both are being measured.
Techniques applied to assess biological function in the corona include: circular dichroism (CD)52,69 and solution NMR86,87 spectroscopy for bound protein structure and conformation; Förster resonance energy transfer (FRET) for measurement of protein–protein interactions61 and conformational changes;7 nano differential scanning fluorimetry (nanoDSF) for protein stability and conformational changes;64 and immunoblotting to evaluate accessibility and function in the corona.63,83,88 To study corona structural organization and functionality at the nanoparticle surface, Herda et al. developed a method to characterize adsorbed protein orientation by exploiting antibody-conjugated gold nanoparticles to map available epitopes.70 When they applied this method towards transferrin proteins covalently conjugated to PEGylated silicon dioxide nanoparticles, they found that only ∼4% of corona proteins adopt the correct orientation to facilitate receptor binding, highlighting the need for more homogenous and controlled protein grafting methodologies. Recently, the Chan lab developed a modified-ELISA workflow to similarly probe protein corona organization and binding functionality when adsorbed from blood serum onto gold nanoparticles, establishing that merely a third of the adsorbed proteins remain functional for binding to their target proteins.89 Imaging advances have led to the development of various techniques to assess protein interactions on surfaces, including single molecule high resolution imaging with photobleaching (SHRImP) by Warning et al. to measure protein conformational changes.6
Methods to study dynamics of protein corona formation include: isothermal titration calorimetry (ITC) to extract thermodynamic binding energies and equilibrium parameters,38,43,90 as reviewed extensively elsewhere,91 and surface plasmon resonance (SPR),16,29,46 biolayer interferometry (BLI),92 and quartz crystal microbalance with dissipation monitoring (QCM-d)52 to determine binding kinetics. Recently, Kari et al. designed a custom biosensor system for in situ determination of protein corona structure and composition by coupling SPR and proteomic MS, enabling differentiation of the hard and soft corona formed on liposomes under physiologically relevant conditions.46 Super-resolution microscopy presents a single-molecule technique with requisite sensitivities to monitor individual protein–nanoparticle binding events, avoiding ensemble-averaged methods of studying corona formation.93 However, it is important to note that application of surface techniques such as SPR and microscopy to study nanoparticles again requires surface immobilization of the nanoparticles. Surface immobilization introduces topographical constraints that affect kinetics and transport, giving rise to sampling artifacts and changing the in-solution nanoparticle properties. Ideally, protein–nanoparticle complexes are studied in solution with physiologically relevant parameters that are known to affect corona formation (including ionic strength, temperature, pH, etc.). Accordingly, in-solution kinetic corona methods have been developed, including fluorescence assays to monitor protein fall-off71 and exchange56 on solubilized nanoparticle surfaces. To expand upon the use of surface charge changes as a proxy for protein corona formation, Zhao and colleagues measured in-solution protein binding dynamics onto nanoparticles using pulsed streaming potential, resulting in knowledge of adsorption rates and equilibria under varying buffer conditions.94 Further, Weiss et al. have developed a microfluidic system to simulate a flow environment, with control over fluid flow and shear applied to nanoparticles and proteins.57 This microfluidic system has elucidated the more complex corona formed in dynamic rather than static conditions.
While prior studies provide insight into bio-corona formation, numerous techniques and model fits are ill-applied and present conclusions not representative of the system. The protein corona is often treated as existing at thermodynamic equilibrium, despite a body of literature providing evidence otherwise.95–97 One frequent manifestation of this equilibrium assumption is the erroneous application of the Langmuir adsorption isotherm to mechanistically describe proteins adsorbing to nanoparticles, despite the fact that many of the model conditions are not satisfied.98 A key point here is that a Langmuir-like binding profile does not necessitate that the binding mechanism is indeed a Langmuir isotherm: this profile shape for protein–surface adsorption processes often emerges as the result of adsorption-induced protein spreading/denaturation, reorientation, and aggregation as a function of bulk protein concentration,7 in contrast to originating from the dynamic equilibrium adsorption process required for Langmuirian adsorption.98 Therefore, while the Langmuir isotherm does provide a simple functional form that may fit data, it should only be applied towards extraction of relative binding affinity measures rather than true thermodynamic parameters or underlying adsorption mechanisms. For instance, ITC is often a method applied with intent to assess protein–nanoparticle binding events. Instead, ITC often measures a convolution of protein binding to individual nanoparticles, to aggregated nanoparticles, and nanoparticle aggregation.26 Aggregation is a kinetically controlled, non-equilibrium process that violates the central assumption of ITC that each titration step is equilibrated, observed as visible aggregation and baseline drifting during the run. Accordingly, the reported free energies and equilibria values must be taken with the perspective that these are whole-system energy changes, often with higher-order processes occurring simultaneously. The suitability of such models and experimental methodologies to describe certain nanoparticle–protein corona formation processes should be carefully considered prior to application.
Finally, molecular dynamics (MD) simulations provide insight into fundamental interactions driving adsorption to surfaces and protein structural changes upon adsorption, as reviewed extensively elsewhere.99–101 Atomistic MD models convey a detailed picture of protein–nanoparticle interactions, including the individual amino acids responsible for association.102 Alternatively, coarse-grained MD models trade such detail for access to longer time and length scales, increasing approximately an order of magnitude from the millisecond and nanometer scales in atomistic models.99 Although coarse-grained models are inherently lower resolution, such as lacking physicochemical details of the nanoparticle surface,103 these models can facilitate the study of protein–protein interactions and adsorption onto smaller nanoparticles, with explicit curvature effects. Both scenarios are unfeasible in atomistic models, which instead highlight interfacial phenomena in dilute protein settings. Some particular MD studies of interest involving protein–nanoparticle systems include: atomistic MD simulations of amyloidogenic peptides on gold nanoparticles (modelled as a gold surface)102 and plasma proteins on model nanomaterials;9,28 hybrid MD simulations, with an atomistic nanoparticle description and a coarse-grained, solvent-explicit protein description;23 and multiscale MD simulations, adopting coarse-grained or meso-scale models for single vs. simultaneous protein adsorption on small gold nanoparticles, respectively.104 MD simulations extend our understanding of dynamic protein–nanoparticle interactions, yet require further refinement and validation against experimental results prior to use as purely predictive tools, due to the underlying complexity of nanoparticles interacting with proteins.99
Towards applied nanoparticle–corona technologies, in vivo studies provide compelling evidence for sustained nanoparticle function or therapeutic efficacy within complex bioenvironments. Animal models such as mice and rats provide the means to study not only function, but also systems-level clearance profiles, bioaccumulation, and toxicity.42,61,83Ex vivo organ slices enable measurement of nanoparticle diffusion by particle tracking studies, providing insight into unfavorable adhesive interactions with the surrounding biological matrix.40 At the cellular level, fluorescence (often confocal) microscopy,40,61,63 immunofluorescence,49,83 and flow cytometry40,41,63 provide information on cellular uptake, spatial localization, cell morphology, and cytotoxicity. As an intermediate between achieving in vitro experimental control and assessing in vivo translatability, transwells offer a useful cellular model for biological barriers such as the blood–brain barrier67 and three-dimensional organoids or tumor spheroids offer a scaled-down organ model for assessing efficacy and toxicity.20,105
4. Development of stealth nanoparticles
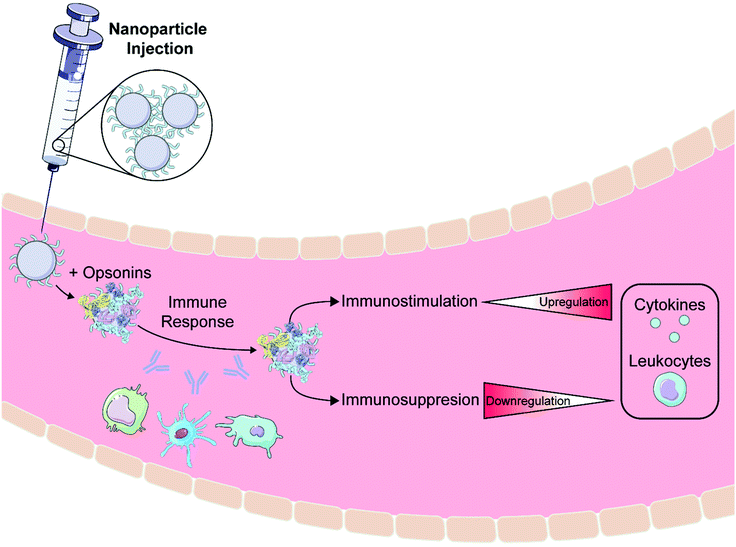
After creation and in vitro characterization of nanoparticle–corona complexes, several challenges still lie between administration and successful use of nanoparticles in vivo. Many such obstacles stem from the recognition of these synthetic nanomaterials by the body. Nanoparticles often trigger an immune response, resulting in immune cell recruitment, antibody and chemokine release, and activation of the mononuclear phagocytic system (MPS) (Fig. 3a). Briefly, the MPS entails the recognition, engulfment, and subsequent clearance of nanoparticles from blood circulation through the action of phagocytic cells, such as Kupffer cells in the liver, dendritic cells in major organs, microglia in the nervous system, and alveolar macrophages in the air spaces of the lungs.106 Consequently, nanoparticles are often found to accumulate in the liver and spleen. Numerous studies have shown that the protein corona plays a critical role in modulating the MPS.3,77,107 Specifically, proteins termed opsonins promote phagocytosis, and include complement proteins and immunoglobulins (IgG, IgA, and IgM). Conversely, dysopsonins are proteins that aid evasion of phagocytosis, and include albumin and apolipoproteins. The protein corona can thus be tuned to mediate challenges that the nanoparticle faces from injection to localization.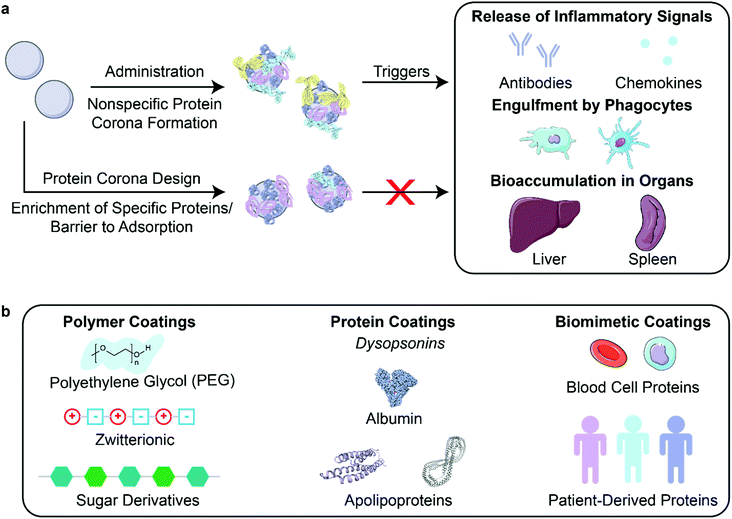 | ||
| Fig. 3 Nanoparticle stealth, strategies and outcomes. (a) Administration of nanoparticles into the body leads to formation of the protein corona that can trigger an immune response or clearance of nanoparticles. Rational design of the protein corona can promote the enrichment of favorable, dysopsonin proteins or mitigate the adsorption of unfavorable, opsonin proteins to promote nanoparticle stealth. (b) Strategies that utilize polymer, protein, or biomimetic coatings have been developed to design the protein corona for better nanoparticle stealth. Protein images (PDB ID 1E7I, 1LE2, and 1AV1)111–113 are reproduced with permission from the RCSB PDB (rcsb.org). Some images in this figure are adapted with permission from Servier Medical Art by Servier (http://smart.servier.com), licensed under a Creative Commons Attribution 3.0 Unported License. | ||
To prevent activation of the immune response and nanoparticle recognition by the body, several strategies can be implemented to provide nanoparticles with stealth properties. In literature, “stealth” is often used to indicate resistance to biofouling, referring to the low nonspecific adsorption of proteins on nanoparticles. Although less adsorption of certain proteins such as opsonins correlates with better biological compatibility, more factors are involved in nanoparticle stealth for biological applications.77 We therefore refer to stealth herein as the ability to evade recognition by the body. In the discussion to follow, we highlight studies that report longer nanoparticle retention time in vivo and lower titer of biomolecules that indicate immune response. Studies have demonstrated this phenomenon through the design of the nanoparticle corona using polymer, protein, or biomimetic coatings (Fig. 3b).
4.1. Polymer coatings for stealth
Attachment of polymers to nanoparticle surfaces provides a facile approach to modify hydrophilicity, size, and other nanomaterial properties, as detailed in section 2.1, that may modify protein corona formation in comparison to the bare nanoparticle and confer stealth in vivo.Recent studies are moving away from the use of PEG as a stealth agent, as fundamental challenges of using PEG-nanoparticle conjugates come to light. The ubiquitous use of PEG in nanomedicine has led to the formation of anti-PEG antibodies in the body and rapid clearance of PEGylated nanoparticles from the body, termed the “accelerated blood clearance” phenomenon.115,116 Furthermore, use of PEG does not necessarily suppress unfavorable protein adsorption onto all nanoparticles,27,117 such as nanosomes with PEG linkers shown to irreversibly aggregate after protein corona formation in whole serum.66 Due to these findings, researchers are investigating other polymer coatings for nanoparticle stealth.109
4.2. Protein coatings for stealth
Another solution towards constructing stealth nanoparticles is to engineer the protein corona itself to avoid triggering the immune system and MPS detection.10 As all nanoparticles are expected to develop coronas in vivo and the existence of these coronas often promotes immune cell association,77 directed adsorption of dysopsonins and/or reduced adsorption of opsonins on the nanoparticle surface can be employed to reduce clearance of nanoparticles.90Nanoparticle surface properties may be altered to direct adsorption of desired proteins or repel unwanted proteins. In a study of peptide-embedded liposomes, it was shown that the adsorption of IgM correlates with rapid clearance through the MPS and accumulation in the lymph nodes.116 By modifying the length of the peptide displayed on the liposome, adsorption of IgM decreased, leading to longer nanoparticle half-life in circulation. To encourage dysopsonin adsorption, nanogels were created using molecular imprinting, a method that templated nanogels to bear a binding site for native dysopsonin protein: albumin.16 Upon injection into a tumor xenograft model, it was shown that the molecularly imprinted nanogels (MIP-NGs) had a higher half-life in blood (6.8 hours), compared to the non-imprinted nanogels (3 hours). Furthermore, these MIP-NGs were observed to circulate in the liver without aggregation or capture for over 10 hours, demonstrating that nanoparticle surface modifications can be utilized to adsorb necessary stealth proteins.
Protein corona shields can be made through the design of the nanoparticle surface a priori. Oh et al. created a protein corona shield for mesoporous silica nanoparticles using a recombinant fusion protein of glutathione-S-transferase genetically combined with Her2-binding affibody.3 This outer corona shield led to the reduction of protein corona formation and subsequent higher retention in plasma. In another study, pre-incubation of charged polystyrene nanoparticles in IgG-depleted plasma formed a corona enriched in vitronectin and fibrinogen on negatively charged polystyrene nanoparticles or enriched in clusterin and hemopexin on positively charged polystyrene nanoparticles.123 These nanoparticle–corona complexes showed reduced uptake by RAW264.7 macrophages and remained stable when reintroduced into whole plasma. Preincubation with the dysopsonin apolipoprotein E on graphene, gold nanoparticles, and iron oxide nanoparticles showed markedly improved blood circulation and better biocompatibility than opsonin IgE-coated nanoparticles.9 Using the growing database of corona proteins, it is increasingly possible to tailor nanoparticle surfaces for avoidance of premature clearance.
4.3. Biomimetic coronas for stealth
Similar to plasma-derived protein coatings for stealth applications, other biomimetic solutions to maintain nanoparticle biocompatibility include employing cell membrane proteins to shield nanoparticles from recognition. To keep nanoparticles in circulation and curtail recognition from immune cells, blood cells are a template for nanoparticle stealth. Corbo et al. have utilized white blood cell (leukocyte) proteins, such as macrophage receptors, to decorate liposomes and produce a new class of nanoparticles called leukosomes.17 They showed that leukosomes have lower accumulation in MPS organs and have reduced uptake by macrophages. Similarly, Meng et al. extract red blood cell membranes to coat immunomagnetic micro and nanoparticles.14 This coating prevents the formation of a protein corona in whole blood for better retention.The use of biomimetic coronas could also accelerate the development of personalized nanomedicine with low immunogenicity. Personalized protein nanoparticles can be made from patient-derived proteins extracted from a variety of human sources such as serum, tears, saliva, or breast milk.24 The proteins are initially cast on a metal nanoparticle core, then extracted to create a biodegradable nanoparticle made up of only proteins. These protein nanoparticles were used in vivo without any inflammation or immune cell recruitment. Biomimetic solutions in creating stealth nanoparticles leverage specific cell-type proteins or personalized proteins to evade detection.
5. Targeting and activation of functional nanoparticles to biological systems
After bypassing recognition and clearance from circulation, nanoparticles must overcome additional barriers towards successful localization and function. As such, targeted delivery of nanoparticles remains a major challenge in the clinical adoption of nanomedicine, and recent literature reveals that efficacious nanoparticles can manipulate protein corona engineering towards this purpose. In this section, we discuss the targeting strategies of nanoparticles and subsequent activation of nanoparticles once they arrive at a biological target of interest.5.1. Challenges and considerations in nanoparticle targeting
Nanoparticle surfaces can be engineered for targeting through the addition of different synthetic and biological ligands, such as small molecules, peptides, and antibodies. However, nanoparticle targeting elements incorporated on bare nanoparticle surfaces and validated in vitro may show different functionality in vivo, where the formation of the protein corona upon administration could inhibit the accessibility of these targeting ligands.13 Some studies show that cellular uptake of nanoparticles is controlled by the outermost protein corona as opposed to the surface ligands meant to target receptor-expressing cells.11,124 A notable exception of this phenomenon was seen in poly(beta-aminoester) polymer nanoparticles with variable terminal targeting peptides.124 The nanoparticles were coated with retinol, a hepatic targeting moiety, and the protein corona formed dictated organ biodistribution, yet cellular uptake was determined by the terminal peptides independent of the corona. For many other cases, however, the in vivo protein corona attenuates the targeting properties of nanoparticles. Serum proteins were shown to decrease association of transferrin-labeled liposomes with glioblastoma cancer cells, although transferrin-labeled liposomes still exhibited better association, tumor uptake, and tumor growth inhibition than unlabeled liposomes.73 There is currently a dearth of literature on the mechanism of these targeting moieties post factum, and work is moving towards understanding the strategies for ab initio nanoparticle design. Certain properties such as size, conformation, and mobility of targeting ligand have come to light as important design parameters for targeting applications.Size of targeting ligands could play a substantial role in targeting potency. An example is transferrin, an 80-kilodalton glycoprotein used in many targeting studies for its well documented ability to promote clathrin-mediated endocytosis, and subsequent intracellular trafficking through recycling pathways.125 Transferrin, peptide LT7 (CHAIYPRH), and DT7 (the D-amino acid analogue of LT7) are all targeting ligands for transferrin receptors that are overexpressed in several cancer types.69,125 Investigation of targeting and uptake of polystyrene nanoparticles functionalized with these ligands revealed that the transferrin-passivated nanoparticles out-performed the peptide-passivated ones.69 Analysis of the protein corona formed around each of these nanoparticles revealed differences in composition, and underscored a size and conformation effect on ligand targeting.
As introduced in section 3.1, the conformation of targeting ligands on nanoparticle surfaces can affect the ability of the nanoparticle to carry out its intended function.23,52 Fibronectin, a protein that binds cell-adhesion receptors called integrins and extracellular matrix components, can undergo pronounced conformational changes when adsorbed onto bare gold nanoparticle surfaces compared to when adsorbed to the nanoparticle surface through protein–protein interactions, leading to loss of function in the former.7,126 The function of targeting modalities on nanoparticles must be preserved in the surface-adsorbed state and during in vivo application. Finally, beyond simply optimizing ligand avidity towards the intended target, Figueroa et al. highlights how increasing mobility of ligands tethered to nanoparticle surfaces drives more cellular uptake.127
5.2. Protein corona strategies in nanoparticle targeting
Several nanoparticle targeting schemes are validated with applications in vivo. Cancer therapies often require targeted approaches because treatments, such as chemotherapy, are cytotoxic to both cancerous and non-diseased cells and have a limited dosing range. Hence, most literature examples of nanoparticle targeting are devoted to designing nanoparticles for cancer therapy. These design principles could be extended to other diseases and tissue types. This section highlights some targeting modalities that can be attached to a variety of nanoparticles through different conjugation chemistries.Targeting strategies that activate transport pathways or bind overexpressed biomarkers are promising for in vivo applications because they increase uptake of the nanoparticles by the target cell. To activate transport pathways, nanoparticle surfaces can be functionalized with ligands that bind to requisite receptors or proteins on the target cell (Fig. 4a). The use of polypeptides is frequently employed to deliver nanomedicine to tumor cells, such as the arginine-glycine-aspartic acid (RGD) peptide motif that binds to integrin transmembrane proteins.110,128,129 Other ligands include synthetically malleable polymers that are readily incorporated through bioconjugation chemistry or layer-by-layer synthesis; a prominent example is hyaluronic acid that binds to CD44 receptors overexpressed in many cancers.105 Additionally, the use of hyaluronic acid is shown to reduce the immunogenicity through the selective adsorption of anti-inflammatory proteins to the formed protein corona.130
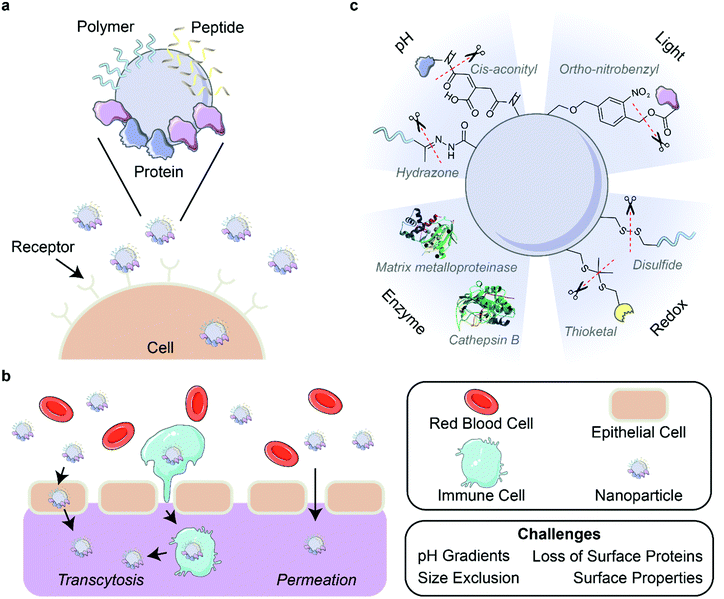 | ||
| Fig. 4 Corona-mediated targeting and activation. (a) Nanoparticles can be targeted to cell receptors through the surface decoration of ligands such as polymers, peptides, and proteins. (b) Delivery of nanoparticles through biological barriers is difficult due to environmental factors such as pH gradients and physical forces that destabilize the outer protein corona. Nanoparticles can pass through barriers by permeation or the targeting of nanoparticles to cells for transcytosis. (c) Strategies for the activation of nanoparticles include the use of pH, light, enzymes, and redox reactions. Protein images (PDB ID 5UE3 and 3AI8)131,132 are reproduced with permission from the RCSB PDB (rcsb.org). Some images in this figure are adapted with permission from Servier Medical Art by Servier (http://smart.servier.com), licensed under a Creative Commons Attribution 3.0 Unported License. | ||
Dual stealth and targeting surface functionalization prevent the adsorption of plasma proteins and thus retains nanoparticle targeting capability. Koide and colleagues designed nanosomes, consisting of core metals covered by an anti-adhesive mixed self-assembled monolayer, capable of preventing protein adsorption while outwardly displaying n-acetylglucosamine (GlcNAc).66 This outer layer triggered uptake through the cancer-specific GlcNAc salvage pathway, and resulted in body circulation, accumulation in the tumor, and reduced tumor size. Similarly, mesoporous silica nanoparticles with a protein shield of glutathione-S-transferase fused with Her2-binding affibodies, as introduced earlier, were shown to adsorb few corona proteins and resulted in increased uptake and growth inhibition of breast cancer in vivo in SK-BR3 xenograft mice.3
Corona proteins adsorbed during nanoparticle transport can also be used for targeting, redirecting accumulation from the liver and spleen leads to better accumulation of nanoparticles in other organs. An interesting case of nanoparticles for cancer therapy is blood-triggered generation of platinum nanoparticles as anti-cancer agents. Platinum, originating from the chemotherapy drug cisplatin, is triggered by albumin to assemble in vivo to function as an anti-cancer agent.83 This native corona of albumin then promotes targeting tumors with better efficacy in leukemia xenograft mice than commercial albumin-platinum conjugates.
5.3. Protein corona strategies in nanoparticle passage across biological barriers
Targeting also aids efficient delivery of nanoparticles through biological barriers such as the blood–brain barrier (BBB), mucous membranes, and epithelial barriers (Fig. 4b). These barriers impose certain limitations on the physical characteristics of the nanoparticles, as introduced in section 2.1. For example, the BBB excludes passage on the basis of size and surface properties including charge and hydrophilicity.13,133 Nanoparticles have been shown to pass the BBB via transcytosis-mediated routes, mediated by immune cells134,135 or the presence of apolipoproteins,15,72 transferrin,49 or other proteins within the adsorbed corona (as reviewed extensively elsewhere133,136,137). Importantly, traversing biological barriers can lead to alterations in the nanoparticle corona. A study of the nanoparticle protein corona was conducted in an in vitro cellular transwell model of the BBB, demonstrating evolution of the protein corona as well as a stabilizing effect after BBB crossing.67 It is shown for gold nanoparticles that only 9 of the 20 most abundant proteins in the corona are retained after passage through this BBB model, where serum albumin and α-2-macroglobulin remain abundant, with enrichment of complement C9. For efficient design of targeting elements to cross biological barriers, it is important to ensure stable attachment such that targeting functionality can be maintained across different environments.Strategies exist at the intersection of nanoparticle surface design and biological environment considerations in creating nanoparticles towards targeted biological barrier crossing. For oral delivery, nanocarriers must withstand acidic pH, enzymatic degradation, and differing surface charge requirements during passage through the mucous membrane and intestinal epithelium. Passage across the negatively charged mucus barrier is best achieved with neutral, hydrophobic molecules, while passage across the intestinal epithelium is optimal with cationic, hydrophobic molecules. With these system constraints in mind, Wang et al. aimed to overcome this issue of ineffective oral administration and uptake of insulin through rational corona design.61 Ultimately, pre-coating albumin on cationic liposomes enabled increased penetration across both mucosal and epithelial barriers: the protein coating is enzymatically hydrolyzed as the liposomes cross the mucus layer, resulting in exposure of the underlying positively charged liposome that subsequently improves transepithelial transport. In vivo experiments show that the uptake amounts and transepithelial permeability of these liposomes carrying insulin were 3.24- and 7.91-fold higher, respectively, than that of free insulin. Continuing this same idea, Zeng et al. suggest that the presence of other proteins such as protease inhibitors in the nanoparticle corona, in addition to pre-loaded albumin, serves the role of protecting albumin from hydrolysis prior to reaching the intended destination (in this case, tumors).83
5.4. Activation of nanoparticle functions upon localization
As discussed in previous sections, nanoparticle systems can be designed to increase their bioavailability, circulation time, and ability to target and localize to desired areas such as specific organs or tumors. However, surface functionalizations that prove beneficial for these purposes can be detrimental once these nanoparticles arrive at their target site.138 It is thus desirable to alter nanoparticle composition in a controlled manner through various cleavable bonds and mechanisms upon nanoparticle localization. Several environmental triggers have been used for this purpose in recent years, with pH, light, enzymes, and redox environments being the most common (Fig. 4c). Other triggers including temperature139 and electrostimulation140 have been demonstrated, but are less common due to the inherent difficulty of applying these external stimuli to in vivo systems in a controlled manner to avoid unintended side effects.In addition to dePEGylation, other modes of pH activation have been leveraged in recent years. Li et al. developed a peptide-assembling nanoparticle system loaded with immune checkpoint inhibitors to both effectively target breast cancer cells and release the cargo once internalized.143 This result was achieved by designing a peptide polymer with cholesterol, a histidine domain for endosomal escape, and a targeting peptide sequence. Once the drug-loaded nanoparticle was endocytosed, the drop in pH led to protonation of the histidine domain, which facilitated endosomal escape and resulted in successful in vivo drug release. Naidu et al. showed different release kinetics of ion channel antagonists from transferrin-functionalized polymeric nanoparticles in various pH environments, finding faster drug release at lower pH.49 This result suggests that pH-responsive systems can be beneficial for treatment of neurotrauma by maintaining drug cargo within the nanoparticles until they enter the acidic (pH ∼5) endosomal environment of damaged central nervous system cells. Overall, these nanoparticle-drug systems benefit from pH activation by controlling drug release to occur at the predetermined location.
Taking advantage of a protein corona stealth effect rather than that of PEG, Yeo et al. evaluated the use of gold nanorods coated with mouse serum proteins and the photosensitizer molecule Chlorin e6 to accumulate in and subsequently eliminate tumors in mice.76 The serum protein corona effectively shielded the nanoparticles from immune system clearance and increased their bioavailability. Once accumulated at the tumor site, visible-light laser irradiation induced the production of ROS by Chlorin e6 which, when combined with the temperature increase of the nanorods themselves, led to complete tumor regression within 19 days and no significant regrowth after 31 days. Also utilizing the nanoparticle-adsorbed protein corona, Fukuda et al. demonstrated the potential of single-walled carbon nanotubes suspended in apolipoprotein A–I to produce ROS under NIR illumination.74 These ROS led to both lower HeLa cancer cell viability and disintegration of the neurotoxic peptide aggregate amyloid beta, which is implicated in neurodegenerative diseases such as Alzheimer's.
6. Biocompatibility
The presence of foreign objects such as bacteria, viruses, and nanoparticles within the body can induce a response by the immune system. Depending on the nanoparticle's properties, this immune response can both render the nanoparticles ineffective and also lead to inflammation and systemic complications.150 Although many advancements have been made in preventing this response as detailed in section 4, it is still important to understand the scope of possible interactions between nanoparticles and the immune system to ensure nanoparticle efficacy and inform rational design of the protein corona (Fig. 5).The human immune system is composed of two branches: the innate immune system and the adaptive immune system. The innate immune system is composed of the complement system and the mononuclear phagocyte system (MPS) working in tandem to identify and eliminate pathogens. This process is activated almost immediately after an infection is detected. The adaptive immune system uses lymphocytes known as T cells and B cells in combination with antibodies to eliminate foreign and native infected cells. In contrast to the innate immune system, the adaptive response requires about one week post-infection to fully develop. Due to this discrepancy in time scale, the innate immune response is the body's first line of defense against perceived foreign invaders and is canonically observed in assaying nanoparticle biocompatibility. Improper nanoparticle design that neglects the innate immune system could lead to untimely clearance, unintended immune response, and complications including systemic toxicity.151 Additionally, over-use of nanoparticle coatings such as PEG can generate anti-PEG antibodies by the adaptive immune system as discussed in section 4.1.1. It is thus of great importance to consider the potential mounted immune responses, particularly the innate response, when designing nanoparticle systems for use in vivo.
6.1. Immune system activation and suppression
The involvement of nanoparticles with the innate immune system can be categorized into immune activation and suppression. Activation is better studied than suppression due to it being easier to elicit with nanoparticles, although both have been demonstrated in recent years.8,152 Immune activation can be further divided into unintended activation via the complement system and intended activation through elevated cytokine levels and leukocyte activation. Immunosuppression aims to mitigate the immune response by depleting inflammatory cytokine levels and limiting leukocyte migration, which can result in longer nanoparticle circulation time from lower MPS clearance and reduced inflammation in hypersensitive systems.153,154Nanoparticle surface properties are key to eliciting immune activation. For instance, Coty et al. demonstrated that dextran-coated poly(isobutylcyanoacrylate) nanoparticles activate different complement pathways depending on the architecture of the dextran coating itself.150 They concluded that the density and length of the dextran coating modulated the ability of different complement proteins such as C3 and mannose-binding lectin to bind to the nanoparticle surface, in turn affecting which pathway was activated. Fülöp et al. explored the complement activation effect of various coating materials on superparamagnetic iron oxide nanoparticles (SPIONs), similarly determining that dextran coating leads to complement activation, in this case by the alternative pathway.151 They incubated SPIONs noncovalently adsorbed to starch, carboxymethyldextran, chitosan, phosphatidylcholine, citric acid, and dextran coatings with human serum samples in vitro and measured the levels of the complement pathway-specific marker SC5b-9. Phosphatidylcholine and chitosan showed no reaction, starch and carboxymethyldextran showed minor effects, and dextran caused massive complement activation. Escamilla-Rivera et al. conducted a similar study, comparing complement activation of iron oxide nanoparticles with bare surfaces, with a polyvinylpyrrolidone (PVP) coating, and with a PEG coating.27 Interestingly, the PEG coating resulted in doubled complement protein adsorption levels in vitro and higher levels of inflammatory cytokines in vivo, while the bare and PVP-coated nanoparticles showed no significant increase in either case. Quach and Kah studied the effect of gold nanoparticle size, shape, and polyelectrolyte ligand on complement activation.25 By detecting the endpoint complement marker SC5b-9 concentration, they determined that polyethyleneimine ligand induced the most complement activation and that there is a negative correlation between nanoparticle surface hydrophilicity and complement activation. Based on the literature, there is little predictability with which nanoparticle materials or coatings will elicit an immune response, necessitating that each be tested individually.12 Due to the importance of the complement pathway in understanding the biocompatibility of nanoparticle systems, more studies are required to fully understand the mechanisms and dependencies of this cascade within the context of nanoparticle activation.
Several nanoparticle systems in recent years have been designed to purposefully leverage the protein corona to activate the immune system. Mo et al. exploited the serum protein corona formed on black phosphorus nanosheets to polarize M0 macrophages into M1 macrophages, stimulating the immune system to eliminate cancer cells.152 Similarly, Kouser et al. investigated the inflammatory response of functionalized carbon nanotubes with adsorbed human properdin, a protein that upregulates the alternative complement pathway.88 The authors adsorbed either the full properdin protein or only the binding domain, thrombospondin type I repeat 4 and 5 (TSR4+5), to cellulose-coated and oxidized carbon nanotubes. Upon in vitro incubation in blood serum, TSR4+5-coated nanotubes inhibited complement activation due to the lack of available surface area for native properdin to adsorb, while properdin-nanotubes maintained complement activation. Furthermore, pre-adsorbed properdin enhanced the uptake of carbon nanotubes by THP-1 macrophages, stimulating the production of pro-inflammatory cytokines, while pre-adsorbed TSR4+5 was ineffective in producing this immunostimulatory response. Taken together, these various studies demonstrate the potential design of nanoparticles to either stimulate immune cell activation directly or engage immunostimulation through the complement system.
Employing nanoparticles that produce ROS can induce an oxidative stress response in cells as another form of immunostimulation. These ROS include hydrogen peroxide, superoxide anion radicals, singlet oxygen, and free hydroxyl radicals, which lead to a variety of oxidative stress responses, including inflammation, apoptosis, DNA damage, and lipid peroxidation.158 While nanoparticles themselves can produce an oxidative stress response, Jayaram et al. demonstrated that the in vitro protein corona also plays a role in the oxidative stress experienced by cells.159 In this study, titanium dioxide nanoparticles were found to produce ROS that caused oxidation of corona proteins, including complement C3, serum albumin, and plasminogen. The oxidized corona proteins subsequently caused downregulation of peroxiredoxin expression, enzymes responsible for clearing peroxide species, thus resulting in an oxidative stress response to the cell. They also concluded that an increase in nanoparticle surface defects exacerbates the oxidative stress effect. Due to the potential of the protein corona to produce an oxidative stress response in cells, it is important to further study nanoparticles in this light in addition to other immunological effects.
6.2. Cytotoxicity assays
The assessment of these various immune activation and suppression functionalities, and more broadly the extent to which a nanoparticle is deemed biocompatible, relies on cytotoxicity assays. These assay outputs depend not only on the nanoparticle properties and interactions, but also on the specific assay used in the study. Common techniques for toxicity assessment include nanoparticle incubation with representative cell systems, such as HeLa cancer cells and THP-1 monocytes to assess cell viability,40 endpoint cytokine level measurements to determine inflammation,25,76 and live mice and rat models to compare in vitro analysis with in vivo efficacy.27,76,160 Although these techniques can be useful tools for characterization, literature increasingly suggests that the adaptation of standardized in vitro toxicity assays to assessing nanoparticle outcomes must be done with care. For example, common cell viability assays include the MTT,142,149,159 LDH,161,162 Trypan Blue,160 and CCK-8 assays.8,74 However, nanoparticles have been shown to interfere with such assays by either adsorbing the reagent or readout molecules, or in the case of colorimetric assays, absorbing or contributing to the output signal being quantified.161–163 This leads to false cytotoxicity or efficacy predictions because the nanoparticle presence alone vastly modulates the assay output, often confirmed by seeing negligible in vitro toxicity yet drastic changes in cell morphology. As such, it is imperative to include the necessary controls for these assays and run multiple, orthogonal assays to avoid reporting misleading results.7. Conclusions
Nanoparticles offer a promising platform towards studying and manipulating biological systems. Yet, formation of the protein corona on nanoparticle surfaces upon introduction into biological environments remains a considerable barrier between in vitro design and in vivo application. Beyond characterizing the protein corona formed on varying nanomaterials, recent developments have sought to elegantly exploit and rationally design the protein corona to achieve improved nanomaterial functionality. In this review, we have described intrinsic nanoparticle and extrinsic biological factors that govern unpredictable protein corona formation, such that these insights may be used towards either mitigation of unfavorable adsorption or enhancement of desired adsorption. Many such factors are co-dependent, therefore work remains in separating out nuances among these variables. To extend experimental data into broader design rules, recent work has involved ensemble machine learning approaches to develop predictive models of protein corona fingerprints formed on nanoparticles based on protein, nanoparticle, and solution characteristics.164 Such models could better inform corona-based design. Moreover, future work should move towards characterization under biologically relevant conditions, such as under flow for intravenous applications, in biofluids most representative of the intended application. Based on these design principles, work has implicated noncovalent adsorption and covalent coupling strategies. Characterization techniques have been adapted from tangential fields and developed anew. Recent advances continue to give insight into minutia of the protein corona, such as the hard vs. soft corona constituents and kinetics. However, particular care must be taken in characterizing individual particle vs. aggregate size, and future characterization should be done primarily in solution rather than in dried or immobilized surface settings. Additionally, models of protein–nanoparticle association should be applied with caution regarding assumptions and limitations, especially if mechanistic or quantitative conclusions are to be made from the data.Corona-mediated nanoparticle functionalities include stealth, targeting, and activation. Stealth continues to be mechanistically explored, as more studies demonstrate the difficulty in fully eradicating protein corona formation. Instead, achieving stealth seems to rely on adsorption of specific proteins to mask the foreign nanoparticle presence. While common strategies rely on hydrophilic, zwitterionic, or carbohydrate shells, new work realizes the promise of protein or biomimetic coatings towards attaining stealth. It is increasingly recognized that controlling, instead of eliminating, protein adsorption will further benefit stealth aims in nanomedicine. After evading recognition during circulation, targeting enables specific localization. Nanoparticle targeting stands to benefit from the remarkable molecular specificity of protein interactions by taking advantage of endogenous protein interactions with their target ligands. Finally, activatable properties have been applied to induce or guide specific nanoparticle function within the targeted area. Activation of nanoparticle function typically relies on biological or externally applied triggers, though future work is required to use such cleavable strategies synergistically with the protein corona. Combining such concepts, recent work has demonstrated modular nanoparticle constructs that are capable of tumor targeting (with cleavable reporters to indicate success) and gene delivery.165 Other exciting work has moved to multiplexed testing of the targeting and function of many nanoparticle chemistries simultaneously via DNA-barcoding.166 Finally, biocompatibility of such nanoparticle–corona constructs has inspired uses in both immune stimulation (both intended and unintended) and immune suppression, where the assays to assess biocompatibility stand to be refined. In sum, design of the protein corona on nanoparticles presents a functional handle to tune construct properties and attain improved outcomes towards in vivo stealth, targeting, and function.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
M. P. L. acknowledges support of Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), the Simons Foundation, NIH NIDA CEBRA award # R21DA044010, Stanley Fahn PDF Junior Faculty Grant with Award #PF-JFA-1760, Beckman Foundation Young Investigator Award, DARPA Young Faculty Award, FFAR New Innovator Award, an IGI award, support from CITRIS and the Banatao Institute, and a USDA award. M. P. L. is a Chan Zuckerberg Biohub Investigator and an Innovative Genomics Institute Investigator. R. L. P. and F. L. acknowledge the support of NSF Graduate Research Fellowships (NSF DGE 1752814), and L. C. acknowledges the support of National Defense Science and Engineering Graduate (NDSEG) Fellowship. We would like to acknowledge the use of medical clipart from Servier Medical Art by Servier (http://smart.servier.com).References
- L. Chio, R. L. Pinals, A. Murali, N. S. Goh and M. P. Landry, Adv. Funct. Mater., 2020, 30, 1910556 CrossRef CAS.
- A. Gupta, R. Das, G. Yesilbag Tonga, T. Mizuhara and V. M. Rotello, ACS Nano, 2018, 12, 89–94 CrossRef CAS PubMed.
- J. Y. Oh, H. S. Kim, L. Palanikumar, E. M. Go, B. Jana, S. A. Park, H. Y. Kim, K. Kim, J. K. Seo, S. K. Kwak, C. Kim, S. Kang and J.-H. Ryu, Nat. Commun., 2018, 9, 4548 CrossRef PubMed.
- M. Hadjidemetriou and K. Kostarelos, Nat. Nanotechnol., 2017, 12, 288–290 CrossRef CAS PubMed.
- P. C. Ke, S. Lin, W. J. Parak, T. P. Davis and F. Caruso, ACS Nano, 2017, 11, 11773–11776 CrossRef CAS PubMed.
- L. A. Warning, Q. Zhang, R. Baiyasi, C. F. Landes and S. Link, J. Phys. Chem. Lett., 2020, 11, 1170–1177 CrossRef CAS PubMed.
- M. Raoufi, M. J. Hajipour, S. M. K. Shahri, I. Schoen, U. Linn and M. Mahmoudi, Nanoscale, 2018, 10, 1228–1233 RSC.
- R. Cai, J. Ren, Y. Ji, Y. Wang, Y. Liu, Z. Chen, Z. Farhadi Sabet, X. Wu, I. Lynch and C. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 1997–2008 CrossRef CAS PubMed.
- X. Lu, P. Xu, H.-M. Ding, Y.-S. Yu, D. Huo and Y.-Q. Ma, Nat. Commun., 2019, 10, 4520 CrossRef PubMed.
- F. Giulimondi, L. Digiacomo, D. Pozzi, S. Palchetti, E. Vulpis, A. L. Capriotti, R. Z. Chiozzi, A. Laganà, H. Amenitsch, L. Masuelli, M. Mahmoudi, I. Screpanti, A. Zingoni and G. Caracciolo, Nat. Commun., 2019, 10, 3686 CrossRef CAS PubMed.
- A. Piloni, C. K. Wong, F. Chen, M. Lord, A. Walther and M. H. Stenzel, Nanoscale, 2019, 11, 23259–23267 RSC.
- T. Ding and J. Sun, Pharm. Res., 2020, 37, 10 CrossRef CAS PubMed.
- P. S. R. Naidu, N. Gavriel, C. G. G. Gray, C. A. Bartlett, L. M. Toomey, J. A. Kretzmann, D. Patalwala, T. McGonigle, E. Denham, C. Hee, D. Ho, N. L. Taylor, M. Norret, N. M. Smith, S. A. Dunlop, K. S. Iyer and M. Fitzgerald, ACS Appl. Mater. Interfaces, 2019, 11, 22085–22095 CrossRef CAS PubMed.
- Q.-F. Meng, Y.-X. Cheng, Q. Huang, M. Zan, W. Xie, Y. Sun, R. Li, X. Wei, S.-S. Guo, X.-Z. Zhao, L. Rao and W. Liu, ACS Appl. Mater. Interfaces, 2019, 11, 28732–28739 CrossRef CAS PubMed.
- Z. Zhang, J. Guan, Z. Jiang, Y. Yang, J. Liu, W. Hua, Y. Mao, C. Li, W. Lu, J. Qian and C. Zhan, Nat. Commun., 2019, 10, 1–11 CrossRef PubMed.
- T. Takeuchi, Y. Kitayama, R. Sasao, T. Yamada, K. Toh, Y. Matsumoto and K. Kataoka, Angew. Chem., Int. Ed., 2017, 56, 7088–7092 CrossRef CAS PubMed.
- C. Corbo, R. Molinaro, F. Taraballi, N. E. Toledano Furman, K. A. Hartman, M. B. Sherman, E. De Rosa, D. K. Kirui, F. Salvatore and E. Tasciotti, ACS Nano, 2017, 11, 3262–3273 CrossRef CAS PubMed.
- W. Perng, G. Palui, W. Wang and H. Mattoussi, Bioconjugate Chem., 2019, 30, 2469–2480 CrossRef CAS PubMed.
- G. Stepien, M. Moros, M. Pérez-Hernández, M. Monge, L. Gutiérrez, R. M. Fratila, M. de las Heras, S. Menao Guillén, J. J. Puente Lanzarote, C. Solans, J. Pardo and J. M. de la Fuente, ACS Appl. Mater. Interfaces, 2018, 10, 4548–4560 CrossRef CAS PubMed.
- Y. Zhu, T. Meng, Y. Tan, X. Yang, Y. Liu, X. Liu, F. Yu, L. Wen, S. Dai, H. Yuan and F. Hu, Mol. Pharmaceutics, 2018, 15, 5374–5386 CrossRef CAS PubMed.
- J. Zhao, S. Wu, J. Qin, D. Shi and Y. Wang, ACS Appl. Mater. Interfaces, 2018, 10, 41986–41998 CrossRef CAS PubMed.
- W. Lai, Q. Wang, L. Li, Z. Hu, J. Chen and Q. Fang, Colloids Surf., B, 2017, 152, 317–325 CrossRef CAS PubMed.
- F. Simonelli, G. Rossi and L. Monticelli, J. Phys. Chem. B, 2019, 123, 1764–1769 CrossRef CAS PubMed.
- J. Lazarovits, Y. Y. Chen, F. Song, W. Ngo, A. J. Tavares, Y.-N. Zhang, J. Audet, B. Tang, Q. Lin, M. C. Tleugabulova, S. Wilhelm, J. R. Krieger, T. Mallevaey and W. C. W. Chan, Nano Lett., 2019, 19, 116–123 CrossRef CAS PubMed.
- Q. H. Quach and J. C. Kah, RSC Adv., 2018, 8, 6616–6619 RSC.
- R. L. Pinals, D. Yang, D. J. Rosenberg, T. Chaudhary, A. R. Crothers, A. T. Iavarone, M. Hammel and M. P. Landry, bioRxiv, 2020, 2020.01.13.905356.
- V. Escamilla-Rivera, A. Solorio-Rodriguez, M. Uribe-Ramirez, O. Lozano, S. Lucas, A. Chagolla-López, R. Winkler and A. De Vizcaya-Ruiz, Int. J. Nanomed., 2019, 14, 2055–2067 CrossRef CAS PubMed.
- H. Lee, Small, 2020, 16, 1906598 CrossRef CAS PubMed.
- H. Mohammad-Beigi, Y. Hayashi, C. M. Zeuthen, H. Eskandari, C. Scavenius, K. Juul-Madsen, T. Vorup-Jensen, J. J. Enghild and D. S. Sutherland, bioRxiv, 2020, 2020.02.05.924480.
- H. Wang, R. Ma, K. Nienhaus and G. U. Nienhaus, Small, 2019, 15, 1900974 CrossRef PubMed.
- D. Chen, N. Parayath, S. Ganesh, W. Wang and M. Amiji, Nanoscale, 2019, 11, 18806–18824 RSC.
- P. Guo, D. Liu, K. Subramanyam, B. Wang, J. Yang, J. Huang, D. T. Auguste and M. A. Moses, Nat. Commun., 2018, 9, 1–9 CrossRef PubMed.
- J. C. Lee, N. D. Donahue, A. S. Mao, A. Karim, M. Komarneni, E. E. Thomas, E. R. Francek, W. Yang and S. Wilhelm, ACS Appl. Nano Mater., 2020, 3, 2421–2429 CrossRef CAS.
- V. Francia, D. Montizaan and A. Salvati, Beilstein J. Nanotechnol., 2020, 11, 338–353 CrossRef CAS PubMed.
- A. Gafur, N. Kristi, A. Maruf, G. Wang and Z. Ye, Biomater. Sci., 2019, 7, 3581–3593 RSC.
- J. F. A. de Oliveira, F. R. Scheffer, R. F. Landis, É. Teixeira-Neto, V. M. Rotello and M. B. Cardoso, ACS Appl. Mater. Interfaces, 2018, 10, 41917–41923 CrossRef PubMed.
- C. Champanhac, J. Simon, K. Landfester and V. Mailänder, Biomacromolecules, 2019, 20, 3724–3732 CrossRef CAS PubMed.
- H. Zhou, Z. Fan, P. Y. Li, J. Deng, D. C. Arhontoulis, C. Y. Li, W. B. Bowne and H. Cheng, ACS Nano, 2018, 12, 10130–10141 CrossRef CAS PubMed.
- L. Kong, Q. Chen, F. Campbell, E. Snaar-Jagalska and A. Kros, Adv. Healthcare Mater., 2020, 1901489 CrossRef CAS PubMed.
- J. G. Dancy, A. S. Wadajkar, N. P. Connolly, R. Galisteo, H. M. Ames, S. Peng, N. L. Tran, O. G. Goloubeva, G. F. Woodworth, J. A. Winkles and A. J. Kim, Sci. Adv., 2020, 6, eaax3931 CrossRef PubMed.
- X. Li, C. Lu, W. Xia, G. Quan, Y. Huang, X. Bai, F. Yu, Q. Xu, W. Qin, D. Liu and X. Pan, AAPS PharmSciTech, 2020, 21, 78 CrossRef CAS PubMed.
- N. Bertrand, P. Grenier, M. Mahmoudi, E. M. Lima, E. A. Appel, F. Dormont, J.-M. Lim, R. Karnik, R. Langer and O. C. Farokhzad, Nat. Commun., 2017, 8, 1–8 CrossRef CAS PubMed.
- N. Gal, M. Schroffenegger and E. Reimhult, J. Phys. Chem. B, 2018, 122, 5820–5834 CrossRef CAS PubMed.
- T. Lima, K. Bernfur, M. Vilanova and T. Cedervall, Sci. Rep., 2020, 10, 1129 CrossRef CAS PubMed.
- P. Shadmani, B. Mehrafrooz, A. Montazeri and R. Naghdabadi, J. Phys.: Condens. Matter, 2020, 32, 115101 CrossRef CAS PubMed.
- O. K. Kari, J. Ndika, P. Parkkila, A. Louna, T. Lajunen, A. Puustinen, T. Viitala, H. Alenius and A. Urtti, Nanoscale, 2020, 12, 1728–1741 RSC.
- C. Yu, Q. Zhou, F. Xiao, Y. Li, H. Hu, Y. Wan, Z. Li and X. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 10481–10493 CrossRef CAS PubMed.
- Y. T. Ho, N. A. Azman, F. W. Y. Loh, G. K. T. Ong, G. Engudar, S. A. Kriz and J. C. Y. Kah, Bioconjugate Chem., 2018, 29, 3923–3934 CrossRef CAS PubMed.
- P. S. R. Naidu, E. Denham, C. A. Bartlett, T. McGonigle, N. L. Taylor, M. Norret, N. M. Smith, S. A. Dunlop, K. S. Iyer and M. Fitzgerald, RSC Adv., 2020, 10, 2856–2869 RSC.
- C. Curtis, D. Toghani, B. Wong and E. Nance, Colloids Surf., B, 2018, 170, 673–682 CrossRef CAS PubMed.
- R. M. Visalakshan, M. N. MacGregor, S. Sasidharan, A. Ghazaryan, A. M. Mierczynska-Vasilev, S. Morsbach, V. Mailänder, K. Landfester, J. D. Hayball and K. Vasilev, ACS Appl. Mater. Interfaces, 2019, 11, 27615–27623 CrossRef CAS PubMed.
- X. Wang, M. Wang, R. Lei, S. F. Zhu, Y. Zhao and C. Chen, ACS Nano, 2017, 11, 4606–4616 CrossRef CAS PubMed.
- R. Palomba, A. L. Palange, I. F. Rizzuti, M. Ferreira, A. Cervadoro, M. G. Barbato, C. Canale and P. Decuzzi, ACS Nano, 2018, 12, 1433–1444 CrossRef CAS PubMed.
- A. C. Anselmo and S. Mitragotri, Adv. Drug Delivery Rev., 2017, 108, 51–67 CrossRef CAS PubMed.
- V. Francia, K. Yang, S. Deville, C. Reker-Smit, I. Nelissen and A. Salvati, ACS Nano, 2019, 13, 11107–11121 CrossRef CAS PubMed.
- R. L. Pinals, D. Yang, A. Lui, W. Cao and M. P. Landry, J. Am. Chem. Soc., 2020, 142, 1254–1264 CrossRef CAS PubMed.
- A. C. G. Weiss, K. Kempe, S. Förster and F. Caruso, Biomacromolecules, 2018, 19, 2580–2594 CrossRef CAS.
- F. Chen, G. Wang, J. I. Griffin, B. Brenneman, N. K. Banda, V. M. Holers, D. S. Backos, L. Wu, S. M. Moghimi and D. Simberg, Nat. Nanotechnol., 2017, 12, 387–393 CrossRef CAS.
- C. Rodriguez-Quijada, H. de Puig, M. Sánchez-Purrà, C. Yelleswarapu, J. J. Evans, J. P. Celli and K. Hamad-Schifferli, ACS Appl. Mater. Interfaces, 2019, 11, 14588–14596 CrossRef CAS PubMed.
- M. Mahmoudi, Trends Biotechnol., 2018, 36, 755–769 CrossRef CAS PubMed.
- A. Wang, T. Yang, W. Fan, Y. Yang, Q. Zhu, S. Guo, C. Zhu, Y. Yuan, T. Zhang and Y. Gan, Adv. Healthcare Mater., 2019, 8, 1801123 CrossRef PubMed.
- E. Nance, in Biomedical Nanotechnology, Humana Press, New York, NY, 2017, pp. 91–104 Search PubMed.
- M. R. Sepand, M. Ghavami, S. Zanganeh, S. Stacks, F. Ghasemi, H. Montazeri, C. Corbo, H. Derakhshankhah, S. N. Ostad, M. H. Ghahremani and M. Mahmoudi, Nanoscale, 2020, 12, 4935–4944 RSC.
- M. Tonigold, J. Simon, D. Estupiñán, M. Kokkinopoulou, J. Reinholz, U. Kintzel, A. Kaltbeitzel, P. Renz, M. P. Domogalla, K. Steinbrink, I. Lieberwirth, D. Crespy, K. Landfester and V. Mailänder, Nat. Nanotechnol., 2018, 13, 862–869 CrossRef CAS.
- J.-G. Piao, F. Gao, Y. Li, L. Yu, D. Liu, Z.-B. Tan, Y. Xiong, L. Yang and Y.-Z. You, Nano Res., 2018, 11, 3193–3204 CrossRef CAS.
- R. Koide and S.-I. Nishimura, Angew. Chem., Int. Ed., 2019, 58, 14513–14518 CrossRef CAS PubMed.
- A. Cox, P. Andreozzi, R. Dal Magro, F. Fiordaliso, A. Corbelli, L. Talamini, C. Chinello, F. Raimondo, F. Magni, M. Tringali, S. Krol, P. Jacob Silva, F. Stellacci, M. Masserini and F. Re, ACS Nano, 2018, 12, 7292–7300 CrossRef CAS PubMed.
- S. Palchetti, D. Pozzi, A. L. Capriotti, G. L. Barbera, R. Z. Chiozzi, L. Digiacomo, G. Peruzzi, G. Caracciolo and A. Laganà, Colloids Surf., B, 2017, 153, 263–271 CrossRef CAS.
- H. Zhang, T. Wu, W. Yu, S. Ruan, Q. He and H. Gao, ACS Appl. Mater. Interfaces, 2018, 10, 9094–9103 CrossRef CAS PubMed.
- L. M. Herda, D. R. Hristov, M. C. Lo Giudice, E. Polo and K. A. Dawson, J. Am. Chem. Soc., 2017, 139, 111–114 CrossRef CAS PubMed.
- W. Zhang, B. Meckes and C. A. Mirkin, ACS Cent. Sci., 2019, 5, 1983–1990 CrossRef CAS.
- R. Dal Magro, B. Albertini, S. Beretta, R. Rigolio, E. Donzelli, A. Chiorazzi, M. Ricci, P. Blasi and G. Sancini, Nanomedicine, 2018, 14, 429–438 CrossRef CAS PubMed.
- A. Jhaveri, P. Deshpande, B. Pattni and V. Torchilin, J. Controlled Release, 2018, 277, 89–101 CrossRef CAS PubMed.
- R. Fukuda, T. Umeyama, M. Tsujimoto, F. Ishidate, T. Tanaka, H. Kataura, H. Imahori and T. Murakami, Carbon, 2020, 161, 718–725 CrossRef CAS.
- M. Di Giosia, F. Valle, A. Cantelli, A. Bottoni, F. Zerbetto, E. Fasoli and M. Calvaresi, Carbon, 2019, 147, 70–82 CrossRef CAS.
- E. L. L. Yeo, P. S. P. Thong, K. C. Soo and J. C. Y. Kah, Nanoscale, 2018, 10, 2461–2472 RSC.
- A. C. G. Weiss, H. G. Kelly, M. Faria, Q. A. Besford, A. K. Wheatley, C.-S. Ang, E. J. Crampin, F. Caruso and S. J. Kent, ACS Nano, 2019, 13, 4980–4991 CrossRef CAS PubMed.
- R. Mout, G. Yesilbag Tonga, L.-S. Wang, M. Ray, T. Roy and V. M. Rotello, ACS Nano, 2017, 11, 3456–3462 CrossRef CAS PubMed.
- F. A. Mann, Z. Lv, J. Großhans, F. Opazo and S. Kruss, Angew. Chem., Int. Ed., 2019, 58, 11469–11473 CrossRef CAS PubMed.
- C. Wang, Z. Wang and L. Dong, Trends Biotechnol., 2018, 36, 661–672 CrossRef CAS PubMed.
- A. Silvestri, D. Di Silvio, I. Llarena, R. A. Murray, M. Marelli, L. Lay, L. Polito and S. E. Moya, Nanoscale, 2017, 9, 14730–14739 RSC.
- M. Carril, D. Padro, P. del Pino, C. Carrillo-Carrion, M. Gallego and W. J. Parak, Nat. Commun., 2017, 8, 1542 CrossRef.
- X. Zeng, J. Sun, S. Li, J. Shi, H. Gao, W. S. Leong, Y. Wu, M. Li, C. Liu, P. Li, J. Kong, Y.-Z. Wu, G. Nie, Y. Fu and G. Zhang, Nat. Commun., 2020, 11, 1–12 Search PubMed.
- M. Kokkinopoulou, J. Simon, K. Landfester, V. Mailänder and I. Lieberwirth, Nanoscale, 2017, 9, 8858–8870 RSC.
- C. Weber, J. Simon, V. Mailänder, S. Morsbach and K. Landfester, Acta Biomater., 2018, 76, 217–224 CrossRef CAS PubMed.
- Y. R. Perera, R. A. Hill and N. C. Fitzkee, Isr. J. Chem., 2019, 59, 962–979 CrossRef.
- A. Ceccon, T. Schmidt, V. Tugarinov, S. A. Kotler, C. D. Schwieters and G. M. Clore, J. Am. Chem. Soc., 2018, 140, 6199–6202 CrossRef CAS PubMed.
- L. Kouser, B. Paudyal, A. Kaur, G. Stenbeck, L. A. Jones, S. M. Abozaid, C. M. Stover, E. Flahaut, R. B. Sim and U. Kishore, Front. Immunol., 2018, 9, 131 CrossRef PubMed.
- Y. Zhang, J. L. Y. Wu, J. Lazarovits and W. C. W. Chan, J. Am. Chem. Soc., 2020, 142, 8827–8836 CrossRef.
- J. Simon, L. K. Müller, M. Kokkinopoulou, I. Lieberwirth, S. Morsbach, K. Landfester and V. Mailänder, Nanoscale, 2018, 10, 10731–10739 RSC.
- D. Prozeller, S. Morsbach and K. Landfester, Nanoscale, 2019, 11, 19265–19273 RSC.
- J. Park, J. E. Park, V. E. Hedrick, K. V. Wood, C. Bonham, W. Lee and Y. Yeo, Small, 2018, 14, 1703670 CrossRef PubMed.
- N. Feiner-Gracia, M. Beck, S. Pujals, S. Tosi, T. Mandal, C. Buske, M. Linden and L. Albertazzi, Small, 2017, 13, 1701631 CrossRef PubMed.
- L. Zhao, L. Zhao, H. Li, P. Sun, J. Wu, K. Li, S. Hu, X. Wang and Q. Pu, Anal. Chem., 2019, 91, 15670–15677 CrossRef CAS.
- T. A. Horbett, J. Biomed. Mater. Res., Part A, 2018, 106, 2777–2788 CrossRef CAS PubMed.
- S. Milani, F. B. Bombelli, A. S. Pitek, K. A. Dawson and J. Rädler, ACS Nano, 2012, 6, 2532–2541 CrossRef CAS PubMed.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- R. A. Latour, J. Biomed. Mater. Res., 2015, 103, 949–958 CrossRef.
- T. Casalini, V. Limongelli, M. Schmutz, C. Som, O. Jordan, P. Wick, G. Borchard and G. Perale, Front. Bioeng. Biotechnol. DOI:10.3389/fbioe.2019.00268.
- V. P. Zhdanov, Curr. Opin. Colloid Interface Sci., 2019, 41, 95–103 CrossRef CAS.
- D. C. Malaspina, L. Pérez-Fuentes, C. Drummond, D. Bastos-González and J. Faraudo, Curr. Opin. Colloid Interface Sci., 2019, 41, 40–49 CrossRef CAS.
- G. Brancolini, L. Bellucci, M. C. Maschio, R. Di Felice and S. Corni, Curr. Opin. Colloid Interface Sci., 2019, 41, 86–94 CrossRef CAS.
- G. Brancolini, H. Lopez, S. Corni and V. Tozzini, Int. J. Mol. Sci., 2019, 20, 3866 CrossRef CAS PubMed.
- F. Tavanti, A. Pedone and M. C. Menziani, Int. J. Mol. Sci., 2019, 20, 3539 CrossRef CAS PubMed.
- S. Correa, N. Boehnke, A. E. Barberio, E. Deiss-Yehiely, A. Shi, B. Oberlton, S. G. Smith, I. Zervantonakis, E. C. Dreaden and P. T. Hammond, ACS Nano, 2020, 14, 2224–2237 CrossRef CAS PubMed.
- D. A. Hume, K. M. Irvine and C. Pridans, Trends Immunol., 2019, 40, 98–112 CrossRef CAS PubMed.
- R. Cai and C. Chen, Adv. Mater., 2019, 31, 1805740 CrossRef CAS PubMed.
- J. T. Huckaby and S. K. Lai, Adv. Drug Delivery Rev., 2018, 124, 125–139 CrossRef CAS PubMed.
- C. Sanchez-Cano and M. Carril, Int. J. Mol. Sci., 2020, 21, 1007 CrossRef PubMed.
- M. Zhou, H. Huang, D. Wang, H. Lu, J. Chen, Z. Chai, S. Q. Yao and Y. Hu, Nano Lett., 2019, 19, 3671–3675 CrossRef CAS PubMed.
- A. A. Bhattacharya, T. Grüne and S. Curry, J. Mol. Biol., 2000, 303, 721–732 CrossRef CAS PubMed.
- C. Wilson, T. Mau, K. H. Weisgraber, M. R. Wardell, R. W. Mahley and D. A. Agard, Structure, 1994, 2, 713–718 CrossRef CAS.
- D. W. Borhani, D. P. Rogers, J. A. Engler and C. G. Brouillette, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 12291–12296 CrossRef CAS PubMed.
- J. Müller, K. N. Bauer, D. Prozeller, J. Simon, V. Mailänder, F. R. Wurm, S. Winzen and K. Landfester, Biomaterials, 2017, 115, 1–8 CrossRef PubMed.
- P. Zhang, F. Sun, S. Liu and S. Jiang, J. Controlled Release, 2016, 244, 184–193 CrossRef CAS PubMed.
- J. Guan, Q. Shen, Z. Zhang, Z. Jiang, Y. Yang, M. Lou, J. Qian, W. Lu and C. Zhan, Nat. Commun., 2018, 9, 2982 CrossRef PubMed.
- M. Papi, D. Caputo, V. Palmieri, R. Coppola, S. Palchetti, F. Bugli, C. Martini, L. Digiacomo, D. Pozzi and G. Caracciolo, Nanoscale, 2017, 9, 10327–10334 RSC.
- S. Li, Y. Cai, J. Cao, M. Cai, Y. Chen and X. Luo, Polym. Chem., 2017, 8, 2472–2483 RSC.
- C. Zhang, J. Lu, Y. Hou, W. Xiong, K. Sheng and H. Lu, ACS Appl. Mater. Interfaces, 2018, 10, 17463–17470 CrossRef CAS PubMed.
- J. Zhao, Z. Qin, J. Wu, L. Li, Q. Jin and J. Ji, Biomater. Sci., 2018, 6, 200–206 RSC.
- K. Zhao, D. Li, W. Xu, J. Ding, W. Jiang, M. Li, C. Wang and X. Chen, Biomaterials, 2017, 116, 82–94 CrossRef CAS PubMed.
- H. Wu, H. Hu, J. Wan, Y. Li, Y. Wu, Y. Tang, C. Xiao, H. Xu, X. Yang and Z. Li, Chem. Eng. J., 2018, 349, 129–145 CrossRef CAS.
- J. Simon, K. N. Bauer, J. Langhanki, T. Opatz, V. Mailänder, K. Landfester and F. R. Wurm, Adv. Sci., 2019, 6, 1901199 CrossRef CAS PubMed.
- C. Fornaguera, M. Guerra-Rebollo, M. Á. Lázaro, A. Cascante, N. Rubio, J. Blanco and S. Borrós, Adv. Healthcare Mater., 2019, 8, 1900849 CrossRef CAS PubMed.
- Y. Shen, X. Li, D. Dong, B. Zhang, Y. Xue and P. Shang, Am. J. Cancer Res., 2018, 8, 916–931 CAS.
- Z. Han and Z.-R. Lu, J. Mater. Chem. B, 2017, 5, 639–654 RSC.
- S. M. Figueroa, D. Fleischmann, S. Beck and A. Goepferich, Macromol. Biosci., 2020, 20, 1900427 CrossRef.
- T. Wang, D. Wang, J. Liu, B. Feng, F. Zhou, H. Zhang, L. Zhou, Q. Yin, Z. Zhang, Z. Cao, H. Yu and Y. Li, Nano Lett., 2017, 17, 5429–5436 CrossRef CAS PubMed.
- H. Han, D. Valdepérez, Q. Jin, B. Yang, Z. Li, Y. Wu, B. Pelaz, W. J. Parak and J. Ji, ACS Nano, 2017, 11, 1281–1291 CrossRef CAS.
- A. Almalik, H. Benabdelkamel, A. Masood, I. O. Alanazi, I. Alradwan, M. A. Majrashi, A. A. Alfadda, W. M. Alghamdi, H. Alrabiah, N. Tirelli and A. H. Alhasan, Sci. Rep., 2017, 7, 10542 CrossRef PubMed.
- R. H. Scannevin, R. Alexander, T. M. Haarlander, S. L. Burke, M. Singer, C. Huo, Y.-M. Zhang, D. Maguire, J. Spurlino, I. Deckman, K. I. Carroll, F. Lewandowski, E. Devine, K. Dzordzorme, B. Tounge, C. Milligan, S. Bayoumy, R. Williams, C. Schalk-Hihi, K. Leonard, P. Jackson, M. Todd, L. C. Kuo and K. J. Rhodes, J. Biol. Chem., 2017, 292, 17963–17974 CrossRef CAS PubMed.
- B. Mirković, M. Renko, S. Turk, I. Sosič, Z. Jevnikar, N. Obermajer, D. Turk, S. Gobec and J. Kos, ChemMedChem, 2011, 6, 1351–1356 CrossRef PubMed.
- V. Agrahari, P.-A. Burnouf, T. Burnouf and V. Agrahari, Adv. Drug Delivery Rev., 2019, 148, 146–180 CrossRef CAS PubMed.
- M. Wu, H. Zhang, C. Tie, C. Yan, Z. Deng, Q. Wan, X. Liu, F. Yan and H. Zheng, Nat. Commun., 2018, 9, 1–13 CrossRef CAS PubMed.
- J. Xue, Z. Zhao, L. Zhang, L. Xue, S. Shen, Y. Wen, Z. Wei, L. Wang, L. Kong, H. Sun, Q. Ping, R. Mo and C. Zhang, Nat. Nanotechnol., 2017, 12, 692–700 CrossRef CAS PubMed.
- L. Tang, Y. Wang and J. Li, Chem. Soc. Rev., 2015, 44, 6954–6980 RSC.
- B. Nabi, S. Rehman, S. Khan, S. Baboota and J. Ali, Brain Res. Bull., 2018, 142, 384–393 CrossRef CAS PubMed.
- Y. Zhu, C. Chen, Z. Cao, S. Shen, L. Li, D. Li, J. Wang and X. Yang, Theranostics, 2019, 9, 8312–8320 CrossRef PubMed.
- Y. Kwon, Y. Choi, J. Jang, S. Yoon and J. Choi, Pharmaceutics, 2020, 12, 204 CrossRef PubMed.
- A. Puiggalí-Jou, L. J. del Valle and C. Alemán, ACS Biomater. Sci. Eng., 2020, 6, 2135–2145 CrossRef.
- J. Yu, X. Chu and Y. Hou, Chem. Commun., 2014, 50, 11614–11630 RSC.
- C. Lim, E.-B. Cho and D. Kim, Korean J. Chem. Eng., 2019, 36, 166–172 CrossRef CAS.
- G. Li, Y. Gao, C. Gong, Z. Han, L. Qiang, Z. Tai, J. Tian and S. Gao, ACS Appl. Mater. Interfaces, 2019, 11, 39513–39524 CrossRef CAS PubMed.
- G. Jin, R. He, Q. Liu, M. Lin, Y. Dong, K. Li, B. Z. Tang, B. Liu and F. Xu, Theranostics, 2019, 9, 246–264 CrossRef CAS PubMed.
- B. Feng, B. Hou, Z. Xu, M. Saeed, H. Yu and Y. Li, Adv. Mater., 2019, 31, 1902960 CrossRef CAS PubMed.
- A. Gao, B. Chen, J. Gao, F. Zhou, M. Saeed, B. Hou, Y. Li and H. Yu, Nano Lett., 2020, 20, 353–362 CrossRef CAS PubMed.
- J. Li, R. Xu, X. Lu, J. He and S. Jin, Int. J. Nanomed., 2017, 12, 8043–8056 CrossRef CAS PubMed.
- Z. Yang, Q. Guo, Y. Cai, X. Zhu, C. Zhu, Y. Li and B. Li, J. Orthop. Transl., 2020, 21, 57–65 CAS.
- H. Yu, F. Jin, D. Liu, G. Shu, X. Wang, J. Qi, M. Sun, P. Yang, S. Jiang, X. Ying and Y. Du, Theranostics, 2020, 10, 2342–2357 CrossRef PubMed.
- J.-B. Coty, E. E. Oliveira and C. Vauthier, Int. J. Pharm., 2017, 532, 769–778 CrossRef CAS PubMed.
- T. Fülöp, R. Nemes, T. Mészáros, R. Urbanics, R. J. Kok, J. A. Jackman, N.-J. Cho, G. Storm and J. Szebeni, J. Controlled Release, 2018, 270, 268–274 CrossRef.
- J. Mo, Y. Xu, X. Wang, W. Wei and J. Zhao, Nanoscale, 2020, 12, 1742–1748 RSC.
- A. Gabizon, D. Tzemach, L. Mak, M. Bronstein and A. T. Horowitz, J. Drug Targeting, 2002, 10, 539–548 CrossRef CAS PubMed.
- B. T. Luk, Y. Jiang, J. A. Copp, C.-M. J. Hu, N. Krishnan, W. Gao, S. Li, R. H. Fang and L. Zhang, Mol. Pharmaceutics, 2018, 15, 3723–3728 CrossRef CAS PubMed.
- K. M. Pondman, M. Sobik, A. Nayak, A. G. Tsolaki, A. Jäkel, E. Flahaut, S. Hampel, B. ten Haken, R. B. Sim and U. Kishore, Nanomedicine, 2014, 10, 1287–1299 CrossRef CAS PubMed.
- G. Wang, F. Chen, N. K. Banda, V. M. Holers, L. Wu, S. M. Moghimi and D. Simberg, Front. Immunol. DOI:10.3389/fimmu.2016.00418.
- Q. Dai, J. Guo, Y. Yan, C.-S. Ang, N. Bertleff-Zieschang and F. Caruso, Biomacromolecules, 2017, 18, 431–439 CrossRef CAS PubMed.
- P. D. Ray, B.-W. Huang and Y. Tsuji, Cell. Signalling, 2012, 24, 981–990 CrossRef CAS PubMed.
- D. T. Jayaram, S. Runa, M. L. Kemp and C. K. Payne, Nanoscale, 2017, 9, 7595–7601 RSC.
- R. F. de Araújo, A. A. de Araújo, J. B. Pessoa, F. P. Freire-Neto, G. R. da Silva, A. L. C. S. Leitão Oliveira, T. G. de Carvalho, H. F. O. Silva, M. Eugênio, C. Sant'Anna and L. H. S. Gasparotto, Pharmacol. Rep., 2017, 69, 119–129 CrossRef PubMed.
- S. Gioria, F. Caputo, P. Urbán, C. M. Maguire, S. Bremer-Hoffmann, A. Prina-Mello, L. Calzolai and D. Mehn, Nanomedicine, 2018, 13, 539–554 CrossRef CAS PubMed.
- K. B. Riaz Ahmed, A. M. Nagy, R. P. Brown, Q. Zhang, S. G. Malghan and P. L. Goering, Toxicol. in Vitro, 2017, 38, 179–192 CrossRef CAS PubMed.
- J. M. Wörle-Knirsch, K. Pulskamp and H. F. Krug, Nano Lett., 2006, 6, 1261–1268 CrossRef PubMed.
- M. R. Findlay, D. N. Freitas, M. Mobed-Miremadi and K. E. Wheeler, Environ. Sci.: Nano, 2018, 5, 64–71 RSC.
- N. Boehnke, S. Correa, L. Hao, W. Wang, J. P. Straehla, S. N. Bhatia and P. T. Hammond, Angew. Chem., 2020, 59, 2776–2783 CrossRef CAS PubMed.
- J. E. Dahlman, K. J. Kauffman, Y. Xing, T. E. Shaw, F. F. Mir, C. C. Dlott, R. Langer, D. G. Anderson and E. T. Wang, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 2060–2065 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |