Nanomaterial-based multiplex optical sensors
Xiaojing
Pei
a,
Guangyu
Tao
b,
Xi
Wu
b,
Yurou
Ma
b,
Rongsheng
Li
 b and
Na
Li
b and
Na
Li
 *b
*b
aCollege of Chemistry and Materials Engineering, Beijing Technology and Business University, Beijing 100048, P. R. China
bBeijing National Laboratory for Molecular Sciences (BNLMS), Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, Institute of Analytical Chemistry, College of Chemistry and Molecular Engineering, Peking University, Beijing, 100871, P. R. China. E-mail: lina@pku.edu.cn; Tel: +86 10 62761187
First published on 11th May 2020
Abstract
The drive for a simultaneous analysis of multiple targets with excellent accuracy and efficiency, which is often required in both basic biomedical research and clinical applications, demands the development of multiplexed bioassays with desired throughput. With the development of nanotechnologies, innovative multiplex optical bioassays have been achieved. Nanomaterials exhibit unique physical and chemical properties such as easily tunable size, large surface-to-volume ratio, excellent catalysis and the desired signal transduction mechanism, which makes them excellent candidates for the fabrication of novel optical nanoprobes. This mini review summarizes nanomaterial-based optical multiplex sensors from the last 5 years. Specific optical techniques covered in this review are fluorescence, surface-enhanced Raman scattering (SERS), localized surface plasmon resonance (LSPR), chemiluminescence (CL), and the multimodality with fundamentals and examples.
Introduction
Multiplexed analysis has attracted considerable interest in the biological and biomedical fields due to its high throughput and detection accuracy.1–3 Compared to singleplexed assays, multiplexed detection allows the acquisition of high-density information with minimal assay time, sample volume and cost, thus being particularly favourable for a clinical diagnosis of fatal human diseases which generally involves a multitude of parameters for decision-making purposes.4 Almost all types of signals, including optical, electrical, thermal, magnetic and mass, have been used for multiplexed detection. Through the light–matter interaction, optical signals such as spectral wavelength and intensity, and lifetime due to absorption, emission and scattering, can be harnessed for multiplexed encoding.With the development of nanotechnology, innovative optical bioassays with high sensitivity and multiplexing capacity have been achieved (Fig. 1). Nanomaterials exhibit unique physical and chemical properties, which makes them excellent candidates for the fabrication of novel optical nanoprobes.5–7 The great potential of such optical labels has paved the way for developing new biomolecule assays with unprecedented analytical performances, including sensitivity, cost-effectiveness and ease of use. Myriad multiplexed bioassay approaches have been developed by employing various kinds of nanomaterials such as semiconductor quantum dots (QDs),8 nanoclusters,9 fluorescent microspheres by incorporating fluorescent nanoprobes or fluorophores,10,11 photon-upconversion nanomaterials,12 carbon dots,13 and plasmonic nanoparticles.14 This mini review intends to provide a broad coverage of the nanomaterial enhanced multiplex optical biosensors with signal transduction and applications based on fluorescence, surface-enhanced Raman scattering (SERS), localized surface plasmon resonance (LSPR), and chemiluminescence (CL). For each signal transduction category, we listed some typical nanomaterials with representative work (Table 1).
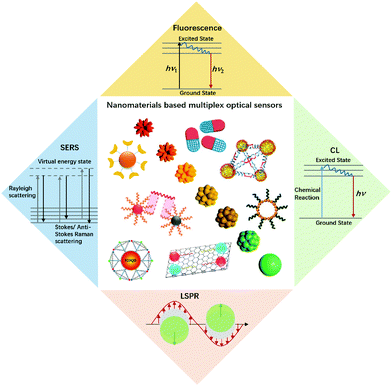 | ||
| Fig. 1 An illustration of nanomaterials and typical optical sensors. Images of nanomaterials were adapted with permission from ref. 14, 33, 41 and 52, Copyright (2017), (2018) and (2019) American Chemical Society; ref. 66, Copyright (2007) American Association for the Advancement of Science. ref. 78, Copyright (2015) WILEY. | ||
| Nanomaterials | Readout | Function | Analytes | Dynamic range factor | Limit of detection | Coding strategies | Multiplexing | Ref. |
|---|---|---|---|---|---|---|---|---|
| a Aflatoxin B1. b Zearalenone. c Vascular endothelial growth factor. d Interleukin-8. e Zika non-structural; protein 1. f Dengue nonstructural protein 1. | ||||||||
| Carbon dots | Fluorescence | Luminescent probes | Bacteria | NA | NA | Spectral | 6 | 23 |
| QDs | MicroRNA | 20 | 0.5 pM | Spectral | 2 | 24 | ||
| AuNCs | Mycotoxins | 2 × 103 | AFB1a: 0.34 pg mL−1 | Spectral | 2 | 25 | ||
| ZENb: 0.53 pg mL−1 | ||||||||
| UCNPs | Bacteria | 2 × 104 | Down to 10 cfu mL−1 | Spectral | 3 | 26 | ||
| Organic dye-doped microspheres | DNA and protein | NA | NA | Spectral, intensity | ∼102 in theory | 27 | ||
| QD-doped microspheres | DNA | 20 | ∼100 pM | Spectral | 3 | 28 | ||
| UCNP-doped microspheres | NA | NA | NA | Spectral, intensity and lifetime | >104 in theory | 29 | ||
| Multifunctional encoded particles | NA | NA | NA | Graphical, spectral | >106 in theory | 30 | ||
| AgNPs | SERS | Enhancement substrate | VEGFc and IL-8d | 104 | ∼1.0 fg mL−1 | Spectral | 2 | 31 |
| Gold nanostars | Virus | 50 | ZIKV NS1e: 0.72 ng mL−1 | Spectral | 2 | 32 | ||
| DENV NS1f: 7.67 ng mL−1 | ||||||||
| Au/Ag assembly | MicroRNA | 103 | Down to 1 pM | Spectral | 3 | 33 | ||
| AgNP embedded silica nanospheres | NA | NA | NA | Spectral, intensity | >10 trillion in theory | 34 | ||
| Stellate gold nanostructures | LSPR | Plasmonic probe | Receptor-mediated endocytosis | NA | NA | Spectral | 7 | 14 |
| Plasmonic nanoparticles | MicroRNA | 102 | 3 pM–10 pM | Spectral and spatial | 9 | 35 | ||
| CuS nanoparticles | CL | Enhancement substrate | Cytokines | 6 × 103 | IFN-γ: 2.9 pg mL−1 | Spatial | 2 | 36 |
| IL-4: 3.2 pg mL−1 | ||||||||
| AuNPs | MicroRNA | 105 | fM level | Spatial | 3 | 37 | ||
Fluorescence
Various nanomaterials with excellent fluorescence properties have been widely used in multiplex optical detection using wavelength, intensity, and lifetime for coding with wavelength being most frequently adopted.15 Due to the limited multiplexing capacity from spectral overlap, emission intensity is often used as the second dimension for multiplexed encoding, which in general is suitable for almost all types of luminescent materials. To fabricate intensity-based codes, a host matrix, such as the silica or polymer microsphere, is usually incorporated with a stoichiometric concentration of the different coloured luminescent materials.16–19 Though being facile, the encoding capacity and accuracy is strongly affected by the fluorescence intensity. Lifetime is emerging as a promising new parameter for multiplexing applications.20,21 Compared to intensity, lifetime is more stable and minimally affected by concentration variations of probes and excitation power. Amongst the nanoprobes for fluorescence-based multiplexed detection, quantum dots, upconversion nanoparticles, and microspheres by incorporating fluorescent nanoprobes or fluorophores are the major probes used with spectral, intensity, lifetime, graphic, or the combined coding strategies. For other nanoprobes, such as fluorescent metal nanoclusters and carbon dots, limited applications have been reported on multiplexed assays,22,23 and thus will not be discussed in detail.Quantum dots
QDs are semiconductor nanocrystals composed of atoms from groups II–VI or III–V in the periodic table. The excellent optical properties, such as the size-tunable absorption and emission wavelengths, as well as narrow emission bands, make QDs the ideal candidates for optical multiplexing. Compared with organic dyes, different emissions of QDs can be simultaneously excited with a single laser for multiplexed assays even in complex biological environments, thus, they are an excellent type of transducer for multiplexed assays. Nie et al. described the multifunctional nanoparticle probes based on QDs for cancer targeting and imaging in living animals.38 Using subcutaneous injection of QD-tagged cancer cells and systemic injection of multifunctional QD probes, sensitive and multicolour fluorescence imaging of cancer cells under in vivo conditions was achieved. These results shed light on the multiplex imaging of molecular targets in vivo.As a type of bright light-emitting nanoprobe, QDs have often been used in the sandwich design using magnetic nanoparticles for easy separation of excess optical probes. With this approach, Pang et al. reported a core–satellite structure assembled with magnetic nanoparticles and QDs for the simultaneous monitoring of miRNAs with femtomolar sensitivity and single-base mismatch specificity.24 Zhang et al. reported a siRNA-directed self-assembled QD biosensor for multiplex microRNA assay with a limit of detection (LOD) of 100 fM.39 For multiplexing, it is convenient to use Förster resonance energy transfer (FRET) design to realize signal switch and facilitate sensitive detection.40 Zhang et al. reported a three-dimensional (3D) DNA nanostructure with the capability of multiplexed detection using a multistep FRET (Fig. 2A).41 The tetrahedron-structured DNA is constructed by four oligonucleotide strands and is subsequently conjugated to a streptavidin-coated QD to obtain a QD-Cy3-Texas Red-Cy5 tetrahedron DNA. This tetrahedral DNA nanostructure has well-defined dye-to-dye spacing and high controllability for energy transfer between intermediary acceptors and terminal acceptors, enabling the generation of a multistep FRET between the QD and three dyes for the simultaneous detection of endonucleases and methyltransferases even in complex biological samples as well as for the screening of multiple enzyme inhibitors. FRET from luminescent lanthanide complexes to several different QD resulted in distinct photoluminescence decays, which were for time-gated photoluminescence coding to facilitate quantification in individual temporal detection windows. This time-gated intensity detection provides an extremely low background and a quick one-step assay format with high-order multiplexing capabilities. With this design, Hildebrandt et al. developed a time-gated multiplexed detection based on Tb complex and QDs for biosensing and imaging.42–44 Although QDs exhibit a narrow emission peak, QDs-dye FRET approaches show limited coding capacity because spectral crosstalk cannot be avoided due to the big full-width at half maximum of the organic dyes.
