Open Access Article
Open Access ArticleFunctional comparison of paper-based immunoassays based on antibodies and engineered binding proteins†
Ki-Joo
Sung
 a,
Yara
Jabbour Al Maalouf
a,
Quinlan R.
Johns
a,
Eric A.
Miller
a,
Yara
Jabbour Al Maalouf
a,
Quinlan R.
Johns
a,
Eric A.
Miller
 a and
Hadley D.
Sikes
a and
Hadley D.
Sikes
 *ab
*ab
aDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA. E-mail: sikes@mit.edu
bSingapore-MIT Alliance for Research and Technology Centre (SMART), 1 CREATE Way, Singapore 138602
First published on 9th March 2020
Abstract
Binding protein scaffolds, such as rcSso7d, have been investigated for use in diagnostic tests; however, the functional performance of rcSso7d has not yet been studied in comparison to antibodies. Here, we assessed the analyte-binding capabilities of rcSso7d and antibodies on cellulose with samples in buffer and 100% human serum.
Zika virus (ZIKV) is a mosquito-borne flavivirus that recently emerged as a global public health issue after being linked to neurological disorders, including microcephaly in infants and Guillain-Barré syndrome in adults.1,2 Rapid, accurate diagnosis of ZIKV is vital to track, control, and prevent the spread of ZIKV. Since clinical symptoms and diagnostic biomarkers for ZIKV are similar to those of other flaviviruses—such as Dengue virus (DENV)—diagnostic tests must minimize cross-reactivity and false-positive results.1–4 The nonstructural protein 1 (NS1) of Zika virus (ZNS1) has been identified as a promising biomarker for ZIKV diagnostics.3–12 Although ZNS1 has similar structure and sequence as other flavivirus NS1 proteins, studies have shown that specific detection of ZIKV is possible using ZNS1-based diagnostics.3–12
The World Health Organization introduced the ASSURED criteria for ideal characteristics of diagnostic tests: Affordable, Sensitive, Specific, User-friendly, Rapid and Robust, Equipment-free, and Delivered to those who need it.13 To address the ASSURED criteria, sandwich immunoassays are often used in diagnostic tests to capture and detect specific biomarkers in patient samples. This format requires a pair of affinity reagents: one is surface-immobilized to capture the target biomarker, and another is labeled to associate a signal to the captured biomarker. Antibodies have been commonly used as the affinity reagents in diagnostic tests; however, in recent years, alternative scaffolds have been investigated for use in diagnostic tests due to their desirable characteristics such as intrinsic thermal stability and ease of production.14,15 In recent years, studies have begun reporting the use of non-antibody scaffolds in full sandwich assays using pairs of nanobodies,16–19 affimers,20,21 DARPins,22 and the reduced-charge Sso7d variant (rcSso7d).23–31 However, a direct functional comparison of antibodies and non-antibody scaffolds in an identical full sandwich immunoassay test format has yet to be conducted.
Here, we investigated the functionality of rcSso7d clones engineered against ZNS1 in a full immunoassay format. We compared the binding capabilities of an rcSso7d-based sandwich assay to antibody-based sandwich assays and rcSso7d/antibody hybrid assays in a cellulose paper-based format. For one hybrid assay, we developed a protein A fusion with a cellulose-binding domain (CBD) to immobilize antibodies on cellulose test strips without requiring chemical functionalization of the cellulose or subsequent bioconjugation reactions. We found that the rcSso7d-based assay had similar limits of detection (LOD) as antibody-based assays. Furthermore, compared to antibody-based assays, the rcSso7d full sandwich assay demonstrated greater improvement in sensitivity when a larger sample volume was applied. Both the rcSso7d full sandwich and an antibody/rcSso7d hybrid assays performed equally well in 100% human serum and in buffer, signifying that rcSso7d is a promising alternative scaffold for use in clinical diagnostic tests.
Results and discussion
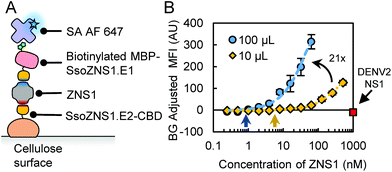
For this study, we used rcSso7d clones previously engineered to bind specifically to ZNS1 (SsoZNS1.E1 and SsoZNS1.E2; E1: binding to epitope 1, E2: binding to epitope 2) with minimal cross-reactivity to a similar non-target biomarker, Dengue-2 virus NS1 (D2NS1).31 To integrate the rcSso7d clones into a paper assay format, we incorporated the clones into genetic fusion constructs that improve their activity as capture and reporter agents. Prior studies required chemical oxidation of cellulose and overnight incubation of the capture proteins on the test zones for covalent immobilization.32 However, we found that a cellulose-binding domain (CBD) fusion construct achieves non-covalent, high-density immobilization of rcSso7d (SsoZNS1.E2-CBD) to cellulose without requiring surface functionalization or overnight incubation.28,29 For the reporter construct, we used the in vivo biotinylated maltose-binding protein (MBP) fusion to associate a biotin moiety with rcSso7d for detection (b-MBP-SsoZNS1.E1).30 A streptavidin–fluorophore conjugate (SA AF647) was used to associate a fluorescent signal to the biotinylated reporter protein.To assess the performance of an rcSso7d-based assay, we incorporated the rcSso7d clones into a full sandwich assay format (Fig. 1A). We tested a range of ZNS1 concentrations in buffer (PBSA: 1× PBS with 1% bovine serum albumin) by incubating 10 μL of the samples for 30 minutes, following conditions established previously27,29–31 (Fig. 1B, yellow diamonds). Since cross-reactivity with other flavivirus NS1 variants can lead to false positive results, we also challenged the system with 1 μM of D2NS1 and confirmed that the engineered rcSso7d clones demonstrated no cross-reactivity this off-target NS1 variant in a cellulose assay format (Fig. 1B, red square).
We also investigated the effect of applying a larger sample volume on assay sensitivity by incrementally applying 100 μL of ZNS1 sample solution over 30 minutes (Fig. 1B, blue circles).33 Previous studies suggest that larger sample volumes can increase sensitivity and reduce the LOD of diagnostic tests.29,33 To quantify the performance of these rcSso7d-based sandwich assays, we conducted quadratic regression on each titration curve (Fig. 1B and S1†) and calculated LODs of 6.1 nM (10 μL) and 0.9 nM (100 μL) (Table 1; Fig. 1B, arrows). We also conducted linear regression on the linear range of the titration curves and compared their slopes to assess the sensitivity34 of the two sample volumes (Fig. S2†) and found approximately a 21-fold improvement in sensitivity by increasing the sample volume 10-fold (Table 1; Fig. 1B).
| Type | LOD, 10 μL (nM) | LOD, 100 μL (nM) | Sensitivity improvement (10 μL to 100 μL) |
|---|---|---|---|
| Sso-CBD + BA-MBP-Sso | 6.1 ± 1.3 | 0.9 ± 0.1 | 21.3 ± 0.3 |
| MouseAb + bAb | 45.0 ± 5.4 | 1.2 ± 0.2 | 8.1 ± 0.2 |
| RabbitAb + bAb | 9.6 ± 0.3 | 0.1 ± 0.1 | 12.2 ± 0.3 |
| ProA-CBD/rabbitAb + BA-MBP-Sso | 21.6 ± 8.4 | 0.9 ± 0.3 | 12.9 ± 0.2 |
| Sso-CBD + bAb | 5.5 ± 3.2 | 0.6 ± 0.2 | 9.4 ± 0.1 |
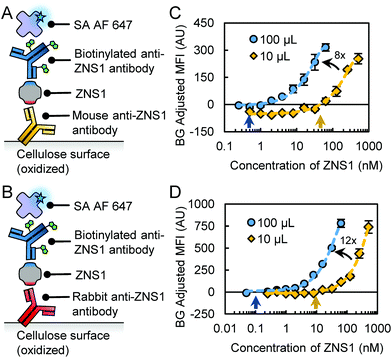
To compare the functionality of the engineered rcSso7d clones to antibodies, we tested commercial anti-ZNS1 antibodies in a cellulose assay format. We assessed two different commercial antibodies for capture: a mouse anti-ZNS1 IgG1 antibody (“mouseAb”; Abcam) and a rabbit anti-ZNS1 IgG antibody (“rabbitAb”; GeneTex) (Fig. 2A and B). Since the antibodies do not intrinsically bind to non-functionalized cellulose fibers, we used oxidized cellulose to covalently immobilize the capture antibodies as described previously.32 We used a biotinylated mouse anti-ZNS1 IgG2a antibody (“bAb”; Arigo Biolaboratories) as the reporter reagent.
After conducting titrations of the ZNS1 biomarker with two different sample volumes (Fig. 2C and D), we found that assay functionality varied depending on the antibodies used in the assay. The mouseAb system had higher background signal, leading to a higher LOD of 45 nM (10 μL) (Table 1; Fig. 2C and S1†). For 100 μL samples, the LOD was more comparable to the rcSso7d assay at 1.2 nM (Table 1; Fig. 2C). The rabbitAb system had improved performance over the mouseAb system, with LODs of 9.6 nM (10 μL) and 0.1 nM (100 μL) (Table 1; Fig. 2D and S1†). Both antibody assays had an 8- to 12-fold sensitivity increase with a 10-fold volume increase (Table 1; Fig. 2C, D and S2†), signifying that excess immobilized antibody molecules were still available for biomarker binding.
Based on these results, the rcSso7d assays perform similarly to antibody assays. However, several intrinsic differences in the assay formats should be noted. The rcSso7d sandwich used non-functionalized cellulose with a CBD-fusion protein and a singly biotinylated rcSso7d as the reporter. In contrast, the antibody sandwiches used oxidized cellulose for immobilization and a bivalent antibody with multiple conjugated biotin moieties as the reporter. These format differences complicate the comparison of diagnostic performance for these binding proteins.
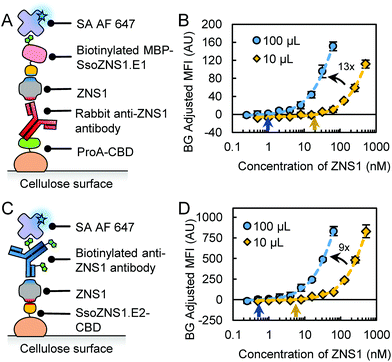
In order to draw a closer comparison between rcSso7d and antibody, we devised a different method to immobilize antibodies on cellulose. Since protein A binds to antibodies, it can be used for the immobilization of antibodies to a surface.35–37 We constructed a fusion protein with protein A and CBD (“ProA-CBD”) to immobilize ProA in high density on non-functionalized cellulose surfaces and bind to compatible capture antibodies (Fig. 3A).35 We used rabbitAb for capture due to the high affinity of protein A to rabbit IgG.38 We also used b-MBP-SsoZNS1.E2 as the reporter species to address the multivalency effects observed with bAb and to minimize non-specific interactions between ProA and an antibody-based reporter molecule. Furthermore, using the ProA-CBD construct, we can reduce the manufacturing and processing burden required from using oxidized cellulose for protein immobilization.
We demonstrated the function of ProA-CBD/Ab in cellulose assays by conducting biomarker titrations and determined LODs of 21.6 nM (10 μL) and 0.9 nM (100 μL) (Table 1; Fig. 3B and S1†). These values are higher than for the antibody full sandwich using rabbitAb, which may be due to different binding interactions (1-step covalent immobilization of antibody vs. 2-step non-covalent immobilization of antibody via protein A binding). Additionally, the use of b-MBP-SsoZNS1.E2 instead of bAb removed multivalency effects that may impact the LOD. However, this format draws a more equitable comparison to the rcSso7d full sandwich by using CBD for immobilization. Compared to the rcSso7d full sandwich, the LODs are similar or slightly higher. The increase in sample volume also led to a 13-fold increase in sensitivity (Table 1; Fig. 3B and S2†).
We tested another hybrid system with rcSso7d-CBD as the capture reagent and bAb as the reporter reagent (Fig. 3C). In this format, we determined LODs of 5.5 nM (10 μL) and 0.6 nM (100 μL) (Table 1; Fig. S1† and Fig. 3D), which were comparable to the LOD in the rcSso7d full sandwich assay. A 10-fold increase in sample volume also led to a 9-fold increase in sensitivity (Table 1; Fig. 3D and S2†). Using the multivalently-labeled bAb as a reporter improved the sensitivity 4-fold (10 μL) and 2-fold (100 μL) relative to the singly biotinylated b-MBP-rcSso7d in the rcSso7d full sandwich assay (Fig. S2†). This increase in sensitivity may be attributed to the multivalency of the biotinylated antibody.
In all of the assay formats, the larger sample volume improved both the LOD and sensitivity of the assays. These results suggest that the amount of free capture molecules are in excess to the biomarker molecules in solution. By introducing a greater molar quantity of biomarker molecules into the assay via a larger sample volume, the capture reagents can continue to bind to free target molecules. Furthermore, the rcSso7d full sandwich assay had a greater increase in sensitivity (21-fold) compared to the antibody full sandwich or hybrid assays (8- to 13-fold). We hypothesize that rcSso7d-CBD is immobilized in higher density than antibodies on oxidized cellulose or antibodies via ProA-CBD; therefore, higher excess of free capture molecules may allow for more efficient capture of target molecules from the sample.
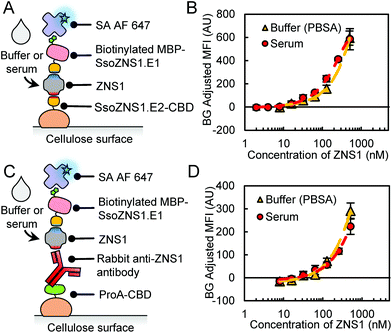
The above studies were all conducted with the ZNS1 biomarker spiked into buffer. Unfortunately, many assays have reduced function in human serum due to matrix effects and often require dilution of the serum sample in buffer to achieve reasonable performance.7,9,12,39 To assess functionality of rcSso7d in a relevant bodily fluid, we conducted a side-by-side comparison titration with ZNS1 spiked into either PBSA or 100% human serum (Fig. 4A). The rcSso7d clones performed equally well in human serum as in buffer (Fig. 4B; Table 2). We also assessed the performance of the ProA-CBD/Ab hybrid assay in serum (Fig. 4C) and found that it retained function in human serum as well (Fig. 4D; Table 2). Both of these assays maintained functionality in 100% human serum with minimal matrix effects.
| Type | LOD, buffer (nM) | LOD, serum (nM) |
|---|---|---|
| Sso-CBD + BA-MBP-Sso | 6.1 ± 1.3 | 3.0 ± 1.8 |
| ProA-CBD/rabbitAb + BA-MBP-Sso | 21.6 ± 8.4 | 19.6 ± 2.7 |
Conclusions
In conclusion, engineered rcSso7d clones against ZNS1 showed comparable functionality to antibodies in cellulose-based assays. We found that, in almost all instances, the LOD of the rcSso7d-based assay was similar to or better than antibody-based assays. Furthermore, larger sample volumes provided greater improvement in the sensitivity of the rcSso7d sandwich assay compared to the antibody-based and hybrid assays. We also developed a method to immobilize antibodies on non-functionalized cellulose surfaces via a protein A fused with a cellulose-binding domain. This construct reduced processing time compared to using oxidized cellulose and can be used to develop hybrid assays when a well-validated capture antibody is available. Finally, the rcSso7d sandwich and ProA-CBD hybrid assays functioned equally well in buffer and 100% human serum without a reduction in LOD as previously reported in other assays using antibodies or aptamers.7,9,12rcSso7d affinity reagents have been demonstrated to yield similar diagnostic performance as antibodies, with the added benefits of thermal stability, inexpensive production, and facile incorporation of new properties via fusion proteins. New rcSso7d variants can also be generated against other target disease biomarkers via straightforward in vitro development processes.31 These non-antibody proteins may be used in rapid diagnostic tests for diseases such as ZIKV. Due to the lack of interest in ZIKV until recent years, the clinically relevant concentration of ZNS1 in patient samples is still largely unknown. However, studies suggest NS1 concentrations in ZIKV patients are lower than in DENV patients,40 which is reported at levels from the high picomolar range to the high micromolar range.41,42 Future work will focus on investigating signal amplification methods to match the LOD of these rcSso7d-based immunoassays to clinically relevant levels. The present finding that rcSso7d-based assays can yield equivalent diagnostic performance to antibody-based assays suggest that rcSso7d can be employed as a promising alternative scaffold to develop rapid diagnostic tests that meet the WHO ASSURED criteria.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
KS acknowledges NSF GRFP (1122374). This work was supported by the Singapore-MIT Alliance for Research and Technology (AMR IRG), MIT Deshpande Center for Technological Innovation, and MIT Tata Center for Technology and Design. YJAM and QRJ acknowledge support from MIT's UROP Program. HDS acknowledges the Esther and Harold E. Edgerton chair at MIT. Authors thank Emma Yee for valuable discussions.Notes and references
- C. Zanluca, C. N. Duarte and D. Santos, Microbes Infect., 2016, 18, 295–301 CrossRef PubMed.
- D. Musso and D. J. Gubler, Clin. Microbiol. Rev., 2016, 29, 487–524 CrossRef CAS PubMed.
- H. Song, J. Qi, J. Haywood and Y. Shi, Nat. Struct. Mol. Biol., 2016, 23, 456–458 CrossRef CAS PubMed.
- A. Balmaseda, K. Stettler, R. Medialdea-Carrera, D. Collado, X. Jin, J. V. Zambrana, S. Jaconi, E. Cameroni, S. Saborio, F. Rovida, E. Percivalle, S. Ijaz, S. Dicks, I. Ushiro-Lumb, L. Barzon, P. Siqueira, D. W. G. Brown, F. Baldanti, R. Tedder, M. Zambon, A. M. Bispo de Filippis, E. Harris and D. Corti, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 8384–8389 CrossRef CAS PubMed.
- F. Bedin, L. Boulet, E. Voilin, G. Theillet, A. Rubens and C. Rozand, J. Med. Virol., 2017, 9999, 1–8 Search PubMed.
- I. Bosch, H. De Puig, M. Hiley, M. Carré-Camps, F. Perdomo-Celis, C. F. Narváez, D. M. Salgado, D. Senthoor, M. O. Grady, E. Phillips, A. Durbin, D. Fandos, H. Miyazaki, C. W. Yen, M. Gélvez-Ramírez, R. V. Warke, L. S. Ribeiro, M. M. Teixeira, R. P. Almeida, J. E. Muñóz-Medina, J. E. Ludert, M. L. Nogueira, T. E. Colombo, A. C. B. Terzian, P. T. Bozza, A. S. Calheiros, Y. R. Vieira, G. Barbosa-Lima, A. Vizzoni, J. Cerbino-Neto, F. A. Bozza, T. M. L. Souza, M. R. O. Trugilho, A. M. B. De Filippis, P. C. De Sequeira, E. T. A. Marques, T. Magalhaes, F. J. Díaz, B. N. Restrepo, K. Marín, S. Mattar, D. Olson, E. J. Asturias, M. Lucera, M. Singla, G. R. Medigeshi, N. De Bosch, J. Tam, J. Gómez-Márquez, C. Clavet, L. Villar, K. Hamad-Schifferli and L. Gehrke, Sci. Transl. Med., 2017, 9, eaan1589 CrossRef PubMed.
- K. H. Lee and H. Zeng, Anal. Chem., 2017, 89, 12743–12748 CrossRef CAS PubMed.
- L. Zhang, X. Du, C. Chen, Z. Chen, L. Zhang, Q. Han, X. Xia, Y. Song and J. Zhang, Viruses, 2018, 10, 1–12 Search PubMed.
- Z. Rong, Q. Wang, N. Sun, X. Jia, K. Wang, R. Xiao and S. Wang, Anal. Chim. Acta, 2019, 1055, 140–147 CrossRef CAS PubMed.
- M. Sánchez-Purrà, M. Carré-Camps, H. De Puig, I. Bosch, L. Gehrke and K. Hamad-Schifferli, ACS Infect. Dis., 2017, 3, 767–776 CrossRef PubMed.
- S. A. Camacho, R. G. Sobral-Filho, P. H. B. Aoki, C. J. L. Constantino and A. G. Brolo, ACS Sens., 2018, 3, 587–594 CrossRef CAS PubMed.
- S. Afsahi, M. B. Lerner, J. M. Goldstein, J. Lee, X. Tang, D. A. Bagarozzi, D. Pan, L. Locascio, A. Walker, F. Barron and B. R. Goldsmith, Biosens. Bioelectron., 2018, 100, 85–88 CrossRef CAS PubMed.
- D. Mabey, R. W. Peeling, A. Ustianowski and M. D. Perkins, Nat. Rev. Microbiol., 2004, 2, 231–240 CrossRef CAS PubMed.
- M. Thaler and P. B. Luppa, Anal. Bioanal. Chem., 2019, 411, 7623–7635 CrossRef CAS PubMed.
- S. Banta, K. Dooley and O. Shur, Annu. Rev. Biomed. Eng., 2013, 15, 93–113 CrossRef CAS PubMed.
- T. Li, S. L. Li, C. Fang, Y. N. Hou, Q. Zhang, X. Du, H. C. Lee and Y. J. Zhao, Anal. Chim. Acta, 2018, 1029, 65–71 CrossRef CAS PubMed.
- J. E. Pinto Torres, J. Goossens, J. Ding, Z. Li, S. Lu, D. Vertommen, P. Naniima, R. Chen, S. Muyldermans, Y. G. J. Sterckx and S. Magez, Sci. Rep., 2018, 8, 9019 CrossRef PubMed.
- L. Xu, H. Cao, C. Huang and L. Jia, Molecules, 2019, 24, 1890 CrossRef CAS PubMed.
- M. Zhu, X. Gong, Y. Hu, W. Ou and Y. Wan, J. Transl. Med., 2014, 12, 352 CrossRef PubMed.
- S. Straw, P. K. Ferrigno, Q. Song, D. Tomlinson and F. Del Galdo, J. Biomed. Sci. Eng., 2013, 6, 32–42 CrossRef.
- E. L. Hesketh, C. Tiede, H. Adamson, T. L. Adams, M. J. Byrne, Y. Meshcheriakova, I. Kruse, M. J. McPherson, G. P. Lomonossoff, D. C. Tomlinson and N. A. Ranson, Sci. Rep., 2019, 9, 7524 CrossRef PubMed.
- S. Gupta and V. Kakkar, Tuberculosis, 2019, 118, 101852 CrossRef CAS PubMed.
- N. Gera, M. Hussain, R. C. Wright and B. M. Rao, J. Mol. Biol., 2011, 409, 601–616 CrossRef CAS PubMed.
- M. W. Traxlmayr, J. D. Kiefer, R. R. Srinivas, E. Lobner, A. W. Tisdale, N. K. Mehta, N. J. Yang, B. Tidor and K. D. Wittrup, J. Biol. Chem., 2016, 291, 22496–22508 CrossRef CAS PubMed.
- N. Zhao, M. A. Schmitt and J. D. Fisk, FEBS J., 2016, 283, 1351–1367 CrossRef CAS PubMed.
- N. Zhao, J. Spencer, M. A. Schmitt and J. D. Fisk, Anal. Biochem., 2017, 521, 59–71 CrossRef CAS PubMed.
- E. A. Miller, M. W. Traxlmayr, J. Shen and H. D. Sikes, Mol. Syst. Des. Eng., 2016, 1, 377–381 RSC.
- E. A. Miller, S. Baniya, D. Osorio, Y. J. Al Maalouf and H. D. Sikes, Biosens. Bioelectron., 2018, 102, 456–463 CrossRef CAS PubMed.
- E. A. Miller, Y. Jabbour Al Maalouf and H. D. Sikes, Anal. Chem., 2018, 90, 9472–9479 CrossRef CAS PubMed.
- K.-J. Sung, E. A. Miller and H. D. Sikes, Mol. Syst. Des. Eng., 2018, 3, 877–882 RSC.
- E. A. Miller, K.-J. Sung, P. Kongsuphol, S. Baniya, H. Q. Aw-yong, V. Tay, Y. Tan, F. M. Kabir, K. Pang-yeo, I. G. Kaspriskie and H. D. Sikes, ACS Comb. Sci., 2020, 22, 49–60 CrossRef CAS PubMed.
- A. K. Badu-Tawiah, S. Lathwal, K. Kaastrup, M. Al-Sayah, D. C. Christodouleas, B. S. Smith, G. M. Whitesides and H. D. Sikes, Lab Chip, 2015, 15, 655–659 RSC.
- S. Kim and H. D. Sikes, ACS Appl. Mater. Interfaces, 2019, 11, 28469–28477 CrossRef PubMed.
- Y. Wu, R. D. Tilley and J. J. Gooding, J. Am. Chem. Soc., 2019, 141, 1162–1170 CrossRef CAS PubMed.
- E. Shpigel, A. Goldlust, A. Eshel, I. K. Ber, G. Efroni, Y. Singer, I. Levy, M. Dekel and O. Shoseyov, Biotechnol. Appl. Biochem., 2000, 31, 197 CrossRef CAS PubMed.
- I. Tsarfati-BarAd, K. Gier, U. Sauer and L. A. Gheber, Sens. Actuators, B, 2019, 284, 289–295 CrossRef CAS.
- M. Iijima and S. Kuroda, Biosens. Bioelectron., 2017, 89, 810–821 CrossRef CAS PubMed.
- D. D. Richman, P. H. Cleveland, M. N. Oxman and K. M. Johnson, J. Immunol., 1982, 128, 2300–2305 CAS.
- D. C. Pawley, M. J. Ricciardi, E. Dikici, S. K. Deo and S. Daunert, ACS Omega, 2019, 4, 6808–6818 CrossRef CAS PubMed.
- J. J. Waggoner, L. Gresh, M. J. Vargas, G. Ballesteros, Y. Tellez, K. J. Soda, M. K. Sahoo, A. Nuñez, A. Balmaseda, E. Harris and B. A. Pinsky, Clin. Infect. Dis., 2016, 63, 1584–1590 CrossRef PubMed.
- S. Alcon, A. Talarmin, M. Debruyne, A. Falconar, V. Deubel and M. Flamand, J. Clin. Microbiol., 2002, 40, 376–381 CrossRef CAS PubMed.
- P. R. Young, P. A. Hilditch, C. Bletchly and W. Halloran, J. Clin. Microbiol., 2000, 38, 1053–1057 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0an00299b |
| This journal is © The Royal Society of Chemistry 2020 |