LMP1 gene detection using a capped gold nanowire array surface plasmon resonance sensor in a microfluidic chip
Chih-Shen
Chuang
 ab,
Chieh-Ying
Wu
a,
Po-Han
Juan
a,
Nai-Cheng
Hou
a,
Yu-Jui
Fan
*bcd,
Pei-Kuen
Wei
e and
Horn-Jiunn
Sheen
ab,
Chieh-Ying
Wu
a,
Po-Han
Juan
a,
Nai-Cheng
Hou
a,
Yu-Jui
Fan
*bcd,
Pei-Kuen
Wei
e and
Horn-Jiunn
Sheen
 *a
*a
aInstitute of Applied Mechanics, National Taiwan University, 1 Roosevelt Rd., Section 4, Taipei 10617, Taiwan. E-mail: sheenh@ntu.edu.tw; Fax: +886-2-3366-5633; Tel: +886-2-3366-5632
bSchool of Biomedical Engineering, Taipei Medical University, 250 Wuxing St., Taipei 11031, Taiwan. E-mail: ray.yj.fan@tmu.edu.tw; Tel: +886-2-2736-1661, ext.7722
cInternational PhD Program for Biomedical Engineering, Taipei Medical University, 250 Wuxing St., Taipei 11031, Taiwan
dGraduate Institute of Biomedical Optomechatronics, Taipei Medical University, 250 Wuxing St., Taipei 11031, Taiwan
eResearch Center for Applied Sciences, Academia Sinica, 128 Academia Road, Section 2, Nankang, Taipei 11529, Taiwan
First published on 25th November 2019
Abstract
Surface plasmon resonance (SPR) nanowire array chips with a microfluidic system are an effective detection method for a rapid test device. This study investigated a capped gold nanowire array and a microfluidic test platform to provide a fundamental understanding of the kinetic binding of SPR nanowires and the surface gold refractive index. The device sensitivity of the SPR nanowire array was 485 nm RIU−1 and the detection limit was 4.1 × 10−5 RIU. Moreover, a kinetic binding analysis also indicated that a peak shift resulted from a specific hybridization of the target molecule with the immobilized probe on the gold nanostructures. The peak shift (red-shift) value of latent membrane protein 1 (LMP1) DNA was 2.21 nm. The results demonstrated that this new method had high sensitivity to detect amplified DNA products without labeling or complex sample treatment. The SPR nanowire chip can detect the PCR products at lower cycle numbers compared to gel electrophoresis due to probe and DNA specificity. Furthermore, the mechanisms of SPR nanowire array fabrication and the detection of the LMP1 gene were studied. The findings can assist in improving the biosensing of DNA-amplified products and in developing rapid detection devices with a small-footprint nanostructured SPR chip.
Introduction
Nasopharyngeal carcinoma (NPC) is one of the epithelial carcinomas characterized by the additional co-expression of latent membrane proteins (LMPs, i.e., LMP1, LMP-2A, and LMP-2B).1,2 Standard methods used to detect LMPs or the Epstein–Barr virus (EBV) are polymerase chain reaction (PCR) and real-time (rt) PCR.3–7 A rapid, label-free and highly sensitive detection method for LMPs would provide a more efficient and economical way for the diagnosis of NPC. Lab-on-a-chip (LOC) with a microfluidic system and surface plasmon resonance (SPR) are two effective methods for the development of next-generation testing devices.8 Recently, cost-effectiveness and measurement sensitivity have been the two significant challenges for the development of next-generation testing devices.8 On the other hand, the cost and handling time of sample pretreatment are also a challenge for rapid detection devices. Kranzfelder et al.9 pointed out that NucleoSpin® Tissue XS kit, DNeasy® Blood and Tissue kit, and QuickExtrat™ DNA extraction solution can provide good results in isolating DNA. The extraction time of QuickExtrat™ DNA extraction solution is less than 5 min, and this kit is easy to handle.9 Therefore, the measurement sensitivity and limit are the important challenges for a rapid detection device.SPR is a resonant oscillation of electrons at the metal and dielectric interface induced by an incident electromagnetic wave. In the past two decades, SPR sensing was found to have advantages of real-time and label-free measurements of biomolecular interactions on metallic surfaces, especially gold surfaces.7,10 The widely used method utilizes an optical prism which can induce SPR on a flat gold surface, known as the Kretschmann configuration. The prism coupling method can also be used to excite metallic nanostructures.11–14 However, the prism coupling method needs a precise control of the incident angle and is too complicated to be integrated with chip-based microfluidic devices.
Recently, transparent types of SPR sensors using extraordinary optical transmission (EOT) in a periodic gold nanohole array or nanoslit to generate SPR have been used for biosensing applications. This method takes advantage of normal incident light and chip-based configuration without the use of an optical prism. Biosensors based on extraordinary transmissions of periodic nanohole arrays and nanoslit arrays in gold have been proposed15–20 and used in medical diagnostics, environmental monitoring, and food safety.21–23 Different from localized surface plasmons (LSPs), which are directly excited by the incident light into metallic nanoparticles, the SPR in a metallic hole array or nanoslit array is based on the coupling of LSPs in nanoapertures and Bloch-wave surface plasmon polariton (BW-SPP) on a periodic metallic surface. The coupling often forms the Fano-like resonance and can enhance the detection of biomolecules due to its sharper resonance profile.23 Periodic gold-nanostructure sensors take advantage of a chip-based and straightforward optical setup. They are easily incorporated into microfluidic devices. The biomolecular interaction causes a change of the refractive index on the metal surface, which can be measured by recording the resonance change in the transmission spectrum.24,25 Compared to the reflective-type SPR sensors, transparent-type SPR sensors benefit from the normal incidence that is capable of being integrated with multiplex microfluidic devices. They also provide a feasible way to achieve chip-based, high-throughput, label-free detection of modern DNA and protein microarrays.
In recent years, several researchers have developed SPR microfluidic devices for microRNA,26–28 protein,29,30 methylation DNA,31,32 and PCR detection.33,34 However, the SPR fabrication process with the Fano-like resonance and the detection of cancer DNA are rarely elucidated. Furthermore, the integration of SPR with LOC-based microfluidic devices for detecting different amplification cycles of cancer DNA remains unclear. DNAs are small biomolecules; therefore the change of the refractive index by a small amount of DNA is very small. The non-specific binding of other substrate biomolecules would lead to a false-positive result. PCR amplification of target DNA can significantly enrich the amount of DNA and thus enhance the specificity of the detection. In this study, the capped gold nanowire array-based SPR and a microfluidic test platform were evaluated. Furthermore, PCR amplification products of LMP1 DNA were investigated to provide a fundamental understanding of the kinetic binding analysis of the SPR nanowire array and the surface gold refractive index. The microfluidic SPR platform is a label-free biosensor with high detection sensitivity. Furthermore, low-cost nanostructures can be achieved by using an automatic nanoimprint technique,35 which makes the SPR-microfluidic devices feasible for various rapid detection applications.
Experimental
Fabrication of a capped gold nanowire SPR chip
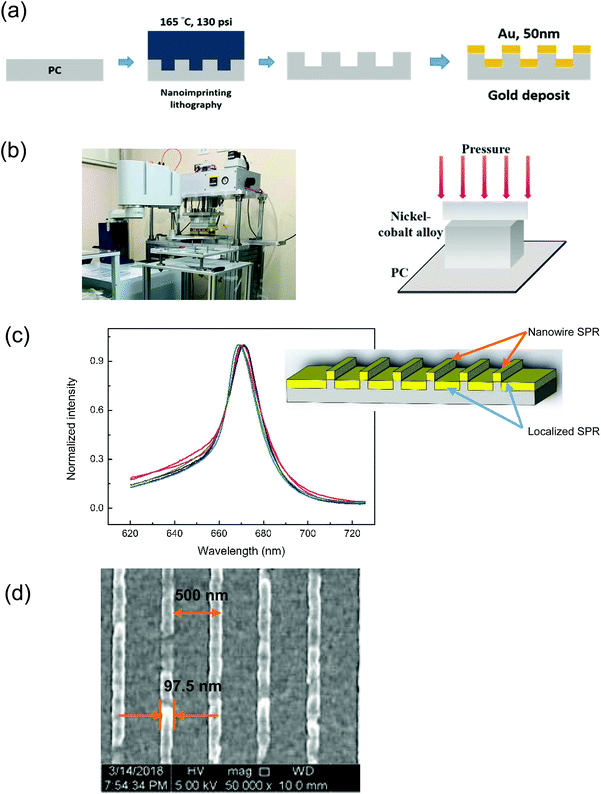
An automatic hot-embossing nanoimprinting machine was used in this study to fabricate the capped gold nanostructure. This fabrication method followed the method used by Lee et al.35 and Lee et al.36Fig. 1(a) and (b) show the fabrication process. The nanowire structure was fabricated using a metal mold for rapid replication. A periodic nanowire structure was defined and fabricated on a silicon wafer by E-beam lithography and reactive ion etching. Then, the structure was transferred onto a nickel–cobalt alloy mold using the electrodeposition method. For the rapid hot-embossing method, the Ni–Co mold is essential. It has low adhesion to plastic films and can withstand the hitting force during the rapid stamping process. The nanowire structures were produced on a polycarbonate (PC) film using an automatic hot-embossing nanoimprinting machine. After imprinting the nanowire structure into a plastic film, the metal mold was peeled off from the replicated plastic film without lowering the temperature of the mold. The time needed for a single stamping process was only 5 s.35 The nanowire structure can be rapidly replicated, and the mold can be used repeatedly. Polycarbonate (PC) was adopted as the substrate material of our sensor owing to its low cost and excellent thermal properties for nanoimprinting fabrication. The nanostructure on the Ni–Co mold was transferred onto a PC film by hot-embossing nanoimprinting lithography, at a temperature of 165 °C and a working pressure of 130 psi. This temperature is much higher than the Tg of PC (135 °C), to rapidly heat the surface of the plastic film. Gold was deposited onto the nanowire structure of PC using a DC sputter, the deposition time was 70 s, and the thickness of gold was 50 nm.In this study, the nanowire array SPR is based on the coupling of LSPs in nanoapertures and Bloch-wave surface plasmon polariton (BW-SPP) on a periodic metallic surface. There were sharp Fano resonances in the spectra. The coupling often forms the Fano-like resonance, as shown in Fig. 1(c). The wavelength sensitivity is comparable with most metallic nanostructure-based sensors using SPR or localized surface plasmon resonance (LSPR). It is noted that the Fano resonance had a very sharp resonance slope. Therefore, the intensity sensitivity (intensity change at a fixed wavelength under a refractive index change) will be much higher compared to conventional SPR sensors. We have checked the resonance quality of the SPR chip. The chips were well reproduced as shown in revised Fig. 1(c). The variations of the resonance wavelength are within 1% for different chips (N = 6). The experimental results of functional reproducibility indicated that the SPR nanowire chip has stable production and functional reproducibility.
A scanning electron microscope (Hitachi S-4800, Japan) was used to evaluate the nanowire width and interval. The voltage of the scanning electron microscope was 5 kV. The SEM image of the SPR chip is shown in Fig. 1(d). The experimental results indicated that the interval of the nanowire was 500 nm, and the nanowire width was 97.5 nm. The surface sensitivity of the gold nanostructure array is optimized using the structures with a 500 nm interval.19 On the other hand, the width of the nanostructure plays an essential role in the resonance linewidth.18 The SPR of the fabricated capped gold nanowire array with an interval of 500 nm was manifested as a transmission spectrum in a wavelength range of 670–690 nm when distilled water was in contact with the gold film.
Fabrication of the integrated microfluidic chip
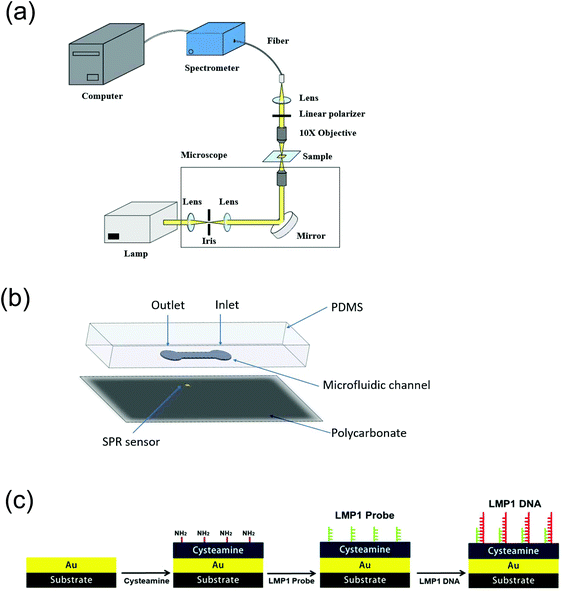
In this experiment, a microfluidic device combined with the transmission (t)-SPR detection technique was used to detect the LMP1 gene. The disposable microfluidic device is a single-layered structure and was fabricated using polydimethylsiloxane (PDMS), as shown in Fig. 2. The microfluidic channel was integrated with the gold capped nanowire array as the biosensor. The Fano resonance is a result of two-mode coupling between the LSP resonance in gold nanowires and the BW-SPP mode on the periodic nanowire structure. The incident light was normally incident on the sample surface. The transmission light was filtered with a linear polarizer with its polarization direction perpendicular to the nanowires because the SPR can only be excited by the transverse magnetic (TM) wave. The TM light was collected through a collimator and passed through a 230 μm optical fiber, which was connected to a spectrometer (model: USB 4000, Ocean Optics, FL, USA) and the optical resolution was 0.3 nm by using a low-cost device. The optical setup and the structure of the microfluidic chip are shown in Fig. 2(a) and (b), respectively. Furthermore, the SPR gold surface needs to be modified to ensure good binding to the probe. In this study, cysteamine was used to modify the SPR gold surface. Immerse 100 μM cysteamine on SPR surface for 2 h. The melting temperature (Tm) of the probe used in this study was 64 °C. This process is shown in Fig. 2(c). The probe was used to react with DNA, and then the SPR chip could be easily detected.Optical setup of the nanowire SPR chip and the test platform assembly
All transmission SPR measurements were carried out by using the custom-built transmission SPR system. The spectral measurement parameters were controlled using a custom-designed LabVIEW program (National Instrument, USA) provided by the manufacturer, and each spectrum recorded consisted of an average of 100 individual spectra (integration time: 6000 ms), which were further processed into transmission spectra using| T (%) = (Sλ − Dλ)/(Rλ − Dλ) × 100%; | (1) |
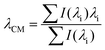 | (2) |
SPR and sensitivity measurement
A TM-polarized wave in the capped gold nanowire array generated sharp and asymmetric Fano resonances in the transmission spectra. The position of the SPR spectrum shifts due to environmental changes around the gold nanostructures. Raw data were obtained and processed using a home-written MATLAB® program. The resonance wavelength of the SPR spectrum was identified by applying the spectral centroid method. The sensor graph represents the change of resonance wavelength against time.The sensitivity test of the SPR nanowire was verified by the refractive index unit (RIU) sensitivity test. Refractive index sensitivities were measured by injecting purified water mixed with various ratios of glycerin into the microfluidic device. The refractive indexes of the mixtures (from 0% to 20% glycerin) ranged from 1.334 to 1.344.
Polymerase chain reaction analysis
Conventional thermal-cycler PCR was also used to amplify the LMP1 DNA templates with a PCR machine (Analytik Jena, Germany). The LMP1 DNA, primer sequences, and probe sequences are given in Table 1. The LMP1 DNA, primer sequence and probe sequence design were those used by Ryan et al.4 and Guo et al.37 and also modified by us. The PCR mixture contained 25 μL of Taq master mix, 1 μL of forward and reverse primers, 1 μL of a DNA template, and 22 μL of nuclease-free water. Amplification reactions were performed for 30 thermal cycles. The temperature conditions required for conventional PCR were set at 95 °C (30 s) for denaturation, 60 °C (30 s) for annealing, and 72 °C (50 s) for extension. Moreover, the initial denaturation and final extension were performed at 95 °C for 5 min and 72 °C for 10 min, respectively. Furthermore, the 4 different PCR thermal cycles (5, 10, 20, and 30 cycles) for LMP1 DNA amplification were also evaluated in this study. Agarose gel electrophoresis based analysis was also employed to confirm the DNA amplicon results. A SPR chip was then used to analyze each of the different PCR products and the results were also compared with gel electrophoresis to investigate the limitation of the different tests.| DNA template: (311 bp) |
| CCATGACCCGCTGCCTCATAACCCTAGCGACTCTGCTGGAAATGATGGAGGCCCTCCAAAATTGACGGAAGAGGTTGAAAACAAAGGAGGTGACCGGGGCCCGCCTTCGATGACAGACGGTGGCGGCGGTCATCCACACCTTCCTACACTGCTTTTGGGTACTTCTGGTTCCGGTGGAGATGATGACGACCCCCACGGCCCAGTTCAGCTAAGCTACTATGACTAACCTTTCTTTACTTCTAGGCATTACCATGTCATAGGCTTGCCTGACTGACTCTCCCTCCATTTACTGGGAATGCCTTAGCTAATCA |
| Primer F (21 bp) |
| AGC GAC TCT GCT GGA AAT GAT |
| Primer R (21 bp) |
| TGA TTA GCT AAG GCA TTC CCA |
| Probe (29 bp) |
| GTC ATA GTA GCT TAG CTG AAC TGG GCC GT |
Results and discussion
The sensitivity of the capped gold nanowire array
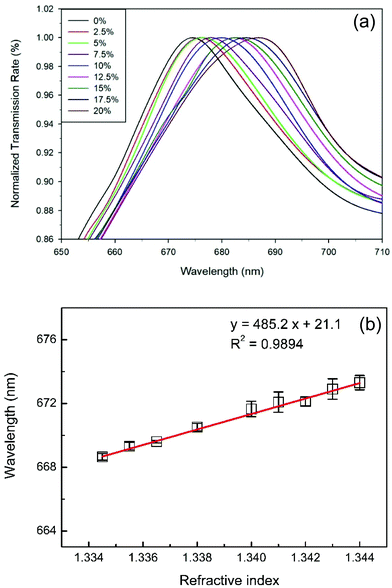
The refractive index unit (RIU) sensitivity test was used to evaluate the sensitivity of the SPR chip by injecting purified water, mixed with various ratios of glycerin, into the microfluidic device. The refractive index of the mixtures (from 0% to 20% glycerin) ranged from 1.334 to 1.344. Fig. 3(a) shows the resonance wavelengths of different glycerin percentages. The experimental results indicated that the resonance wavelength of 0% glycerin was 674 nm. The resonance wavelengths of 5%, 10%, 15%, and 20% glycerin were 676, 679, 682, and 686 nm, respectively. The noise level determined the detection limit of the chip and the system in the measurement. Fig. 3(b) shows the wavelength sensitivity of the 500 nm period SPR nanowire array. The peak wavelength was 674 nm, and the wavelength sensitivity was 485 nm RIU−1. With this resolution, the current nanostructure and simple system achieved a detection limit of 4.1 × 10−5 RIU. Furthermore, the wavelength at 674 nm was visible and was easily covered by the LED light source. Cennamo et al.38 pointed out that the SPR sensor showed high sensitivity for measuring low concentration glycerin and the sensor could be used in biochemical applications.In contrast, the deviations of each wavelength are also shown in Fig. 3(b). The deviations were obtained by measuring the sample 10 times (N = 10). From the experimental results, the deviations of Δλ/λ0 were found to be below 0.1%. Compared to other SPR sensors, the current SPR nanowire chip demonstrated cost-effectiveness and good reproducibility of production and measurement. The SPR nanowire chip with a visible light source can be used to develop a portable real-time detection device for biological binding reactions. Furthermore, the SPR nanowire chip can be integrated with the on-chip PCR device in further studies.
Kinetic binding analysis of the SPR nanowire chip
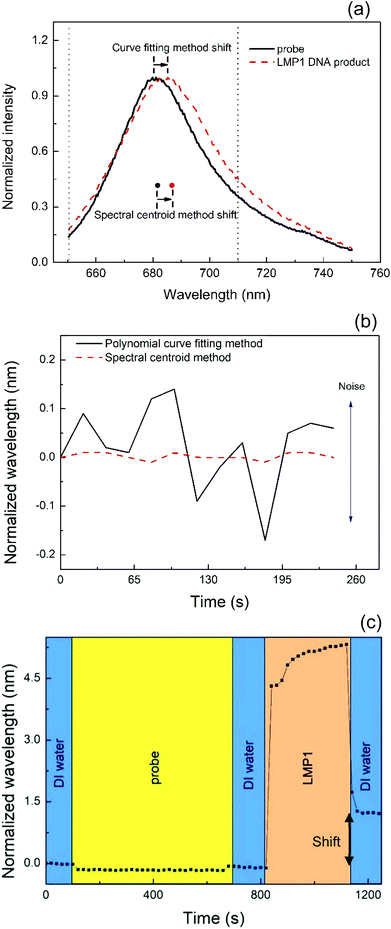
Two different kinetic binding analyses were employed to evaluate the wavelength change of the probe and the LMP1 DNA products. Fig. 4(a) shows the red-shift spectra with normalized wavelengths of two different kinetic binding analyses. The curve fitting results showed two peaks at different wavelengths. Moreover, the spectral centroid method gave two central points in the range from 650 nm to 710 nm. The results indicated that the LMP1-amplified DNA was detected by the SPR nanowire chip. The experimental results depicted that the SPR chip could be used to detect different DNA-based molecules without any labeling. The current results were in good agreement with those of Mousavi et al.27 and Lee et al.35Fig. 4(b) shows the stability of the resonance wavelength in pure water as a function of time. The resonance wavelength was obtained by the spectral centroid method. To compare the performance of the analytic method, we also calculated the peak resonance wavelength using a curve fitting method. It can be seen that the wavelength resolution is greatly improved using the spectral centroid method. The standard deviation of the wavelength measurement is 0.02 nm.Furthermore, the real-time progression reaction of the probe and LMP1 DNA was first detected in this study. Fig. 4(c) shows the kinetic binding analysis of the entire test. The SPR chip was first treated with cysteamine to form thiol bonding. After that, the spectrum of the SPR chip was stabilized by using water. After 2 min, the probe was injected into the microchannel for 10 min. After 10 min of immersion, the excess probe was washed away for 2 min. After that, the amplified LMP1 DNA products were first denatured, and then linked to the probe at the probe Tm (64 °C). After another 5 min of immersion, the excess DNA which had not been hybridized was washed away. In Fig. 4(c), the spectrum exhibited a sharp peak shift at 1140 s when the amplified DNA products reacted with the probe. Moreover, compared to the spectrum of the probe, the difference was a red-shift caused by the DNA product.
For the single-stranded DNA (ss-DNA) molecule detection, owing to the low molecular weight and low wavelength sensitivity of the LSPR, the DNA detection limit and specificity have to be improved. A widely used method to improve the specificity is to use the sandwich-type (capture DNA probe–target DNA–reporter DNA probe) method. To enhance the detection limit, the reporter DNAs are usually conjugated with gold nanoparticles.39–41 On the other hand, methylation of DNA can also be used to improve the microfluidic-based SPR detection.31,32 In this study, we used the low-cost hot-embossing nanoimprint method to fabricate a 97 nm wide, 500 nm period nanowire array on the plastic substrate. After coating a 50 nm gold film on the nanowire array, we can generate the Fano-like resonance on the gold surface. Such Fano resonance is the coupling between the LSPR in the nanowires and the propagation SPR on the continuous gold surface. It has a narrower bandwidth and higher intensity sensitivity.26,27 From the experimental results, the immobilized probe on the capped gold nanowire array and the target molecule leads to a red-shift in the SPR resonance wavelength. Therefore, the reporter DNA probe is not required, and the unmethylated DNA can be detected during the test.
LMP1 DNA detection by the SPR nanowire chip
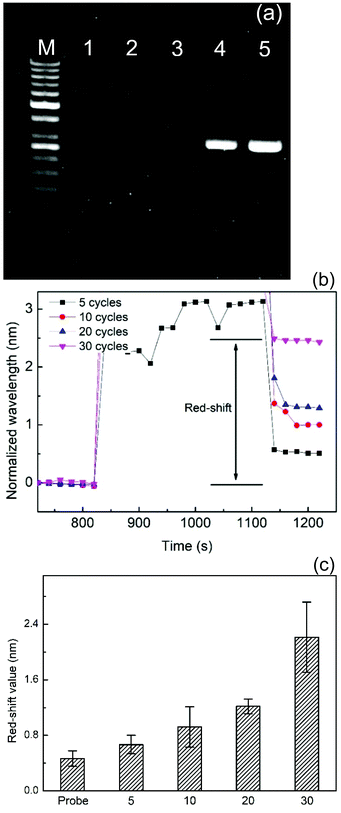
As mentioned earlier, the SPR chip combined with a microchannel can be used to detect DNA molecules without the reporter DNA probe. Therefore, the LMP1 DNA amplified by using the conventional PCR machine can also be used to understand the wavelength change mechanism and the detection limit of the SPR nanowire chip. To evaluate the limitation of the different LMP1 DNA amplification products, agarose gel electrophoresis based analysis was employed in this study. Agarose gel electrophoresis of 4 different PCR thermal cycles (5, 10, 20, and 30 cycles) of the LMP1 DNA amplification is shown in Fig. 5(a). Lane M was the DNA size marker, lane 1 was the negative control, and lanes 2–5 denoted the LMP1 DNA amplification cycle numbers of 5, 10, 20, and 30, respectively. The experimental results indicated that the fluorescence band was not clearly visible at the cycle numbers of 5 and 10 (lanes 2 and 3). However, the fluorescence band can be easily observed at the PCR cycle numbers of 20 and 30 (lanes 4 and 5).The kinetic binding analysis of 4 different PCR thermal cycles of the LMP1 DNA amplification is shown in Fig. 5(b). In the figure, the red-shift of 4 different PCR amplified products was observed by using the spectral centroid method. The results indicated that the lower cycle numbers of the PCR products could be detected by the SPR nanowire chip. In Fig. 5(c), the experimental results demonstrated that the red-shift values were 0.67, 0.92, 1.22 and 2.21 nm for the PCR cycle numbers of 5, 10, 20, and 30, respectively. The red-shift values showed a linear increase from 5 to 30 cycles.
Furthermore, the SPR nanowire chip combined with a microfluidic device may ensure the detection of the PCR products at lower cycle numbers, i.e., 5 and 10, which could not produce a visible brightness band by the agarose gel electrophoresis method. The observed results, Fig. 5(a) and (c), were in good agreement with Nguyen et al.,33 Trinh et al.,34 and Zhang et al.42 In this study, the variations of the resonance wavelengths are within 1% for different chips. Moreover, we used the same primer ratio for the PCR process. However, Nagai et al.43 pointed out that the accurate value for the DNA concentration should be obtained from the optimal primer ratio. Zhang et al.42 also reported the high sensitivity of the DNA based on the powerful amplification and the excess primer removal. Because of the variation of the amplified DNAs, the measured SPR shift has a significant standard deviation.
Gold nanowire arrays have been applied to detect biomolecular interactions by using localized surface plasmon resonance (LSPR) in the nanowires. In this work, we applied the PCR method instead of the sandwich-type measurement. PCR enhances explicitly the number of DNA molecules for the target DNA. Therefore, it does not need the reporter DNA probes and the conjugation of gold nanoparticles. Using the PCR product and the proposed chip modified with the capture DNA probe, we can achieve DNA detection in a lower DNA copy number.
Commercial PCR machines only amplify specific DNAs. For detection, the widely used method is electrophoresis analysis. It needs additional fluorescent labeling to detect the DNAs. The electrophoresis process takes time and requires the preparation of running gels. In this work, the detection is performed by using the low-cost gold-coated nanowire chip. The chip was first modified with a capture DNA probe. When the amplified DNAs are captured explicitly by the DNA probe, the change in the surface refractive index results in a significant change in the SPR peak and this can be detected using a spectrometer. Since there is no requirement for DNA labeling, the detection is much simpler and quicker.
As shown in Fig. 5, the measurement sensitivity is better than the fluorescence electrophoresis method. Because the sensing chip has a low price and the measurement system is simple, the chip can also be used to develop a new portable device in the future. However, sample pretreatment is another crucial issue. There are some commercial kits for DNA sample pretreatment, which can provide good results in isolating the DNA. The extraction time of QuickExtrat™ DNA extraction solution is less than 5 min and it is easy to handle.9 After the sample treatment, the extracted DNA can be individually amplified using the PCR machine and then detected using the gold nanowire array chip modified with the capture DNA probe. It is noted that the specific DNA amplification can also be performed by using the PCR microfluidic chip.44
In other words, this work is promising for biosensing applications using the LMP1 gene. In comparison with the SPR nanowire chip with the microfluidic device, the red shift can be easily observed at the PCR cycle numbers of 5 to 30. SPR nanowire chip can detect lower cycle numbers of the DNA amplification due to probe and DNA specificity. To summarize this study, the SPR nanowire chip coated with the probe had a lower detection limit compared to agarose gel electrophoresis. Furthermore, our gold nanowire array sensing chip can be integrated into the PCR chip. The detection would be more convenient by directly injecting the extracted DNA into the on-chip PCR/SPR device in the future.
Conclusions
In this study, LMP1 gene detection was achieved by using a microfluidic device with a SPR chip consisting of a gold-capped nanowire array. A transmission-type SPR sensing platform with high sensitivity and low background noise was established to measure the SPR wavelength by the spectral centroid method. The SPR nanostructure with a 500 nm period can be used to detect DNA and genes without labeling. With the PCR amplification, the specificity and detection signals can be significantly improved. The current SPR nanowire chip is proved to ensure the detection of the PCR products at lower cycle numbers. In contrast, using the agarose gel electrophoresis method, the visible brightness band could not be produced. The SPR nanowire chip can detect lower cycle numbers of the DNA amplification due to probe and DNA specificity. The detection systems used in this study have the advantages of cost-effectiveness and good measurement and production repeatability. In summary, the results can assist in improving the biosensing of DNA-amplified products and in developing rapid detection devices with small-footprint nanostructured SPR chips. Meanwhile, the SPR chip can be integrated into the online PCR device platform in the future.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
Financial support (MOST 106-2221-E-002-127, 107-2221-E-002-131 and 107-2622-E-002-016-CC2) from the Ministry of Science and Technology of Taiwan is acknowledged.Notes and references
- M. L. Gulley and W. Tang, J. Mol. Diagn., 2008, 10, 279–292 CrossRef PubMed.
- T. Riedel, C. Rodriguez-Emmenegger, A. de los Santos Pereira, A. Bědajánková, P. Jinoch, P. M. Boltovets and E. Brynda, Biosens. Bioelectron., 2014, 55, 278–284 CrossRef CAS.
- X. Liu, J. Tang, M. Wang, Q. Ma and Y. Wang, Int. J. Mol. Sci., 2013, 14, 23922–23940 CrossRef.
- J. L. Ryan, H. Fan, S. L. Glaser, S. A. Schichman, N. Raab-Traub and M. L. Gulley, J. Mol. Diagn., 2004, 6, 378–385 CrossRef CAS.
- D. E. Tsai, L. Douglas, C. Andreadis, D. T. Vogl, S. Arnoldi, R. Kotloff, J. Svoboda, R. D. Bloom, K. M. Olthoff, S. C. Brozena, S. J. Schuster, E. A. Stadtmauer, E. S. Robertson, M. A. Wasil and V. N. Ahya, Am. J. Transplant., 2008, 8, 1016–1024 CrossRef CAS.
- M. N. Islam, S. Yadav, M. H. Haque, A. Munaz, F. Islam, M. S. A. Hossain, V. Gopalan, A. K. Lam and N. T. Nguyen, Biosens. Bioelectron., 2017, 92, 668–678 CrossRef.
- H. Nagai, K. Tomioka and S. Okumura, Appl. Biochem. Biotechnol., 2019, 187, 323–337 CrossRef CAS PubMed.
- S. K. Vashist, P. B. Luppa, L. Y. Yeo, A. Ozcan and J. H. T. Luong, Trends Biotechnol., 2015, 33, 692–705 CrossRef CAS.
- P. Kranzfelder, T. Ekrem and E. Stur, Mol. Ecol. Resour., 2016, 16, 353–363 CrossRef CAS PubMed.
- S. A. Maier, Plasmonics: Fundamentals and Applications, Springer Science, New York, 2007, pp. 188–192 Search PubMed.
- M. E. Stewart, C. R. Anderton, L. B. Thompson, J. Maria, S. K. Gray, J. A. Rogers and R. G. Nuzzo, Chem. Rev., 2008, 108, 494–521 CrossRef CAS.
- H. J. Lezec, A. Degiron, E. Devaux, R. A. Linke, L. Martin-Moreno, F. J. Garcia-Vidal and T. W. Ebbesen, Science, 2002, 297, 820–822 CrossRef CAS.
- J. N. Anker, W. P. Hall, O. Lyandres, N. C. Shah, J. Zhao and R. P. Van Duyne, Nat. Mater., 2008, 7, 442–453 CrossRef CAS PubMed.
- A. G. Brolo, R. Gordon, B. Leathem and K. L. Kavanagh, Langmuir, 2004, 20, 4813–4815 CrossRef CAS PubMed.
- K. A. Tetz, L. Pang and Y. Fainman, Opt. Lett., 2006, 31, 1528–1530 CrossRef PubMed.
- J. C. Yang, J. Ji, J. M. Hogle and D. N. Larson, Nano Lett., 2008, 8, 2718–2724 CrossRef CAS PubMed.
- K. L. Lee, C. W. Lee, W. S. Wang and P. K. Wei, J. Biomed. Opt., 2007, 12, 044023 CrossRef PubMed.
- K. L. Lee, S. H. Wu and P. K. Wei, Opt. Express, 2009, 17, 23104 CrossRef CAS PubMed.
- K. L. Lee and P. K. Wei, Appl. Phys. Lett., 2011, 99, 083108 CrossRef.