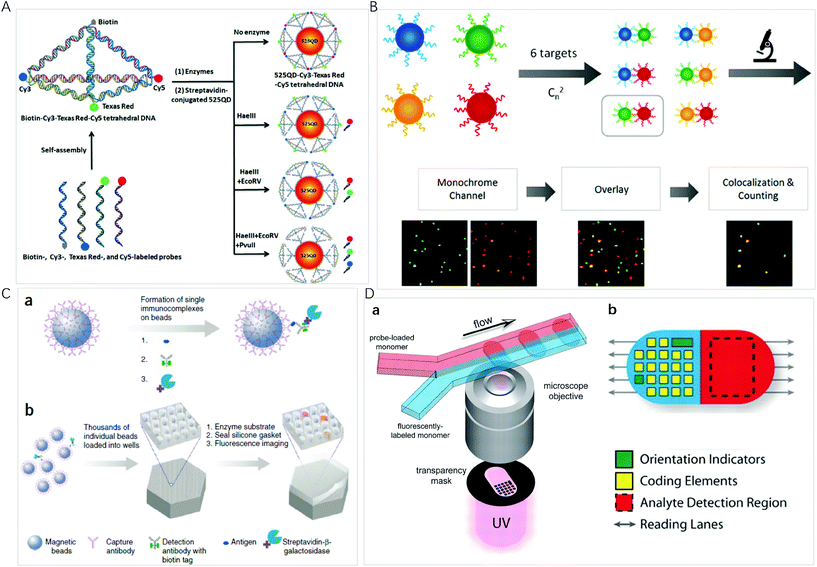 | ||
| Fig. 2 (A) Schematic illustration of the assembly of the 525 QD-Cy3-Texas Red-Cy5 tetrahedron DNA nanostructure for multiple enzymes assay. Reprinted with permission from ref. 41, Copyright (2019) American Chemical Society; (B) schematic illustration of the principle for multiplexed detection and workflow of the colocalized particle counting platform. Reprinted with permission from ref. 11, Copyright (2020) American Chemical Society; (C) the single-molecule arrays of femtoliter-sized wells. (a) Single protein molecules are captured and labeled on beads, and (b) beads with or without a labeled immune conjugate are loaded into femtoliter-volume well arrays for isolation and detection of single molecules by fluorescence imaging. Reprinted with permission from ref. 3, Copyright (2019) American Chemical Society. (D) (a) Schematic diagram of dot-coded particle synthesis showing polymerization across two adjacent laminar streams to make single-probe, (b) diagrammatic representation of particle features for encoding and analyte detection. Reprinted with permission from ref. 66, Copyright (2007) American Association for the Advancement of Science. | ||
Upconversion nanomaterials
The coding capacity based on fluorescent colour and intensity is limited by spectral overlap and background interference. Lanthanide-doped upconversion nanoparticles (UCNPs) exhibit anti-Stokes luminescence or upconversion luminescence, holding great promise as novel photonic elements: the near-infrared excitation presents much better penetration depth on biological samples and does not generate strong autofluorescence;18 the spectra of UCNPs are typically multiple and narrow emission lines, which minimize spectral crosstalk; the intensity of multiple emission lines can be either tuned individually or combinatorially for ratiometric encoding; the long decay times create a temporal coding dimension in a wide microsecond-to-millisecond range which facilitates easy time-gated measurements and signals independent of penetration depth in fluorescence microscopic imaging.45 Wang et al. employed multicolour UCNPs and aptamers to selectively capture and simultaneously quantify the three target bacteria on the basis of independent peaks. Improved by the magnetic separation, a LOD of 25, 10, and 15 cfu mL−1 for S. aureus, V. parahemolyticus, and S. typhimurium, respectively, were achieved.26 Huang et al. reported the synthesis and fine control of sodium scandium fluoride (NaxScF3+x) nanocrystals doped with different lanthanides by tuning of the ratio of oleic acid to 1-octadecene. The nanocrystals changed from the pure monoclinic phase (NaxScF3+x) to the pure hexagonal phase (NaScF4) when the polarity was increased, and the resulting strong red emission (660 nm) to strong green upconversion emission (520–540 nm) can be employed for optical coding.46 By incorporating the UCNPs into porous polystyrene or polydimethylsiloxane microspheres, a new matrix of 3D encoded (emission, intensity and lifetime) microspheres can be generated.21,29,47,48 With the emission-intensity-lifetime combined coding strategy, these microspheres show improved potential encoding capacity compared to the encoding methods based on the emission and intensity, and have been used in biosensing and imaging.49,50 On the other hand, time-resolved spectral measurements and imaging generally require laser sources, multichannel detectors, and time-gated instrumentation, increasing the cost in instrumentation and lab staff training.Fluorescent microspheres
Fluorescent microspheres carry luminescent materials, such as organic fluorophores, lanthanide complexes, QDs, or UCNPs encoded internally by entrapment within polymer or silica microspheres, or externally by covalent conjugation to the surface. By concentrating highly fluorescent nanoprobes in the microsphere, the bright fluorescence with intensity up to 104 times of that of an organic fluorophore can ensure that a biomolecule recognition event can be signalled by one particle, facilitating ultrasensitive bioassays and monitoring of rare biological events.51 For multiplexing, the incorporation of multiple fluorescent probes into a microsphere provides a way to use spectral, intensity, lifetime, graphic, or the combined coding strategies to exponentially increase the number of codes.The high brightness of fluorescent microspheres makes it possible to develop multiplexing methods at the single-particle level, using the spectrum, the brightness/intensity or lifetime for the coding of different targets of interest. It is an ideal platform for bioassays due to the high sensitivity, small amount of sample and reagent consumption, and rapid response time. Li et al. developed an automatic fluorescent nanoparticle (FNP) counting platform with a standard fluorescence microscopic imaging setup for amplification-free multiplexed detection of attomoles of nucleic acids.52 Moreover, combining the masking tactic with FNP counting, the simultaneous detection of multiple single-nucleotide variants was realized with both ultrahigh specificity and low-abundance sensitivity.53 Very recently, this research group presented a colocalized particle counting platform that can realize the separation-free multiplexed detection of 6 nucleic acid targets with a zeptomole sensitivity and a dynamic range up to 5 orders of magnitude (Fig. 2B).11 The sequence specific coding is realized by an arbitrary combination of two fluorescence channels with different emitting colours. The presence of a target induces the formation of a sandwich nanostructure via hybridization, and thus the occurrence of colocalization of two different coloured microspheres.
Miniaturization of reaction volumes and confinement of analytes of interest into ultrasmall containers can greatly enhance sensitivity. Microwell arrays allow a large number of reactions to be performed in parallel and observed simultaneously at the single-molecule or single-cell level.54 Using an optical fiber glass bundle to generate femtoliter-sized microwell arrays for preparing small volume containers to confine a single bead or target molecule in one microwell, Walt et al. have done comprehensive work on a digital enzyme-linked immunosorbent using single-molecule arrays (Simoa), which has been commercialized by Quanterix Corp (Fig. 2C).3,55 Sensitive quantification is achieved by counting the number of fluorescent microwells. For multiplexed detection, dyes of different colours or intensities are attached to the beads to generate optically distinct bead subpopulations; the same experimental procedure as singleplexed detection is then performed. Simoa immunoassays have been widely used for multiplexed detection of proteins,56,57 nucleic acids,58 and small biomolecules.59 To simplify experimental protocols, this research group designed the cyclic olefin polymer array using easily accessible materials as an alternative to optical fibers.60 Warren C. W. Chan et al. fabricated 3-micron diameter wells on a glass slide and used a similar strategy to develop a simple and low-cost chip-based wireless multiplexed diagnostic device by combining the QD-based barcode technology with smartphones and isothermal amplification.61
A library of fluorescence coded microspheres can be made by the use of two fluorophores at distinct ratios. The coded microspheres can be further conjugated with a reagent specific to a particular bioassay, e.g. the antibody, the oligonucleotides, enzyme substrates, or receptors. Taking the immunoassay as an example, the antibody-conjugated microspheres bind with the target of interest, and the second antibody to the analyte is attached to a reporter dye label. The use of different coloured beads enables multiplexed detection in a single sample. This coding strategy and experimental design makes it convenient to use a dual detection flow cytometer for multiplexed detection, with one channel decoding the microspheres and another channel measuring fluorescence intensity for quantification. This type of assay has been commercialized by Luminex Corp.27 The Luminex xMAP array can simultaneously detect up to 100 tests by incorporating 5.6 μm polystyrene microspheres internally dyed with two or three spectrally distinct organic fluorophores, and has been widely used for multiplexed miRNAs,62 drugs63 and gene64 detection. Its novel optical properties such as size-tunable emission and simultaneous excitation of QD render it an ideal fluorophore for spectral and intensity multiplexing. By trapping QDs with different emission spectra at different concentrations, numerous barcodes can be obtained. Theoretically, n intensity levels with m colours generate (nm − 1) unique codes.65 For example, 10 intensity levels and 6 colours can generate more than one million barcodes. However, the actual coding capabilities are likely to be substantially lower due to spectral overlapping, intensity variations, and signal-to-noise requirements. The limitation in practical application would be that expensive instrumentation is needed for the dual channel detection of single microspheres of different colours.
The microsphere-based multiplexed assay still suffers from the limited achievable barcodes due to spectral overlap. Graphical codes are the patterning of optical elements on a microcarrier, such as striped rods, ridged particles and dot-patterned particles. Doyle et al. presented a method based on continuous-flow lithography that integrates particle synthesis, encoding and probe incorporation into a single process to generate multifunctional particles bearing over a million unique codes (Fig. 2D).66 In the coding preparation, one fluorescently labelled monomer stream and one probe-loaded monomer stream flow through a spot where polymerization occurs across an adjacent laminar stream to produce encoded single-probe, half-fluorescent particles. Each particle is an extruded 2D shape whose morphology is determined by a photomask that is inserted into the field-stop position of the microscope and whose chemistry is determined by the content of the coflowing monomer streams. To further broaden the multiplexing capacity, this research group combined spatial patterning with rare-earth upconversion nanocrystals (UCNs) by flow lithography with exponentially scalable encoding capacities (>106 particles).30 They synthesized a palette of 9 spectrally distinct UCNs by adjusting the relative stoichiometries of the lanthanide ions Yb3+, Er3+ and Tm3+ in the reaction premix, resulting in narrow emission bands centred at 470, 550 and 650 nm. By embedding different UCNs within barcoded microparticles consisting of up to 6 stripes, an encoding capacity of greater than 1 million is easily achieved. With this concept, a high-throughput and accurate quantification of biomarkers ranging in size from large mRNAs to small miRNAs were achieved.67,68
Surface-enhanced Raman scattering
Surface-enhanced Raman scattering (SERS) methods present outstanding properties, including high sensitivity, fingerprinting capability, and insusceptibility to photobleaching; in particular, much narrower scatter peaks enable a high spectral coding capacity.69 Though not used as optical probes, metallic nanostructures are used to enhance the electromagnetic field around Raman reporters to achieve sensitive multiplexed detection with a LOD down to femtomolar concentrations. The morphology and the surface modification of the enhancement substrates play a very important role in the sensitivity of assays through the local electromagnetic field and electron transfer.70Nobel metals such as gold and silver have been of particular interest for SERS substrates because of their tunable plasmon resonance by varying nanoparticle size and shape. Using the Raman-active reporter modified silver nanoparticles with the convenience of magnetic bead collection, Xu et al. presented a SERS-microfluidic droplet platform for the simultaneous detection of the vascular endothelial growth factor and interleukin-8 secreted by a single cell with a LOD of 1.0 fg mL−1 in one droplet.71 By coadsorption of multiple dyes onto gold nanorods, Alastair W. Wark et al. created the SERS tag that can be detected across a much wider range of excitation wavelengths (514–1064 nm).72 The ability to colocalize and track individual optically encoded nanoparticles across a wide range of wavelengths simultaneously is promising in multiplexed tracking and sensing applications in biological environments. Gold nanostar increased surface-to-volume ratios, allowing for more Raman reporters to be attached onto the nanostar surface and the electromagnetic field coupling between tips of adjacent nanostars could further enhance SERS signals. With 1,2-bis(4-pyridyl)ethylene and 4-mercaptobenzoic acid attached to gold nanostars, Hamad–Schifferli et al. reported a novel SERS-based lateral flow assays platform that can distinguish between Zika and dengue, with LOD lower than colorimetric readout.32 SERS-encoded gold nanostars were conjugated to specific antibodies for Zika NS1 and dengue NS1 diseases for the dipstick immunoassay, which exhibited 15-fold and 7-fold improved detection limits for Zika NS1 and dengue NS1, respectively. The lateral flow assay stripe format combined with portable hand-held Raman readers can find a breadth of applications.
When the distance between two metal micro/nanoparticles is relatively close (1–10 nm), the electromagnetic field between the particles superimposes to form hot spots where high SERS enhancement factors (107–108) can be achieved. Hot spots can be fabricated between nanoparticles and nanoparticles, nanoparticles and caves, nanoarrays and nanoarrays, and nanoparticles and nanowires.73 For example, a Au/Ag assembly with high density can produce sizable coupled hot spots that largely contribute to the enhancement factor. Using Cy5- and Cy5.5-labelled silver-coated gold nanostars (AuNS@Ag), Vo-Dinh et al. designed a “turn-on” SERS sensor for multiplexed detection of miRNAs.74 Compared to uncoated gold nanostars, AuNS@Ag was demonstrated to offer over an order of magnitude of signal enhancement, rendering it an excellent SERS substrate. Using the patterned Au nanowire-on-film SERS sensor to generate hot spots to enhance the Cy5 tag, Kim et al. reported an ultrasensitive, multiplex pathogen DNA detection method combined with an exonuclease III-assisted target DNA recycling reaction with the LOD down to 100 fM.75 3D Ag microspheres with hollow structures possess superior optical properties and SERS activity compared with their corresponding solid counterparts. With 5,5′-dithiobis(2-nitrobenzoic acid), 4-aminothiophenol, and 4,4′-dipyridyl as specific Raman labels, Ye et al. fabricated an ultrasensitive and reliable SERS sensor with micrometer-sized hollow silver microspheres and hollow silver microspheres for the simultaneous detection of multiple microRNA biomarkers with a LOD of 10 fM.33
Alternatively, SERS hot spots could be generated through the formation of “nanogaps”. The greatly enhanced local electromagnetic field in the nanogap leads to strongly enhanced Raman signals. By mediating poly-adenine and encoding Raman reporters, Song et al. synthesized nanogap-based universal SERS nanotags.76 The universal nanotags were then functionalized by different types of biological probes and used as SERS nanoprobes to recognize various bioactive molecules. The enhancement ability and the reproducibility of randomly distributed hot spots for SERS-based assays are always limited, and it is still necessary to construct more controllable and orderly hot spots. The hot spots of interior nanogaps compared with exterior nanogaps between metal nanostructures are usually more controllable. Song et al. reported a DNA-mediated Au/Ag nanomushroom with interior nanogaps fingerprinted with 4-nitrothiophenol (NBT), ROX and ABT for the simultaneous SERS detection of various DNA and RNA targets.77 The DNA involved in the nanostructures can act as not only a gap DNA (mediated DNA) but also a probe DNA (hybridized DNA). Furthermore, the DNA tetrahedron structure can be used to attach gold/silver nanoparticles and precisely locate the Raman tag to improve the SERS effect, the reproducibility of the assay, and biocompatibility with biological samples. Kuang et al. designed the DNA-frame driven Ag-pyramids, bearing multiple aptamers and triple Raman reporters that enabled the simultaneous detection of biomarkers at the attomolar level.78 Similarly, Xu et al. prepared the tetrahedron probe with triple Raman reporter modified gold nanoparticles and the complementary nucleic acid aptamer for the simultaneous ratiometric detection of telomerase and the epithelial cell-adhesion molecule in living cells.79
The spectral fingerprint region of most organic Raman reporters is in the range of 1000–1700 cm−1, which overlaps with that of endogenous biological molecules (such as peptide, protein, and cytochrome C). To minimize the interference from the endogenous components of biological samples, Raman labels including alkyne, azide, nitrile, deuterium, and metal-carbonyl with vibration bands located in the Raman silent region of biomolecules (1800–2800 cm−1) were used. By a rational selection of 3 different molecules with single narrow Raman bands in the biosilent region with peaks at 2105, 2159, and 2227 cm−1, Tang et al. designed three new bioorthogonal SERS nanoprobes and applied the cocktail for MDA-MB-231 and MCF-7 breast cancer multiplex phenotype detection (Fig. 3A).80 Three bioorthogonal Raman reporters were anchored on the surface of gold nanoflowers (AuNF) which served as the Raman signal enhancer. Then the surface of AuNF was modified by silica and polyethylene glycol to improve stability. The mature double antibody sandwich allowed the formation of a microscale satellite assembly structure between a magnetic bead and single SERS tags, and therefore a pure and single SERS emission can be observed under the routine excitation laser spot. Using three triple bonds (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N, C
N, C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C) containing Raman reporters contributing separate SERS emissions located at 2105, 2159, and 2227 cm−1 and with AuNPs serving as the Raman signal enhancer, Zeng et al. reported an interference-free mixing SERS emission readout to simultaneously indicate 3 specific liver cancer antigens in 39 clinical serum samples.81 Applying a series of CN-bridged coordination polymer encapsulated AuNPs as the optical label, Hu et al. developed a universal and interference-free optical label through a facile and auxiliary agent free self-assembly route.82 The Raman shift of the CN-bridge can be flexibly tuned with a simple substitution of iron ions in the synthetic process. About 2n − 1 optical labels can be achieved by only using n single emissions in the Raman-silent region of biomolecules.
C) containing Raman reporters contributing separate SERS emissions located at 2105, 2159, and 2227 cm−1 and with AuNPs serving as the Raman signal enhancer, Zeng et al. reported an interference-free mixing SERS emission readout to simultaneously indicate 3 specific liver cancer antigens in 39 clinical serum samples.81 Applying a series of CN-bridged coordination polymer encapsulated AuNPs as the optical label, Hu et al. developed a universal and interference-free optical label through a facile and auxiliary agent free self-assembly route.82 The Raman shift of the CN-bridge can be flexibly tuned with a simple substitution of iron ions in the synthetic process. About 2n − 1 optical labels can be achieved by only using n single emissions in the Raman-silent region of biomolecules.
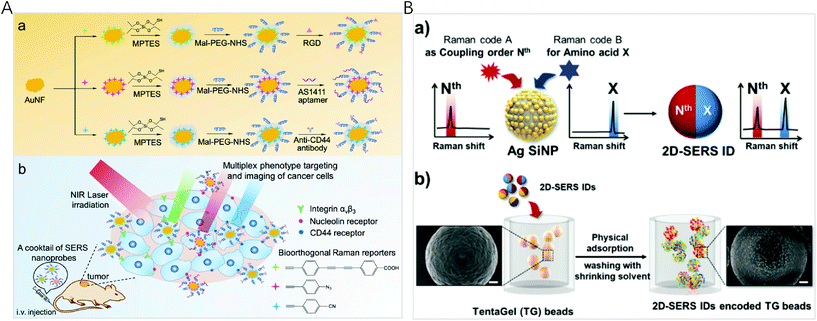 | ||
| Fig. 3 (A) (a) Fabrication of bioorthogonal SERS nanoprobes with three specific targeting moieties and (b) cocktail of bioorthogonal SERS nanoprobes for simultaneous multiplex cancer phenotype targeting and diagnosis in mice after intravenous injection. Reprinted with permission from ref. 80, Copyright (2019) American Chemical Society. (B) (a) Schematic illustration of the two-dimensional SERS encoding method: (b) design principle encoding procedure on polymer beads and (c) decoding procedure by Raman mapping. Reprinted with permission from ref. 34, Copyright (2019) The Royal Society of Chemistry. | ||
Although Raman spectra have narrow bands, spectral window still gets crowded when multiple reporters are used simultaneously, inevitably resulting in spectral overlap. To establish a simple and efficient encoding method for high-throughput bioassays, Jeong et al. reported two-dimensional SERS nano-identifiers (2D-SERS IDs) for high-throughput screening with an on-bead compound library (Fig. 3B).34 The 2D-SERS IDs were concurrently labelled with two kinds of Raman label compounds having well separated spectral bands, with one for a coupling step code and another for a code of amino acid (AA) at that coupling step. On the basis of this 2D-SERS encoding strategy, in theory, by using only 30 kinds of Raman labels (20 AA codes and 10 coupling step codes), all possible combinations of decamer peptide (over 10 trillion peptides) could be completely and uniquely encoded. After synthesizing a compound library with this encoding process, conducting bioassays and decoding for screening lead to an easy identification of each peptide sequence on the beads with an only single Raman scanning on the surface of a 2D-SERS ID encoded polymer micro-bead without prior preparation.
Localized surface plasmon resonance
Localized surface plasmon resonance (LSPR) occurs when the frequency of the incident light matches the natural frequency of surface electron oscillating of metal nanoparticles with sizes much smaller than the incident wavelength. For gold and silver nanoparticles, their plasmonic bands are generally located in the visible region, making them suitable for sensing applications. In the year 2000, Mirkin et al. reported the scanometric array based on the use of spherical nucleic acid–gold nanoparticle conjugates (SNA–AuNPs).83 Due to the cooperative melting transitions over the narrow temperature ranges of the SNA–AuNPs, a significantly higher target discrimination capability can be achieved with stringent conditions. This group later on presented a high density scanometric microRNA array for the profiling of prostate cancer markers using spherical nucleic acid–gold nanoparticle conjugates with higher sensitivity and selectivity than fluorophore based, high-density array techniques.84 Recently, by positioning two different sizes of gold nanocubes on gold surfaces with precisely defined locations for each particle via template-confined, DNA-mediated nanoparticle assembly, this research group reported a multiplexed surface encoding.85 This multiplexed encoding platform dramatically increased the sophistication and density of codes that can be written using colloidal nanoparticles, which may enable high-security, high-resolution encoding applications.Although the high extinction cross-section of the plasmonic nanoparticles enables the colorimetric assays, suspension-based multiplexed colorimetry finds limited applications due to the limited spectral tunability based on the interparticle distance adjustment.86,87 Single particle based dark-field microscopic imaging provides a sensitive technique for multiplexed detection, because light-scattering of nanoparticles do not have the problem of a saturated emission rate or photobleaching. Huang et al. applied the LSPR light-scattering of single nanoparticles and sandwich-type immunoassays to the simultaneous imaging and quantification of AFP and CEA.88 Combining magnetic beads with three colour-encoded plasmonic nanocrystals of AuNP, AuNR and gold/silver nanoparticle (Au/AgNP) to signal three dissimilar virus-related protective antigen genes, Ebola virus, Variola virus, and Bacillus anthracis, Sun et al. developed a novel digital triplex DNA assay method with a LOD range of 0.5–3 fM.89 Nam et al. reported a multiplexed detection strategy with optokinetically coded plasmonic nanoprobes (NPs) on a supported lipid bilayer (Fig. 4A).35 The nanoprobes were coded optically (combinatorial plasmonic couplings) and kinetically (particle mobility) to generate highly multiplexed detection of targets. In situ single-particle monitoring and normalized RGB analysis of the optokinetically combinatorial assemblies among three M-NPs and three I-NPs with dark-field microscopy (DFM) allowed for differentiating and quantifying 9 different miRNA targets in one sample.
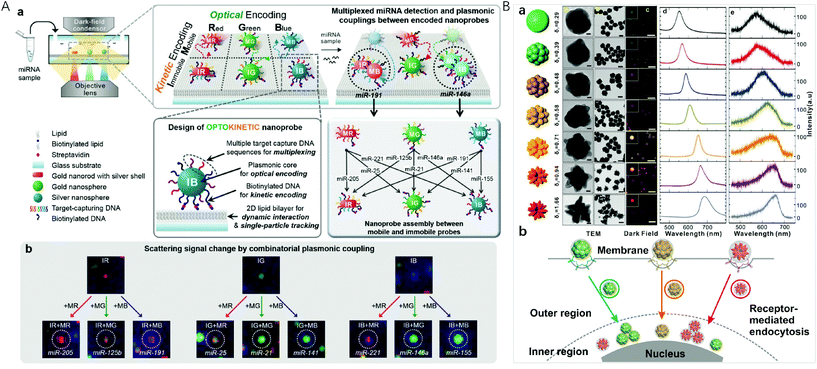 | ||
| Fig. 4 (A) Optokinetically encoded nanoprobe-tethered supported lipid bilayer assay, (a) six types of NPs are prepared by kinetic [mobile (M) and immobile (I)] and optical [red (R), green (G), and blue (B)] coding methods. NPs are composed of a plasmonic nanoparticle core, multiple target capture DNAs, and biotinylated DNA (bottom left). The nineplexing strategy using combinatorial assemblies between OK and NPs relies on each assembly mode, which is mediated by 9 different target miRNAs (bottom right). (b) Nine different scattering signal changes due to combinatorial plasmonic couplings induced by M-NPs binding to I-NPs. Reprinted with permission from ref. 35, Copyright (2017) American Chemical Society; (B) (a) synthesis of uniform-yet-color-resolvable fractal gold nanostructure plasmonic probes and schematic illustration of internalization and intracellular transport of NPs; (b) schematic illustration of internalization and intracellular transport of NPs. Reprinted with permission from ref. 14, Copyright (2019) American Chemical Society. | ||
Plasmonic imaging with metal nanoparticle labels has recently emerged as a long-term cellular imaging method due to the ultrahigh light-scattering and photostability. However, direct and simultaneous monitoring of multiple molecular or cellular species remains a hurdle due to the lack of uniform-yet-colour resolvable plasmonic NP labels. Fan et al. synthesized DNA-engineered fractal nanoplasmonic labels with ultrahigh light-scattering and photostability for super multiplexed imaging in single cells (Fig. 4B).14 These colour-resolvable nanoplasmonic labels have a uniform size of ∼50 nm with an inner hollow gap of ∼1 nm, presenting distinct plasmonic colours and scattering wavelengths under DFM. This Au nanostructure consists of three components: a solid Au core of ∼15 nm, a hollow gap of ∼1 nm, and a stellate Au shell. The control on the outer shell morphology of the stellate gold nanostructures allowed the tuning of plasmonic colours. By labelling intracellular proteins, the colour-resolvable plasmonic labels enabled a simultaneous dynamic imaging of different types of endocytosis in single cells. This super multiplex-plasmonic image method sheds light on elucidating complex nanoparticle–protein interactions and facilitates the development of novel nanomedicines for diagnosis and therapy. Flow cytometry has the ability to measure intracellular AuNPs by collecting the scattered light from a large population of live cells through efficient single cell analysis. El-Sayed et al. designed red-shifted excitation lasers which greatly enhanced the optical signal needed for the flow cytometry based detection of AuNP (26 nm in diameter) and AuNR (67 × 33 nm) uptake in triple negative breast cancer cells.90 This method with its ability to sort cells based on specific nanoparticle content, was useful in detecting cellular AuNPs.
Chemiluminescence
Using a chemical reaction to produce excited luminophores instead of an excitation source, chemiluminescence (CL) has become an attractive multiplex method in spatial-resolved arrays due to its simplicity, low cost and the close to zero background signal.91 However, the generated CL signal is relatively weak, and, therefore, sensitivity needs to be improved. A number of approaches have been explored for the amplification and enhancement of the signal by employing catalysts including the usual peroxidase enzyme and surface-active nanomaterials such as metal nanoparticles.Gold nanoparticles are often employed as an enhancement substrate to improve CL sensitivity. AuNPs have a large specific surface area, and, therefore, antibody (Ab) and enzymes such as horseradish peroxidase (HRP) can be labelled onto the surface of AuNPs to facilitate recognition and CL signal production. Jiang et al. developed a dual-readout CL-gold lateral flow test for the multiplex detection of disease biomarkers at the picomolar level (Fig. 5A).92 In this assay, the colour of the Ab–Au–HRP conjugate in the test line is used for the visual readout, and HRP on the surface of AuNPs is employed to amplify the signals to generate CL that allows for quantitative and highly sensitive detection. With the recognition event, the emission product produced by HRP-catalysis on the surface of AuNPs is enhanced by the AuNPs. Compared with the conventional gold lateral flow test, this method can dramatically increase sensitivity by three orders of magnitude. By combining with a disposable protein array, Ju et al. designed a multilayer hemin/G-quadruplex DNAzyme wrapped AuNP (M-DNAzyme/AuNP) tag for ultrasensitive imaging for the simultaneous detection of 4 cancer biomarkers.93 The M-DNAzyme/AuNP tag was prepared by assembling a high ratio of alkylthiol-capped signal DNA containing multiple G-quadruplex sequences to biotinylated DNA on AuNPs and then reacting with hemin to form multilayer hemin/G-quadruplex DNAzyme units. It could be bound to the biotinylated secondary antibody of a sandwich immune complex by biotin–streptavidin conjugation to catalyze the CL reaction on a protein array, which produced strong CL emission. Li et al. introduced a dual-signal amplification strategy by employing primary antibody functionalized AuNPs (Ab1-AuNPs) immobilized on the detection zone as amplified capture probes, and Co(II) catalyst, secondary antibody, luminol multifunctionalized AuNPs (Co(II)-Ab2-luminol-AuNPs) with excellent CL activity as amplified signal probes.94 Auto separated CL signals with temporal resolution were obtained by time delayed transport of H2O2 to different detection zones for multiplexed analysis. The CL signal improved about 20-fold higher than that when horseradish peroxidase labelled antibody modified luminol-AuNPs were used as a signal probe with the detection limits down to 0.06 pg mL−1, 0.3 pg mL−1 and 0.4 pg mL−1 for heart-type fatty acid-binding protein, cardiac troponin I and copeptin detection, respectively.
 | ||
| Fig. 5 (A) Scheme of the C-mode GLFT for the simultaneous, quantitative detection of AFP and CEA. Reprinted with permission from ref. 92, Copyright (2016) The Royal Society of Chemistry; (B) schematic illustration of a multianalyte immunoassay based on the synthesized encoding carriers using a sandwich structure concept. Reprinted with permission from ref. 97, Copyright (2012) American Chemical Society. | ||
Some nanoparticles exhibit peroxidase-like catalytic activity, and have been used as nanozymes for colorimetric and CL detection. Yang et al. reported a CL imaging nanozyme immunoassay, in which CuS nanoparticles act as the catalytic tag for the simultaneous multiplexed detection of cytokines.36 The strategy provided a universal nanozyme-labelled multiplex immunoassay for the high-throughput detection of relevant biomarkers and further disease diagnosis. Han et al. reported the intrinsically selective CL switching at the surface of catalytic Fe3O4 nanoparticles for the sensitive detection and simultaneous determination of various pesticides.95 The CL signals can be quenched by the addition of ethanol, and the quenching effect can be inhibited through the specific binding of target molecules on Fe3O4 nanoparticles, leading to CL “turn-on” in the presence of ethanol. Fe3O4 nanoparticles with different surface groups can generate a unique CL response pattern for the simultaneous determination of various pesticides with the LOD down to 0.1 nM. The very simple and versatile strategy opened a new window of interest in the application of magnetic nanoparticles and the development of CL sensors.
Multimodality
A few works have reported the use of multimodality for expanding the coding dimension. A composite nanoparticle has been used for bioassays by incorporating SERS and fluorescence labels simultaneously into it. Using the fluorescence mode for fast screening of biomolecules while the SERS mode for quantitative analysis, Cui et al. reported a SERS-fluorescence dual-mode nanoprobe based on multilayer core–shell nanoparticles.96 In the nanoprobe, Raman-reporter-labelled nanoparticles provide SERS signals, while quantum dots provide fluorescence signals. The dual mode nanoprobe could be used for rapid and sensitive bioanalysis and imaging. Furthermore, this research group demonstrated this dual mode using organic-metal-QD hybrid nanoparticles (OMQ NPs) with a nanolayered structure (Fig. 5B).97 This nanostructure enabled a new coding based on a joint SERs-fluorescence spectrum. This structure assembled SERS reporters and fluorescent agents onto different layers of OMQ NPs, leading to an easy fabrication protocol when a large number of agents need to be involved into encoding carriers. When m kinds of QDs and n kinds of SERS reporters are employed, the number of available codes dramatically increases to (2m+n − 1), which is ((2m+n − 1)/(2m − 1)) or ((2m+n − 1)/(2n − 1)) times of coding capacity obtained by using only fluorescence or SERS spectra. Furthermore, this group designed a SERS and fluorescence dual-encoded magnetic nanoprobe for multiplex cancer cell separation.98 Silica-coated magnetic nanobeads (MBs) were used as the inner core. Au core–Ag shell nanorods (Au@Ag NRs) were employed as the SERS generators and attached on the silica-coated MBs. After burying Au@Ag NRs with another silica layer, CdTe QDs were anchored onto the silica layer. Finally, antibodies were covalently linked to CdTe QDs. This method facilitated multiplex, rapid, and accurate cancer cell separation and has great potential in high-throughput analysis and cancer diagnosis.The presence of metallic structures near fluorescent dyes is known to improve photophysical properties. Noble metals such as gold and silver are frequently used to enhance fluorescence signals due to near-field photophysical interactions. Core–shell NPs can be designed to position precisely fluorophore molecules at a distance from the NP surface to maximize fluorescence enhancement and avoid metallic quenching. Meunier et al. developed gold–silver alloy core silica shell nanoparticles (Au/Ag@SiO2 NPs) as the contrast agent for multiplex cell-imaging applications based on their distinctive combination of scattering and fluorescent colours.99 This multiplatform identification strategy was successfully applied for the detection of four different nanoparticular architectures in cell-imaging.
More multimodality such as photo-heat conversion100,101 and photo-electrochemistry102 has enabled the development of innovative methodologies for multiplexed sensing. Photoactive material, such as QDs, which possess unique photophysical properties and regulated optoelectronic characteristics, has taken the development of photoelectrochemical (PEC) techniques with bioanalysis to new heights.103–105 UCNP-based PEC biosensors substitute ultraviolet and visible light source with infrared excitation to maintain strong penetration depth and near-zero photobleaching,106,107 possessing high advantages of cost effectiveness, fast response, high sensitivity, and easy miniaturization for potential applications in multiplexed detection.
Conclusions
This mini review highlights the role of nanomaterial-based optical sensors for multiplexed analysis, and discusses recent representative progress and applications. Although great progress has been made with attractive physicochemical features of nanomaterials, several challenges still remain to be addressed. The majority of the currently proposed methods are still in the proof of concept stage, and only a few are proved to be ready for practical samples. In the light of this, efforts are desperately needed to improve analytical performances such as reproducibility and ease of implementation. The encoding and decoding modality of signals lays the foundation for multiplexing capabilities, and specialist equipment with a multi-wavelength detector and time-gated function is necessary for optical sensors. Multiplex capabilities are also compromised by nonspecific binding and cross-reactivity. Therefore, efforts may also have to be made to reduce the background noise and nonspecific binding by changing the size, shape or composition to optimize the properties of nanomaterials. A portable low-cost multiplex instrument is also indispensable in clinical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21535006 and 21974006).Notes and references
- H. Peter, J. Wienke and F. F. Bier, in Multiplex Biomarker Techniques: Methods and Applications, ed. P. C. Guest, 2017, vol. 1546, ch. 25, pp. 283–294 Search PubMed.
- S. Laing, K. Gracie and K. Faulds, Chem. Soc. Rev., 2016, 45, 1901–1918 RSC.
- L. Cohen and D. R. Walt, Chem. Rev., 2019, 119, 293–321 CrossRef CAS PubMed.
- T. Tsujikawa, S. Kumar, R. N. Borkar, V. Azimi, G. Thibault, Y. H. Chang, A. Balter, R. Kawashima, G. Choe, D. Sauer, E. El Rassi, D. R. Clayburgh, M. F. Kulesz-Martin, E. R. Lutz, L. Zheng, E. M. Jaffee, P. Leyshock, A. A. Margolin, M. Mori, J. W. Gray, P. W. Flint and L. M. Coussens, Cell Rep., 2017, 19, 203–217 CrossRef CAS PubMed.
- X. Fu, L. Chen and J. Choo, Anal. Chem., 2017, 89, 124–137 CrossRef CAS PubMed.
- Y. Han, T. Chen, Y. Li, L. Chen, L. Wei and L. Xiao, Anal. Chem., 2019, 91, 11146–11153 CrossRef CAS PubMed.
- W. Zhou, X. Gao, D. Liu and X. Chen, Chem. Rev., 2015, 115, 10575–10636 CrossRef CAS PubMed.
- R. Gupta, W. J. Peveler, K. Lix and W. R. Algar, Anal. Chem., 2019, 91, 10955–10960 CrossRef CAS PubMed.
- Y. Jiang, Y. Tang and P. Miao, Nanoscale, 2019, 11, 8119–8123 RSC.
- X. Pei, X. Wu, J. Xiong, G. Wang, G. Tao, Y. Ma and N. Li, Analyst, 2020, 145, 3612–3619 RSC.
- G. Tao, T. Lai, X. Xu, Y. Ma, X. Wu, X. Pei, F. Liu and N. Li, Anal. Chem., 2020, 92, 3697–3706 CrossRef CAS PubMed.
- Z. Zhang, S. Shikha, J. Liu, J. Zhang, Q. Mei and Y. Zhang, Anal. Chem., 2019, 91, 548–568 CrossRef CAS PubMed.
- J. Kim, S. J. Park and D. H. Min, Anal. Chem., 2017, 89, 232–248 CrossRef CAS PubMed.
- J. Shen, L. Liang, M. Xiao, X. Xie, F. Wang, Q. Li, Z. Ge, J. Li, J. Shi, L. Wang, L. Li, H. Pei and C. Fan, J. Am. Chem. Soc., 2019, 141, 11938–11946 CrossRef CAS PubMed.
- Y. Fan, S. Wang and F. Zhang, Angew. Chem., Int. Ed., 2019, 58, 13208–13219 CrossRef CAS PubMed.
- L. N. Song, S. Ahn and D. R. Walt, Anal. Chem., 2006, 78, 1023–1033 CrossRef CAS PubMed.
- Y. Leng, K. Sun, X. Chen and W. Li, Chem. Soc. Rev., 2015, 44, 5552–5595 RSC.
- H. H. Gorris and O. S. Wolfbeis, Angew. Chem., Int. Ed., 2013, 52, 3584–3600 CrossRef CAS PubMed.
- H. Zhang, J. Luo, N. Beloglazova, S. Yang, S. De Saeger, G. M. Mari, S. Zhang, J. Shen, Z. Wang and X. Yu, J. Agric. Food Chem., 2019, 67, 6041–6047 CrossRef CAS PubMed.
- Y. Lu, J. Lu, J. Zhao, J. Cusido, F. M. Raymo, J. Yuan, S. Yang, R. C. Leif, Y. Huo, J. A. Piper, J. Paul Robinson, E. M. Goldys and D. Jin, Nat. Commun., 2014, 5, 3741 CrossRef CAS PubMed.
- Y. Fan, P. Wang, Y. Lu, R. Wang, L. Zhou, X. Zheng, X. Li, J. A. Piper and F. Zhang, Nat. Nanotechnol., 2018, 13, 941–946 CrossRef CAS PubMed.
- P. Shah, P. W. Thulstrup, S. K. Cho, Y. J. Bhang, J. C. Ahn, S. W. Choi, M. J. Bjerrum and S. W. Yang, Analyst, 2014, 139, 2158–2166 RSC.
- L. Zheng, P. Qi and D. Zhang, Sens. Actuators, B, 2019, 286, 206–213 CrossRef CAS.
- J.-J. Wang, C. Zheng, Y.-Z. Jiang, Z. Zheng, M. Lin, Y. Lin, Z.-L. Zhang, H. Wang and D.-W. Pang, Anal. Chem., 2020, 92, 830–837 CrossRef CAS PubMed.
- I. M. Khan, S. Niazi, Y. Yu, A. Mohsin, B. S. Mushtaq, M. W. Iqbal, A. Rehman, W. Akhtar and Z. Wang, Anal. Chem., 2019, 91, 14085–14092 CrossRef CAS PubMed.
- S. Wu, N. Duan, Z. Shi, C. Fang and Z. Wang, Anal. Chem., 2014, 86, 3100–3107 CrossRef CAS PubMed.
- S. A. Dunbar, C. A. Vander Zee, K. G. Oliver, K. L. Karem and J. W. Jacobson, J. Microbiol. Methods, 2003, 53, 245–252 CrossRef CAS PubMed.
- J. Hu, C. Y. Wen, Z. L. Zhang, M. Xie, J. Hu, M. Wu and D. W. Pang, Anal. Chem., 2013, 85, 11929–11935 CrossRef CAS PubMed.
- Y. Lu, J. Zhao, R. Zhang, Y. Liu, D. Liu, E. M. Goldys, X. Yang, P. Xi, A. Sunna, J. Lu, Y. Shi, R. C. Leif, Y. Huo, J. Shen, J. A. Piper, J. P. Robinson and D. Jin, Nat. Photonics, 2014, 8, 33–37 Search PubMed.
- J. Lee, P. W. Bisso, R. L. Srinivas, J. J. Kim, A. J. Swiston and P. S. Doyle, Nat. Mater., 2014, 13, 524–529 CrossRef CAS PubMed.
- D. Sun, F. Cao, W. Xu, Q. Chen, W. Shi and S. Xu, Anal. Chem., 2019, 91, 2551–2558 CrossRef CAS PubMed.
- M. Sanchez-Purra, M. Carre-Camps, H. de Puig, I. Bosch, L. Gehrke and K. Hamad-Schifferli, ACS Infect. Dis., 2017, 3, 767–776 CrossRef CAS PubMed.
- W. Zhou, Y. F. Tian, B. C. Yin and B. C. Ye, Anal. Chem., 2017, 89, 6120–6128 CrossRef CAS PubMed.
- S. Jeong, H. Kang, M. G. Cha, S. G. Lee, J. Kim, H. Chang, Y. S. Lee and D. H. Jeong, Chem. Commun., 2019, 55, 2700–2703 RSC.
- S. Kim, J. E. Park, W. Hwang, J. Seo, Y. K. Lee, J. H. Hwang and J. M. Nam, J. Am. Chem. Soc., 2017, 139, 3558–3566 CrossRef CAS PubMed.
- Y. Zhong, X. Tang, J. Li, Q. Lan, L. Min, C. Ren, X. Hu, R. M. Torrente-Rodriguez, W. Gao and Z. Yang, Chem. Commun., 2018, 54, 13813–13816 RSC.
- S. Yue, T. Zhao, S. Bi and Z. Zhang, Biosens. Bioelectron., 2017, 98, 234–239 CrossRef CAS PubMed.
- X. H. Gao, Y. Y. Cui, R. M. Levenson, L. W. K. Chung and S. M. Nie, Nat. Biotechnol., 2004, 22, 969–976 CrossRef CAS PubMed.
- F. Ma, S. Jiang and C.-Y. Zhang, Biosens. Bioelectron., 2020, 157, 112177 CrossRef CAS PubMed.
- D. Geissler, L. J. Charbonniere, R. F. Ziessel, N. G. Butlin, H. G. Lohmannsroben and N. Hildebrandt, Angew. Chem., Int. Ed., 2010, 49, 1396–1401 CrossRef CAS PubMed.
- J. Hu, M.-h. Liu and C.-y. Zhang, ACS Nano, 2019, 13, 7191–7201 CrossRef CAS.
- X. Qiu and N. Hildebrandt, ACS Nano, 2015, 9, 8449–8457 CrossRef CAS PubMed.
- C. Chen, L. Ao, Y. T. Wu, V. Cifliku, M. Cardoso Dos Santos, E. Bourrier, M. Delbianco, D. Parker, J. M. Zwier, L. Huang and N. Hildebrandt, Angew. Chem., Int. Ed., 2018, 57, 13686–13690 CrossRef CAS.
- C. Chen, B. Corry, L. Huang and N. Hildebrandt, J. Am. Chem. Soc., 2019, 141, 11123–11141 CrossRef CAS PubMed.
- K. Huang, N. M. Idris and Y. Zhang, Small, 2016, 12, 836–852 CrossRef CAS.
- X. Teng, Y. Zhu, W. Wei, S. Wang, J. Huang, R. Naccache, W. Hu, A. I. Y. Tok, Y. Han, Q. Zhang, Q. Fan, W. Huang, J. A. Capobianco and L. Huang, J. Am. Chem. Soc., 2012, 134, 8340–8343 CrossRef CAS PubMed.
- L. Zhou, Y. Fan, R. Wang, X. Li, L. Fan and F. Zhang, Angew. Chem., Int. Ed., 2018, 57, 12824–12829 CrossRef CAS PubMed.
- S. O. Kelley, C. A. Mirkin, D. R. Walt, R. F. Ismagilov, M. Toner and E. H. Sargent, Nat. Nanotechnol., 2014, 9, 969–980 CrossRef CAS PubMed.
- Y. Zhang, W. Zhang, K. Zeng, Y. Ao, M. Wang, Z. Yu, F. Qi, W. Yu, H. Mao, L. Tao, C. Zhang, T. T. Y. Tan, X. Yang, K. Pu and S. Gao, Small, 2020, 16, 1906797 CrossRef CAS PubMed.
- X. Lin, Y. Wang, X. Chen, R. Yang, Z. Wang, J. Feng, H. Wang, K. W. C. Lai, J. He, F. Wang and P. Shi, Adv. Healthcare Mater., 2017, 6, 1700446 CrossRef PubMed.
- W. Zhao, L. Wang and W. Tan, in Bio-Applications of Nanoparticles, ed. W. C. W. Chan, 2007, vol. 620, pp. 129–135 Search PubMed.
- X. Pei, H. Yin, T. Lai, J. Zhang, F. Liu, X. Xu and N. Li, Anal. Chem., 2018, 90, 1376–1383 CrossRef CAS PubMed.
- X. Pei, T. Lai, G. Tao, H. Hong, F. Liu and N. Li, Anal. Chem., 2018, 90, 4226–4233 CrossRef CAS PubMed.
- L. Cohen and D. R. Walt, Annu. Rev. Anal. Chem., 2017, 10, 345–363 CrossRef CAS PubMed.
- D. M. Rissin and D. R. Walt, J. Am. Chem. Soc., 2006, 128, 6286–6287 CrossRef CAS.
- D. Wu, M. D. Milutinovic and D. R. Walt, Analyst, 2015, 140, 6277–6282 RSC.
- D. M. Rissin, C. W. Kan, T. G. Campbell, S. C. Howes, D. R. Fournier, L. Song, T. Piech, P. P. Patel, L. Chang, A. J. Rivnak, E. P. Ferrell, J. D. Randall, G. K. Provuncher, D. R. Walt and D. C. Duffy, Nat. Biotechnol., 2010, 28, 595–599 CrossRef CAS PubMed.
- L. Cohen, M. R. Hartman, A. Amardey-Wellington and D. R. Walt, Nucleic Acids Res., 2017, 45, e137 CrossRef CAS PubMed.
- X. Wang, L. Cohen, J. Wang and D. R. Walt, J. Am. Chem. Soc., 2018, 140, 18132–18139 CrossRef CAS PubMed.
- B. K. Duan, P. E. Cavanagh, X. Li and D. R. Walt, Anal. Chem., 2018, 90, 3091–3098 CrossRef CAS PubMed.
- K. Ming, J. Kim, M. J. Biondi, A. Syed, K. Chen, A. Lam, M. Ostrowski, A. Rebbapragada, J. J. Feld and W. C. W. Chan, ACS Nano, 2015, 9, 3060–3074 CrossRef CAS PubMed.
- D. Li, Y. Wang, C. Lau and J. Lu, Anal. Chem., 2014, 86, 10148–10156 CrossRef CAS PubMed.
- P. Carl, D. Sarma, B. J. R. Gregorio, K. Hoffmann, A. Lehmann, K. Rurack and R. J. Schneider, Anal. Chem., 2019, 91, 12988–12996 CrossRef CAS PubMed.
- C. Parisi, A. Markou, A. Strati, S. Kasimir-Bauer and E. S. Lianidou, Anal. Chem., 2019, 91, 3443–3451 CrossRef CAS PubMed.
- M. Y. Han, X. H. Gao, J. Z. Su and S. Nie, Nat. Biotechnol., 2001, 19, 631–635 CrossRef CAS PubMed.
- D. C. Pregibon, M. Toner and P. S. Doyle, Science, 2007, 315, 1393–1396 CrossRef CAS PubMed.
- H. Lee, S. J. Shapiro, S. C. Chapin and P. S. Doyle, Anal. Chem., 2016, 88, 3075–3081 CrossRef CAS PubMed.
- M. B. Nagarajan, A. M. Tentori, W. C. Zhang, F. J. Slack and P. S. Doyle, Anal. Chem., 2018, 90, 10279–10285 CrossRef CAS PubMed.
- M. Fleischmann, P. J. Hendra and A. J. McQuillan, Chem. Phys. Lett., 1974, 26, 163–166 CrossRef CAS.
- Z. Wang, S. Zong, L. Wu, D. Zhu and Y. Cui, Chem. Rev., 2017, 117, 7910–7963 CrossRef CAS PubMed.
- D. Sun, F. H. Cao, W. Q. Xu, Q. D. Chen, W. Shi and S. P. Xu, Anal. Chem., 2019, 91, 2551–2558 CrossRef CAS PubMed.
- A. McLintock, C. A. Cunha-Matos, M. Zagnoni, O. R. Millington and A. W. Wark, ACS Nano, 2014, 8, 8600–8609 CrossRef CAS PubMed.
- J. Wu, L. Zhang, F. Huang, X. Ji, H. Dai and W. Wu, J. Hazard. Mater., 2020, 387, 121714 CrossRef CAS PubMed.
- H. N. Wang, B. M. Crawford, A. M. Fales, M. L. Bowie, V. L. Seewaldt and T. Vo-Dinh, J. Phys. Chem. C, 2016, 120, 21047–21050 CrossRef CAS.
- S. M. Yoo, T. Kang, H. Kang, H. Lee, M. Kang, S. Y. Lee and B. Kim, Small, 2011, 7, 3371–3376 CrossRef CAS.
- B. Zhao, J. Shen, S. Chen, D. Wang, F. Li, S. Mathur, S. Song and C. Fan, Chem. Sci., 2014, 5, 4460–4466 RSC.
- J. Su, D. Wang, L. Noerbel, J. Shen, Z. Zhao, Y. Dou, T. Peng, J. Shi, S. Mathur, C. Fan and S. Song, Anal. Chem., 2017, 89, 2531–2538 CrossRef CAS PubMed.
- L. Xu, W. Yan, W. Ma, H. Kuang, X. Wu, L. Liu, Y. Zhao, L. Wang and C. Xu, Adv. Mater., 2015, 27, 1706–1711 CrossRef CAS PubMed.
- X. Guo, X. Wu, M. Sun, L. Xu, H. Kuang and C. Xu, Anal. Chem., 2020, 92, 2310–2315 CrossRef CAS PubMed.
- J. Wang, D. Liang, J. Feng and X. Tang, Anal. Chem., 2019, 91, 11045–11054 CrossRef CAS PubMed.
- X. R. Bai, L. H. Wang, J. Q. Ren, X. W. Bai, L. W. Zeng, A. G. Shen and J. M. Hu, Anal. Chem., 2019, 91, 2955–2963 CrossRef CAS PubMed.
- M.-Y. Gao, Q. Chen, W. Li, A. G. Shen and J. M. Hu, Anal. Chem., 2019, 91, 13866–13873 CrossRef CAS PubMed.
- T. A. Taton, C. A. Mirkin and R. L. Letsinger, Science, 2000, 289, 1757–1760 CrossRef CAS PubMed.
- A. H. Alhasan, D. Y. Kim, W. L. Daniel, E. Watson, J. J. Meeks, C. S. Thaxton and C. A. Mirkin, Anal. Chem., 2012, 84, 4153–4160 CrossRef CAS PubMed.
- Q.-Y. Lin, E. Palacios, W. Zhou, Z. Li, J. A. Mason, Z. Liu, H. Lin, P.-C. Chen, V. P. Dravid, K. Aydin and C. A. Mirkin, Nano Lett., 2018, 18, 2645–2649 CrossRef CAS PubMed.
- Y. Y. Wu, P. Huang and F. Y. Wu, Food Chem., 2020, 304, 125377 CrossRef CAS PubMed.
- J. H. Heo, G. S. Yi, B. S. Lee, H. H. Cho, J. W. Lee and J. H. Lee, Nanoscale, 2016, 8, 18341–18351 RSC.
- J. Ma, L. Zhan, R. S. Li, P. F. Gao and C. Z. Huang, Anal. Chem., 2017, 89, 8484–8489 CrossRef CAS PubMed.
- G. Li, L. Zhu, Y. He, H. Tan and S. Sun, Anal. Bioanal. Chem., 2017, 409, 3657–3666 CrossRef CAS PubMed.
- Y. Wu, M. R. K. Ali, K. Dansby and M. A. El-Sayed, Anal. Chem., 2019, 91, 14261–14267 CrossRef CAS PubMed.
- N. Siraj, B. El-Zahab, S. Hamdan, T. E. Karam, L. H. Haber, M. Li, S. O. Fakayode, S. Das, B. Valle, R. M. Strongin, G. Patonay, H. O. Sintim, G. A. Baker, A. Powe, M. Lowry, J. O. Karolin, C. D. Geddes and I. M. Warner, Anal. Chem., 2016, 88, 170–202 CrossRef CAS PubMed.
- Y. Chen, J. Sun, Y. Xianyu, B. Yin, Y. Niu, S. Wang, F. Cao, X. Zhang, Y. Wang and X. Jiang, Nanoscale, 2016, 8, 15205–15212 RSC.
- C. Zong, J. Wu, J. Xu, H. Ju and F. Yan, Biosens. Bioelectron., 2013, 43, 372–378 CrossRef CAS PubMed.
- F. Li, L. Guo, Y. Hu, Z. Li, J. Liu, J. He and H. Cui, Talanta, 2020, 207, 120346 CrossRef CAS PubMed.
- G. Guan, L. Yang, Q. Mei, K. Zhang, Z. Zhang and M.-Y. Han, Anal. Chem., 2012, 84, 9492–9497 CrossRef CAS PubMed.
- S. Zong, Z. Wang, R. Zhang, C. Wang, S. Xu and Y. Cui, Biosens. Bioelectron., 2013, 41, 745–751 CrossRef CAS PubMed.
- Z. Wang, S. Zong, W. Li, C. Wang, S. Xu, H. Chen and Y. Cui, J. Am. Chem. Soc., 2012, 134, 2993–3000 CrossRef CAS PubMed.
- Z. Wang, S. Zong, H. Chen, C. Wang, S. Xu and Y. Cui, Adv. Healthcare Mater., 2014, 3, 1889–1897 CrossRef CAS PubMed.
- S. Tu, D. Rioux, J. Perreault, D. Brouard and M. Meunier, J. Phys. Chem. C, 2017, 121, 8944–8951 CrossRef CAS.
- G. Cai, Z. Yu, P. Tong and D. Tang, Nanoscale, 2019, 11, 15659–15667 RSC.
- L. Huang, J. Chen, Z. Yu and D. Tang, Anal. Chem., 2020, 92, 2809–2814 CrossRef CAS PubMed.
- J. Shu and D. Tang, Anal. Chem., 2020, 92, 363–377 CrossRef CAS PubMed.
- J. Shu and D. Tang, Chem. – Asian J., 2017, 12, 2780–2789 CrossRef CAS PubMed.
- G. Cai, Z. Yu, R. Ren and D. Tang, ACS Sens., 2018, 3, 632–639 CrossRef CAS PubMed.
- Z. Qiu, J. Shu and D. Tang, Anal. Chem., 2017, 89, 5152–5160 CrossRef CAS PubMed.
- S. Lv, K. Zhang, L. Zhu and D. Tang, Anal. Chem., 2020, 92, 1470–1476 CrossRef CAS PubMed.
- Z. Luo, Q. Qi, L. Zhang, R. Zeng, L. Su and D. Tang, Anal. Chem., 2019, 91, 4149–4156 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |






