Mitochondria- and nucleolus-targeted copper(I) complexes with pyrazole-linked triphenylphosphine moieties for live cell imaging†
Mayank‡
a,
Richa
Rani‡
a,
Ashutosh
Singh‡
b,
Neha
Garg
*b,
Navneet
Kaur
 *c and
Narinder
Singh
*c and
Narinder
Singh
 *a
*a
aDepartment of Chemistry, Indian Institute of Technology Ropar, India. E-mail: nsingh@iitrpr.ac.in
bSchool of Basic Sciences, Indian Institute of Technology Mandi, India. E-mail: neha@iitmandi.ac.in
cDepartment of Chemistry, Punjab University Chandigarh, India. E-mail: navneetkaur@pu.ac.in
First published on 21st October 2019
Abstract
The labelling and imaging of mitochondria and nucleolus have attracted great attention because of the involvement of these cellular organelles in critical cellular activities. Therefore, a large number of mitochondria- or nucleolus-labelling probes have been developed throughout the world. However, in the current study, we successfully developed two pyrazole-based, copper-linked triphenylphosphine-coupled emissive metallo-complexes (C1 and C2) for the simultaneous visualization of mitochondria and nucleolus in a single run. These complexes were very inexpensive and could be synthesized by a simple one-pot multicomponent reaction scheme. The complexes were very specific, and only a small concentration of 5 μM was found to be sufficient to probe both the organelles efficiently. Additionally, even under a shorter incubation period (half hour), the fluorescence intensity from the cells was appreciable. Also, both the compounds were found to be photostable when torched with 10% of a 100 mW laser for up to 10 min. All these results indicate that both the complexes may contribute towards the future development of cell imaging tools. To the best of our knowledge, this is the first report on the development of multifunctional live cell imaging tools for simultaneous mitochondria and nucleolus imaging and within the shortest incubation time of about 30 minutes.
Introduction
Mitochondria, which are membrane-bound cellular organelles, are responsible for synthesizing ATPs within the cells.1 They take part in regulating the membrane potential and also coordinate the apoptosis-programmed cell death phenomenon.2,3 A “disturbed mitochondrial membrane” is an indicating factor of abnormalities that may include malignancies and neurodegenerative diseases.4 The visualization of mitochondrial morphology by live-cell imaging techniques is therefore considered important for the diagnosis of various diseased conditions under the cellular level.5 Therefore, various types of mitochondria-selective fluorescent probes have been developed and used successfully to observe the morphology of mitochondria via cell imaging techniques. However, the photo-bleaching of these probes under continuous light irradiation is a major drawback associated with these entities.6The nucleolus is yet another subcellular structure that is present within the nucleus of the eukaryotic cells. The process of ribosome synthesis occurs within this organelle.7 The activities within the nucleolus are critical for the development, growth, and proliferation of living cells.8 Thus, along with mitochondria, monitoring the morphology of the nucleolus is also important for distinguishing the cells under normal and diseased states.8–10 STRO RNASelect is a commercially available fluorescent probe that is applied commonly to visualize the morphological changes within the nucleolus of the cellular system.6 Working with this probe is not easy because of its very specific storage conditions.6 Therefore, serious efforts are still ongoing to develop an ideal fluorescent probe for visualizing the nucleolus within living cells. The nucleolus and mitochondria are the sites for rRNA synthesis. Also, the rRNA transcription processes of both the organelles are controlled by a group of common proteins; for example, the MYC transcription factor regulates gene-encoding proteins.11 Therefore, the scientific community is highly interested in studying the mitochondria-nucleolus crosstalk and for that, the simultaneous visualization of mitochondria and nucleolus is very important. Considering all these facts, we intended to develop a photostable, easy-to-synthesize and cost-effective fluorescent probe for simultaneously tracking mitochondria and nucleolus via live cell imaging techniques.
We thus designed two copper(I) complexes with pyrazole-linked triphenylphosphine moieties (C1 and C2), and these probes were used successfully in the simultaneous visualization of mitochondria and nucleolus in living cells. The triphenylphosphine moiety's selective binding ability towards mitochondria is well-known to us. Therefore, as a way to impart important mitochondria-binding capabilities, a triphenylphosphine moiety was incorporated in our design.12 Cisplatin- and PT-ACRAMTU-based metallodrugs are known to target the nucleolus efficiently as an additional mechanism to kill cancer cells.13 Notably, both of the metallo-complexes, i.e., cisplatin and PT-ACRAMTU are nucleic acid-binding molecules. Moreover, as per a recently published report, the nucleic acid-binding capabilities within a molecule are found very useful in imparting a nucleolus-binding potential within the probe.14 Therefore as a way to make it nucleolus-specific, nucleic acid-binding capabilities were imparted in our design.
Finally, to optimize lipophilic characteristics, anthracene- and pyrene-based large hydrophobic groups were considered in the design. The optimum lipophilic properties so produced enabled the complexes to penetrate through the cell, nucleus, and nucleolus membranes and facilitated these entities towards nucleolus binding. Finally, the designed C1/C2 metallic complexes were successfully synthesized and fully characterized. These complexes were then utilized successfully for the simultaneous visualization of mitochondria and nucleolus via the confocal cell imaging technique. Simultaneous mitochondria-nucleolus imaging and high photostability of the C1/C2 probes are the typical characteristics of the current study.
Results and discussion
Synthesis and characterization of C1/C2 probes
The molecules 1,1′-(anthracen-9-ylmethylene) bis(1H-pyrazole) (1) and 1,1′-(pyren-4-ylmethylene) bis(1H-pyrazole) (2) were synthesized by synthetic organic chemistry reactions. By using these compounds, C1/C2 were subsequently synthesized. The synthetic procedures of (1) and (2) and the C1/C2 complexes are provided in the Experimental section of the manuscript. The structural features of (1), (2) and C1/C2 and the characterization data are available in the ESI (Tables ST1, ST2 and Fig. SF1–SF6†). Notably, the 31P NMR spectra in both cases revealed only a single signal corresponding to the metallo-complex-associated triphenylphosphine moiety and no additional 31P NMR signal for free triphenylphosphine was observed (Fig. SF5, ESI†). Therefore, the complete incorporation of triphenylphosphine within the complex was confirmed. The melting points of C1 and C2 were found to be 183 and 179 °C, respectively.The structural features and photophysical properties of probes
The optimized geometry and electronic parameters of C1/C2 were calculated by density functional theory (DFT) calculations. For DFT optimization, B3LYP as a method, GENECP as a basic set, QC to SCF, 6-311G** for all atoms and LAND2Z for Cu were provided and calculations were performed. The ground state optimized structure of C1/C2 revealed Cu(I) as the metal center and possessed distorted tetrahedral geometry (Fig. 1A and B). The 3d10 Cu(I) center was found to bind to two nitrogen atoms of a ligand molecule, phosphorus of PPh3, and one chloride atom. Fig. 1C and D reveal the positioning of various HOMO–LUMO, respectively, for the C1 and C2 complexes. HOMO, HOMO−1, HOMO−2, and HOMO−3 were found to be situated in the vicinity of central Cu(I) in case of both the complexes. Besides this, an extensive spread of HOMO towards the aromatic part of triphenylphosphine was visible in both the cases. LUMO and LUMO+1 of C1 were found to be situated on the anthracene ring, and LUMO+2 and LUMO+3 of C1 were found to be situated on the aromatic rings of the triphenylphosphine moiety. However, in case of C2, LUMO, LUMO+1, and LUMO+2 were found to be situated on the pyrene ring, and LUMO+3 was found to be situated on the aromatic part of the triphenylphosphine moiety of the complex. The important bond lengths and bond angles of C1 and C2 are represented in ESI Tables ST3 and ST4,† respectively. The energy of each HOMO–LUMO is further represented in Table ST5, ESI.†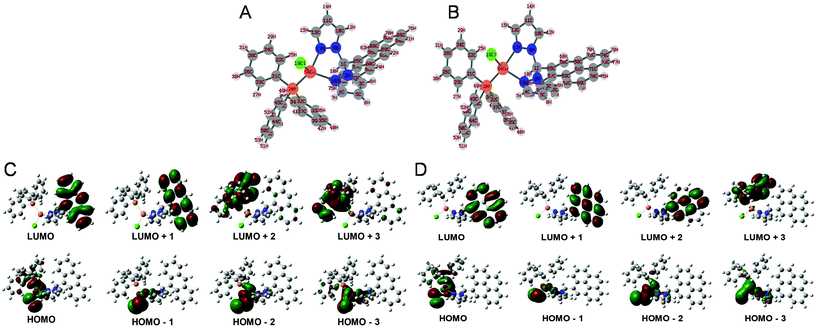 | ||
| Fig. 1 Optimized structures of C1 (A), C2 (B); the positions of various orbitals for C1 (C) and C2 (D) as obtained from DFT studies. | ||
Subsequently, we further explored the photophysical properties of the C1/C2 complexes. In the case of C1, an excitation maximum wavelength of 373 nm and an emission maximum wavelength of 425 nm were observed. Moreover, the molar absorption coefficient and quantum yield in case of C1 were found to be 2.6 × 104 L mol−1 cm−1 and 0.62, respectively. Similarly, in the case of the C2 complex, an excitation maximum wavelength of 344 nm and an emission maximum wavelength of 377 nm were observed. The molar absorption coefficient and quantum yield in this case were found to be 1.4 × 104 L mol−1 cm−1 and 0.51, respectively. The excitation and emission spectra of C1 and C2 are represented in Fig. 2, and Table 1 represents the photophysical properties of both the complexes.
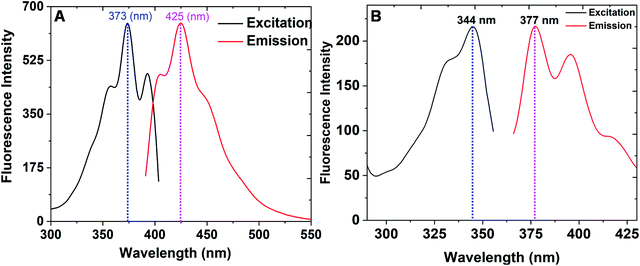 | ||
| Fig. 2 (A) The excitation and emission spectra of C1. (B) The excitation and emission spectra of C2. | ||
| Property | C1 complex | C2 complex |
|---|---|---|
| Excitation maximum wavelengths | 373 nm | 344 nm |
| Emission maximum wavelengths | 425 nm | 377 nm |
| Molar absorption coefficients | 2.6 × 104 L mol−1 cm−1 | 1.4 × 104 L mol−1 cm−1 |
| Fluorescence quantum yields | 0.62 | 0.51 |
Nucleic acid-binding studies
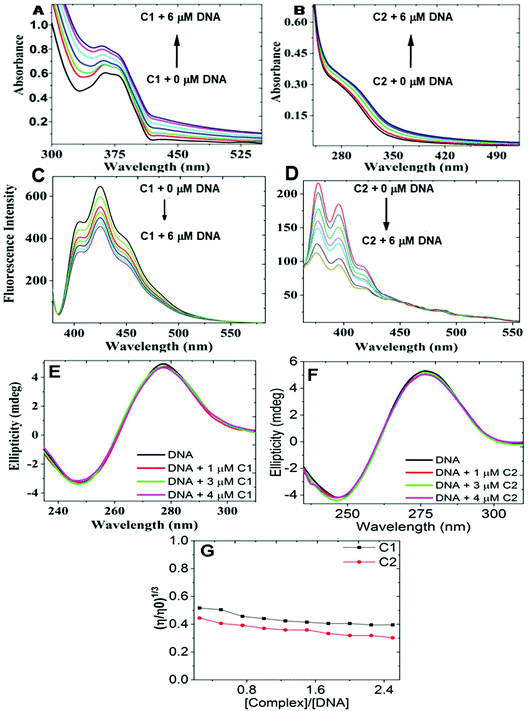
As per earlier reports, the nucleic acid-binding potential is useful in imparting nucleolus-binding properties in ligand molecules. Therefore, studies were conducted to explore the nucleic acid-binding potential of the C1/C2 complexes (Fig. 3). The UV-Vis absorption titration study of C1/C2 with continuously increasing concentrations of the nucleic acid (0–10 μM) revealed a hyperchromic shift. These observations suggested that C1/C2 accomplished binding by electrostatic or groove binding interactions.15 Two phenomena can explain the hyperchromic shift observed during the titration studies. The first case arises when a large surface area and planer aromatic chromophores of the ligand facilitate its strong interaction with the nucleic acid molecule, therefore causing the partial insertion of aromatic moieties between the stacking base pairs, which ultimately results in groove binding.15The second case arises because of the presence of a transition metal ion within the molecule. Because of the enormous nucleic acid uncoiling ability, these complexes can uncoil it to expose the hidden base-pair sequences. Consequently, bond formation occurs between the transition metal and the exposed base, causing strong groove binding. Therefore, the results obtained from the UV-Vis titration study suggested the groove-binding abilities of both the complexes.15–17 Subsequently, to get a deeper insight, fluorescence titration studies of C1/C2 with increasing concentrations of the nucleic acid were also performed. Herein, continuous quenching in the fluorescence emission profile of C1/C2 was observed in the titration study.18 To explore the mechanism of quenching, the fluorescence titration experiment was further conducted at 296, 300, and 303 K. The Stern–Volmer plots were then plotted by taking F0/F on the Y-axis and concentration on the X-axis (Fig. SF7, ESI†). The slope of the Stern–Volmer plots was found to decrease with the increase in temperature. Therefore, a static quenching mechanism was concluded for the complexes. For determining the binding constants of DNA with C1 and C2, log[(F0/F) − 1] vs. log[Q] was plotted (Fig. SF8, ESI†). From the intercept of the current plot, the binding constant Kb was calculated in both cases. In the case of C1-DNA, Kb was found to be 2.07 × 104 mol−1 and for C2-DNA, Kb was found to be 12.0 × 106 mol−1.19
The results obtained from the above titration studies sufficiently described the C1/C2-nucleic acid binding, but additional proofs are still required to confirm the groove binding properties.18 Therefore, circular dichroism (CD) titration studies of the nucleic acid with increasing concentrations of C1/C2 were performed. The CD spectra of deoxyribonucleic acid revealed two bands: a positive band at 275 nm and a negative band at 245 nm.18 The changes that occur in the DNA's CD signals are caused by the major structural changes within the DNA. Such structural changes are very common in intercalative DNA binding and therefore, substantial changes in the CD signals are also expected under the intercalative binding mechanism. However, in groove binding, no major change in the structural feature of DNA is expected and therefore, no major change in the CD signals is expected in this case. The result obtained in our case revealed a slight effect on both the bands and therefore, groove binding was further realized.18 Upon intercalative DNA binding, there occurred base pair separation and insertion of the molecule within the separated base pairs. These structural changes are only expected in case of intercalation and are responsible for increasing the viscosity of the DNA sample. Therefore, no viscosity change is expected in the case of groove binding. C1/C2's groove binding was therefore confirmed additionally from deoxyribonucleic acid's viscosity change experiment as per a method already reported in the literature.18 The results obtained by us revealed slight decrease in viscosity upon increasing the concentration of the C1/C2 complexes.18 Therefore, the groove-binding abilities of the C1/C2 complexes were confirmed by these studies.
Simultaneous mitochondria and nucleolus imaging by C1/C2 probes
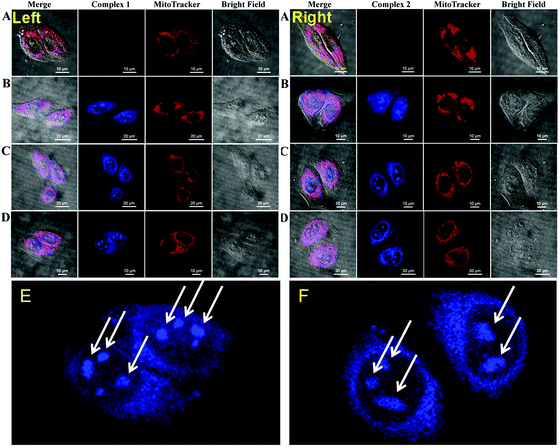
Finally, after realizing the optimum nucleic acid-binding properties of the C1/C2 complexes, we then investigated the biological relevance of the designed moieties. To examine the mitochondria binding of the C1/C2 metallo-complexes, the MitoTracker dye was used. Part A of Fig. 4 (left for C1 and right for C2) indicates a control experiment, where the cells are not treated with the C1/C2 complexes. In the case of the control experiment, the confocal red channel staining in MitoTracker-treated cells revealed the positioning of the mitochondria inside the cells, and no staining was observed under the blue channel. Parts B–D of Fig. 4 reveal the blue and red channel confocal images of the HeLa cells simultaneously treated with C1 (left) and C2 (right) along with MitoTracker in both the cases.Herein, the red channel reveals the MitoTracker staining and the blue channel shows staining due to the C1/C2 complexes. The red-blue merged channel images reveal the exact overlapping of the red and blue staining of the cells and confirm the mitochondria binding of our complexes. All the observations, viz, at 30 min (B), 1 h (C) and 2 h (D) revealed the mitochondria-binding ability of the C1/C2 complexes. The complex C1 showed early localization in mitochondria after 30 min of incubation (part B of Fig. 4, left). However, after the same incubation time, C2 was found localized in the nuclear space along its binding to the mitochondria (part B of Fig. 4, right). However, the mitochondrial localization of C2 was observed after 1 h of incubation (part C of Fig. 4, right). Fig. 4(E) and (F) reveal the enlarged views of the HeLa cells stained with C1 and C2, respectively. These results revealed the nucleolus binding of both the complexes (white arrows). After confirming the simultaneous mitochondria and nucleolus imaging potential of C1/C2, their MTT cytotoxicity assay was performed. Both the complexes were found to be the least cytotoxic to HeLa cells and even at a 20 μM of treatment concentration, about 90% of cell viability was observed in both the cases (Fig. SF9, ESI†). However, commercially available MitoTracker was reported to show high cytotoxicity under similar experimentation conditions.20 Thus, along with multifunctional cell imaging properties, the least cytotoxicity was yet another excellent feature that was found to be associated with both the complexes.
Photostability of C1/C2 probes
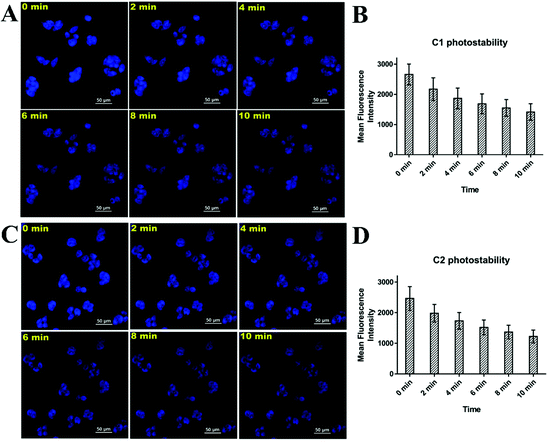
Finally, the photostability of both complexes was investigated. C1/C2 also showed excellent photostability when torched with 10% of 100 mW DAPI laser of a Nikon Eclipse Ti-U inverted microscope continuously for 10 min (Fig. 5). Although the mean fluorescence intensity of C1/C2 gradually decreased with the exposure time, still, the cells showed an admirable blue color even after 10 min of exposure with an extremely powerful laser source. Consequently, the C1/C2 complexes also provided us with an economical cell imaging tool, with admirable photostability for the simultaneous detection of the mitochondria and nucleolus inside the living cells. The photostability results of commercially available MitoTracker were also explored (Fig. SF10, ESI†). The photostability of MitoTracker was found to be comparatively better, but the simultaneous mitochondria and nucleolus imaging potential and the least cytotoxicity are the superior features of our C1/C2 complexes.Finally, we compared the performances of the C1/C2 complexes with that of the commercially available mitochondria probing agent “MitoTracker Red” and other agents reported in the literature (Table ST6, ESI†). Under the same concentration of 5 μM and at the time interval of only 30 min, the mitochondrial imaging results obtained with the C1/C2 complexes were found to be very excellent, i.e., they were similar to that of the commercially available MitoTracker Red dye. Moreover, with MitoTracker Red, we can only probe mitochondria; however, the C1/C2 complexes enabled the simultaneous visualization of mitochondria and nucleolus in a single run and just within a timeframe of 30 min. Furthermore, compared to the already reported complexes, the C1/C2 complexes were found to be far better in terms of simultaneous mitochondria and nucleolus tracking, a shorter incubation period of 30 min, quick results and low concentration requirement during each experiment (Table ST6, ESI†). Moreover, the C1/C2 complexes possessed very high photostability and were easy to synthesize. The synthesis process of the C1/C2 complexes only included a single-step one-pot reaction and therefore, these complexes were very inexpensive compared to commercially available probes. Hence, soon, we may expect the commercial applicability of the same or similar probes towards mitochondria/nucleolus tracking.
Conclusion
The C1/C2 complexes designed by us are valuable tools with tremendous cell imaging potentials. Because of the simultaneous mitochondria and nucleolus staining capabilities along with high photostability, the C1/C2 complexes can be considered as exceptional tools for simultaneously tracking mitochondria and nucleolus within the living cells. Therefore, the C1/C2 metallo-complexes developed by us are multifunctional diagnostic tools and seem to be highly useful in cell imaging techniques.Materials and methods
Reagents and instruments used
Pyrazole, 9-anthracenecarboxaldehyde, 1-pyrenecarboxaldehyde, thionyl chloride, copper(I) chloride and sodium hydride used for the synthesis were obtained from Sigma-Aldrich and used as such without any further treatment. A 400 MHz nuclear magnetic resonance spectrometer (NMR) by JEOL, XEVO G2-XS QTOF HR-Mass instrument by Waters, Shimadzu 2400 UV-Vis absorption spectrophotometer, LS-55 fluorescence spectrophotometer by PerkinElmer, circular dichromism instrument J-1500 from JASCO, IR spectrophotometer from Bruker and BD LSRFortessa™ cell analyzer were used for our study. HeLa and MCF-7 cancer cell lines were procured from the National Centre for Cell Sciences, Pune, India. Dulbecco's Modified Eagle's media (DMEM), penicillin–streptomycin antibiotic, fetal bovine serum (FBS), and phosphate buffer saline (7.4 pH, 1×) were obtained from Thermo scientific India. Gaussian-09 software package in association with GaussView-05 was used for DFT study, and Schrodinger Maestro-18.2 release was used for molecular docking study.The synthetic procedure of compounds
1,1′-(Anthracen-9-ylmethylene) bis(1H-pyrazole) and 1,1′-(pyren-4-ylmethylene) bis(1H-pyrazole) required for producing C1/C2 probes were synthesized by synthetic organic reactions. First, 50 ml THF containing NaH (48.00 mmol, 1.16 g) was added to 15 ml of THF solution containing pyrazole (48.00 mmol, 3.27 g) with continuous stirring under a nitrogen-based inert atmosphere. The mixture was further stirred at 0 °C for 30 additional minutes to produce a yellow-colored solution. Subsequently, the dropwise addition of SOCl2 (24 mmol, 2.91 g) was done to get a yellowish-white suspension. The resulting suspension was further stirred for 40 min and afterward, 4.85 mmol of 9-anthracenecarboxaldehyde/1-pyrenecarbaldehyde along with 1.46 mmol of anhydrous CoCl2 was added; the resulting reaction mixture was refluxed for 10 h. After the completion of the reaction, extraction was done using a chloroform/water mixture. The chloroform fraction so obtained was dried, and products in their pure form were recovered by column chromatography, obtaining 45% and 50% yields of (1) and (2), respectively.21 The resulting compounds were characterized by 1H NMR, 13C NMR, and melting point analysis techniques.The synthetic procedure of C1/C2 metallo-complexes
For producing C1/C2 metallo-complexes, 100 mg each of 1,1′-(anthracen-9-ylmethylene) bis(1H-pyrazole) and 1,1′-(pyren-4-ylmethylene) bis(1H-pyrazole) were individually taken in 20 ml of HPLC-grade acetonitrile, and the resulting mixture was stirred at room temperature to form a clear solution. Then, 27.44 mg (0.28 mmol) of CuCl and 72.8 mg (0.28 mmol) of triphenylphosphine were solubilized in sufficient amount of HPLC-grade acetonitrile. The resulting solutions were then mixed, and the reaction mixture was stirred continuously to separate the precipitates of the required C1/C2 complexes. The complexes were then characterized by 31P NMR- and HRMS-based analytical techniques, and the melting points of both the complexes were also recorded.Density functional theory calculations
As a way to investigate the optimized geometry and electronic structural features of C1/C2, DFT calculations were performed. Gaussian-09 software package in association with GaussView-05 was used to perform all the quantum mechanical calculations. The structures of C1/C2, initially required for calculations, were drawn using a builder option available in the software package. The initial structures were then passed through the clean-up process to get the structures ready for calculations. For ground-state DFT calculations, Gaussian calculation setup wizard was used, and optimization as a job type, DFT along with B3LYP as a method and GENECP as a basic set were selected. Finally, by providing QC to SCF, 6-311G** for all other atoms and LAND2Z for Cu, calculations were performed.Nucleic acid-binding studies
The solution of C1/C2 required for DNA-binding studies was prepared in DMSO. The stock solution of DNA was prepared in phosphate buffer saline (PBS). The stock solutions so obtained were subsequently diluted by PBS to prepare the required concentration. For UV-Vis studies, a cuvette of 1 cm path length was used. Titration was conducted by taking a 20 μM solution of C1/C2 in the cuvette and continuously increasing the concentration of DNA from 0 to 6 μM. For the fluorescence study, a quartz cell of 1 cm path length was used. Titration was done by continuously increasing the concentration of DNA. To record circular dichromism spectrometer readings, 20 μM of DNA was taken, and titration with increasing concentration of C1/C2 was done. For viscosity measurements, an Ostwald's viscometer was used, and experimentations were conducted at a temperature of 25 °C. A digital stopwatch was used to measure the flow time, and data obtained were plotted by taking (η/η0)1/3 on the Y-axis and [complexes]/[DNA] ratio on the X-axis.Cell culture and MTT assay
HeLa cells required for our study were cultured in DMEM media. To achieve sufficient growth, 10% of fetal bovine serum (FBS) and 1% penicillin–streptomycin were also incorporated into DMEM while doing the culturing experiments. The cells were then incubated at 37 °C, and a continuous supply of 5% CO2 was provided. For MTT cytotoxicity assay, the methodology already reported by us was used.22Mitochondria/nucleolus binding assay
For investigating the mitochondria/nucleolus binding of the C1/C2 complexes, the confocal cell imaging technique was used. A simultaneous treatment with C1/C2 (5 μM) and MitoTracker Red dye (5 μM) was given to the HeLa cells, and confocal imaging was done after 30 min, 1 h and 2 h of the incubation period. In a control experiment, cell treatment with only MitoTracker Red was given, and no C1/C2 were used. The MitoTracker dye bound selectively to mitochondria and helped us to locate the mitochondria within the cell. Confocal cell imaging was done by using the red channel (for locating mitochondria) and the blue channel (for locating C1/C2 staining). The red-blue merged channel was used to confirm the mitochondria binding of both the complexes.Photostability assay of C1/C2 complexes
To evaluate the photostability of the C1/C2 complexes in the intracellular environment, the HeLa cells cultured on a coverslip were incubated with complex C1 and C2 (5 μM) for 30 min. The mounted glass slide for imaging was prepared as stated in the above section. The cells on the mounted slides were directly exposed to 10% of 100 mW DAPI laser of a Nikon Eclipse Ti-U inverted microscope for 10 min, and the image was captured at an interval of 2 min. The fluorescence intensity of each image at different times was quantified using the NIS element software provided with the Nikon Eclipse Ti-U inverted microscope.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Mayank thanks Indian Institute of Technology Ropar for the Senior Research Fellowship, RR Thanks DST-SERB India for the National Post-Doc Fellowship (Grant Number NPDF2017/002440), and AS thanks UGC India for the research fellowship. NG acknowledges Ramanujan SERB grant, India (SB/S2/RJN-072/2015). NG and Ashutosh Singh acknowledge Advanced Materials Research Center (AMRC) and BioX center, Indian Institute of Technology Mandi for providing all the necessary facilities.References
- K. Han, J.-Y. Zhu, H.-Z. Jia, S.-B. Wang, S.-Y. Li, X.-Z. Zhang and H.-Y. Han, ACS Appl. Mater. Interfaces, 2016, 8, 25060–25068 CrossRef CAS PubMed.
- Y. B. Chen, M. A. Aon, Y. T. Hsu, L. Soane, X. Teng, J. M. McCaffery, W. C. Cheng, B. Qi, H. Li, K. N. Alavian, M. Dayhoff-Brannigan, S. Zou, F. J. Pineda, B. O'Rourke, Y. H. Ko, P. L. Pedersen, L. K. Kaczmarek, E. A. Jonas and J. M. Hardwick, J. Cell Biol., 2011, 195, 263–276 CrossRef CAS PubMed.
- D. Kessel and Y. Luo, Cell Death Differ., 1999, 6, 28–35 CrossRef CAS PubMed.
- F. Burte, V. Carelli, P. F. Chinnery and P. Yu-Wai-Man, Nat. Rev. Neurol., 2015, 11, 11–24 CrossRef CAS PubMed.
- A. C. Shaikh, M. E. Varma, R. D. Mule, S. Banerjee, P. P. Kulkarni and N. T. Patil, J. Org. Chem., 2019, 84, 1766–1777 CrossRef CAS PubMed.
- Y. Chris, W. Zhang, R. T. Kwok, C. W. Leung, J. W. Lam and B. Z. Tang, J. Mater. Chem. B, 2016, 4, 2614–2619 RSC.
- M. M. Yusupov, G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. Cate and H. F. Noller, Science, 2001, 292, 883–896 CrossRef CAS PubMed.
- R. Y. Tsai and R. D. McKay, Genes Dev., 2002, 16, 2991–3003 CrossRef CAS PubMed.
- S. Khan, N. C. Verma and C. K. Nandi, ACS Appl. Nano Mater., 2018, 1, 483–487 CrossRef CAS.
- C. Ge, H. Huang, Y. Wang, H. Zhao, P. Zhang and Q. Zhang, ACS Appl. Bio Mater., 2018, 1, 1587–1593 CrossRef CAS.
- S. Rossetti, A. J. Wierzbicki and N. Sacchi, Oncotarget, 2018, 9, 5016 CrossRef PubMed.
- J. Liu, Y. Chen, G. Li, P. Zhang, C. Jin, L. Zeng, L. Ji and H. Chao, Biomaterials, 2015, 56, 140–153 CrossRef CAS.
- A. J. Pickard and U. Bierbach, ChemMedChem, 2013, 8, 1441–1449 CrossRef CAS.
- K. Singh, S. Singh, P. Srivastava, S. Sivakumar and A. K. Patra, Chem. Commun., 2017, 53, 6144–6147 RSC.
- C. Gurudevaru, M. Gopalakrishnan, K. Senthilkumar, H. Hemachandran, R. Siva, T. Srinivasan, D. Velmurugan, S. Shanmugan and N. Palanisami, Appl. Organomet. Chem., 2018, 32, e3998 CrossRef.
- T. Afrati, A. A. Pantazaki, C. Dendrinou-Samara, C. Raptopoulou, A. Terzis and D. P. Kessissoglou, Dalton Trans., 2010, 39, 765–775 RSC.
- E. Jayanthi, S. Kalaiselvi, V. V. Padma, N. S. Bhuvanesh and N. Dharmaraj, Dalton Trans., 2016, 45, 1693–1707 RSC.
- M. Verma, N. Kaur and N. Singh, Langmuir, 2018, 34, 6591–6600 CrossRef CAS PubMed.
- J. Sindhu, A. K. Bhasin, N. Kaur, N. Singh and K. Bhasin, New J. Chem., 2019, 43, 2065–2076 RSC.
- S. Zhang, T. Wu, J. Fan, Z. Li, N. Jiang, J. Wang, B. Dou, S. Sun, F. Song and X. Peng, Org. Biomol. Chem., 2013, 11, 555–558 RSC.
- B.-F. Ruan, Y.-Z. Zhu, W.-D. Liu, B.-A. Song and Y.-P. Tian, Eur. J. Med. Chem., 2014, 72, 46–51 CrossRef CAS.
- Mayank, A. Singh, N. Kaur, N. Garg and N. Singh, Comput. Biol. Chem., 2019, 83, 107104 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9an01513b |
| ‡ Equal Contribution. |
| This journal is © The Royal Society of Chemistry 2020 |