- D. Zhang, Y. Yan, Q. Li, T. Yu, W. Cheng, L. Wong, H. Ju and S. Ding, J. Biotechnol., 2012, 160, 123–128 CrossRef CAS PubMed.
- O. Tokel, U. H. Yildiz, F. Inci, N. G. Durmus, O. O. Ekiz, B. Turker, C. Cetin, S. Rao, K. Sridhar, N. Natarajan, H. Shafiee, A. Dana and U. Demirci, Sci. Rep., 2015, 5, 9152 CrossRef CAS PubMed.
- H. Shi, K. Nie, B. Dong, M. Long, H. Xu and Z. Liu, Chem. Eng. J., 2019, 361, 635–650 CrossRef CAS.
- K. L. Lee, C. C. Chang, M. L. You, M. Y. Pan and P. K. Wei, Sci. Rep., 2018, 8, 9762 CrossRef PubMed.
- J. Sambles, G. Bradbery and F. Yang, Contemp. Phys., 1991, 32, 173–183 CrossRef CAS.
- J. Homola, S. S. Yee and G. Gauglitz, Sens. Actuators, B, 1999, 54, 3–15 CrossRef CAS.
- M. Z. Mousavi, H. Y. Chen, S. H. Wu, S. W. Peng, K. L. Lee, P. K. Wei and J. Y. Cheng, Analyst, 2013, 138, 2740–2748 RSC.
- M. Z. Mousavi, H. Y. Chen, K. L. Lee, H. Lin, H. H. Chen, Y. F. Lin, C. S. Wong, H. F. Lin, P. K. Wei and J. Y. Cheng, Analyst, 2015, 140, 4097–4104 RSC.
- W. K. Yeung, H. Y. Chen, J. J. Sun, T. H. Hsieh, Z. M. Mansoureh, H. H. Chen, K. L. Lee, H. Lin, P. K. Wei and J. Y. Cheng, Analyst, 2018, 143, 4715–4722 RSC.
- K. L. Lee, M. L. You, C. H. Tsai, E. H. Lin, S. Y. Hsieh, M. H. Ho, J. C. Hsu and P. K. Wei, Biosens. Bioelectron., 2016, 75, 88–95 CrossRef CAS PubMed.
- J. Burger, C. Ruth, J. Woehrle, P. A. Meyer, N. B. Ammar, N. Kilb, T. Brandstetter, F. Proll, G. Proll, G. Urban and G. Roth, SLAS Technol., 2017, 22, 437–446 Search PubMed.
- A. De, W. Sparreboom, A. Berg and E. T. Carlen, Biomicrofluidics, 2014, 8, 054119 CrossRef PubMed.
- R. Kurita, H. Yanagisawa, K. Yoshioka and O. Niwa, Anal. Chem., 2015, 87, 11581–11586 CrossRef CAS PubMed.
- T. T. Nguyen, K. T. L. Trinh, W. J. Yoon and N. Y. Lee, Sens. Actuators, B, 2017, 242, 1–8 CrossRef CAS.
- K. T. L. Trinh, Y. Zhang and N. Y. Lee, Anal. Chim. Acta, 2018, 1040, 63–73 CrossRef CAS PubMed.
- K. L. Lee, T. Y. Wu, H. Y. Hsu, S. Y. Yang and P. K. Wei, Sensors, 2017, 17, 1548 CrossRef PubMed.
- K. L. Lee, H. Y. Hsu, M. L. You, C. C. Chang, M. Y. Pan, X. Shi, K. Ueno, H. Misawa and P. K. Wei, Sci. Rep., 2017, 7, 44104 CrossRef PubMed.
- Q. Guo, I. W. K. Tham, S. Lin, Y. Su, Z. Chen, J. Lin, L. Han, Q. Lin, J. Pan and J. J. Lu, Head Neck Oncol., 2012, 4, 81–87 Search PubMed.
- N. Cennamo, L. Zeni, E. Catalano, F. Arcadio and A. Minardo, Appl. Sci., 2018, 8, 1172 CrossRef.
- X. Hong and E. A. H. Hall, Analyst, 2012, 137, 4712–4719 RSC.
- Q. Wang, R. Liu, X. Yang, K. Wang, J. Zhu, L. He and Q. Li, Sens. Actuators, B, 2016, 223, 613–620 CrossRef CAS.
- N. Yaakov, Y. Chaikin, E. Wexselblatt, Y. Tor, A. Vaskevich and I. Rubinstein, Chem. – Eur. J., 2017, 23, 10148–10155 CrossRef CAS PubMed.
- D. Zhang, Y. Yan, Q. Li, T. Yu, L. Wang, H. Ju and S. Ding, J. Biotechnol., 2012, 160, 123–128 CrossRef CAS PubMed.
- H. Nagai, K. Tomioka and S. Okumura, Appl. Biochem. Biotechnol., 2019, 187, 323–337 CrossRef CAS PubMed.
- C. Zhang, J. Xu, W. Ma and W. Zheng, Biotechnol. Adv., 2006, 24, 243–284 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |