Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Hollow polymer dots: nature-mimicking architecture for efficient photocatalytic hydrogen evolution reaction†
Aijie
Liu
a,
Cheuk-Wai
Tai
 b,
Kateřina
Holá
a and
Haining
Tian
b,
Kateřina
Holá
a and
Haining
Tian
 *a
*a
aDepartment of Chemistry-Ångström Lab., Uppsala University, Box 523, SE 751 20, Sweden. E-mail: haining.tian@kemi.uu.se
bDepartment of Materials and Environmental Chemistry, Arrhenius Laboratory, Stockholm University, SE 106 91 Stockholm, Sweden
First published on 31st January 2019
Abstract
Mimicking nature is always beneficial for improving the performance of artificial systems. Artificial photosynthesis for hydrogen production is one of the examples, where we can derive significant inspiration from nature. In this study, polymer dots (Pdots) prepared using photoactive polymer PFODTBT and amphiphilic co-polymer under ultra-sonication exhibited a hollow structure mimicking a photosynthetic bacterial, which was highly beneficial for hydrogen evolution. A systematic study of this structure showed that the polymer shell acts as a biological membrane that maintains a slightly higher pH inside the cavity (ΔpH 0.4) compared to the bulk solution. More importantly, a fast proton diffusion across the porous polymer shell was detected. The photocatalytic activity of hollow nanostructure shows 50 times enhancement of initial hydrogen evolution reaction (HER) rate as compared to solid nanoparticles. Further optimization of the photocatalytic performance was achieved by verifying the decrease in Pdots size from 90 nm to 50 nm, showing a significant increase in the photocatalytic performance of the system. This study reveals nature-mimicking hollow Pdots with porous shells as can be a type of promising photocatalysts in the application of solar energy conversion and storage.
Introduction
Clean and renewable energy is highly required nowadays to meet the increasing global energy demand and prevent environmental pollution. Solar energy is one of the renewable energy resources showing great potential to replace fossil fuels and meet the worldwide energy demand. For this reason, the development of highly efficient and cost-effective photocatalysts in the application of converting solar energy into storable fuels is desirable.1,2 In nature, photosynthetic organelles have well-organized functional proteins between bilayers for photocatalytic functions to convert light energy into chemical energy.3,4 To date, bioinspired artificial photocatalysts have been widely studied; thus, highly efficient and robust photocatalyst systems for solar fuel production can be designed and synthesized based on brilliant natural photosynthesis strategies.5,6 To date, inorganic photocatalysts have been extensively studied owing to their long lifetime and high energy-conversion efficiency.7–10 However, owing to the low cost, low toxicity and facilely tunable band gaps, organic semiconductors have recently gained considerable attention in sustainable energy conversion fields.11–13Graphitic carbon nitride (g-C3N4), which was reported by Antonietti et al. for the first time in 2008, is one of the most popular organic photocatalysts for hydrogen generation.14 Recently, other semiconducting polymers, such as poly(azomethine) networks,15 poly[(9H-carbazole-2,7-diyl)-1,4-phenylene] and its derivatives,16 and covalent organic frameworks, have been investigated as promising photocatalyst systems.17–19 Coopers et al. have studied the importance of porous organic polymers for photocatalytic activities.20–23 However, most systems have been reported to suffer from low dispersibility and solubility of the conjugated polymers in water; therefore, certain amount of organic solvent is required for the photocatalytic experiments. Our previous study revealed that organic conjugated polymer dots (Pdots) based on poly[(9,9′-dioctylfluoreny-2,7-diyl)-co-(1,4-benzo-(2,1′,3)thiadiazole)] (PFBT) with excellent water dispersibility could dramatically improve photocatalytic performance.24 Experimental and theoretical studies of organic Pdots with various polymer structures suggested that benzothiodiazole (BT) units in polymers play an essential role in the photocatalytic process. From these polymers, the polymer PFODTBT25 with extra thiophene units, compared to the polymer PFBT, yielded impressive rates of hydrogen evolution reactions (HERs).26 Taking advantages of the tunable polymer back-bone for efficient photocatalysts, a metal complex-linked conjugated polymer formed Pdots reached a satisfactory hydrogen evolution rate.27 Most recently, Kosco et al. reported that residual Pd in polymers should work as a co-catalyst for photocatalysis.28
Undoubtedly, the structure of polymers has a significant effect on photocatalytic performance. A majority of previous studies have been focusing on the modification of polymer backbones at the molecular level. However, excellent photocatalytic performance can be determined by finely tuning the nanostructures of the used catalysts.29,30 We have previously shown that Pdots exhibited excellent HER as compared to its pristine polymer.24 In addition to the polymer backbone, properties that facilitates the photocatalytic activity,26 nano-sized Pdots with good water dispersibility and large hydrophilic surface area might contribute to enhanced performance. However, the study of linearly conjugated polymer-based porous (or hollow) nanostructures in photocatalysis is almost an uncharted chapter. To better understand the role of Pdots' morphology in photocatalysis, PFODTBT-based Pdots were used as a model system in this study (see Scheme 1). PFODTBT-based Pdots possess a hollow nanostructure under ultrasonication with a polymer shell mimicking the membrane of photosynthetic bacteria, which was integrated using photocatalytic conducting polymer and amphiphilic PEG–COOH–grafted polystyrene (PS). After a systematic study of physiochemical environment of polymer vesicles, the results revealed an existence of a small pH gradient between the inside and outside of the vesicles. Note that hollow nanostructures remained stable over a large pH range, and nanoscale porous polymer shell containing hydrophilic channels allows rapid proton diffusion, which benefits photocatalytic proton reduction. We also proved that production limited reaction can be mostly reactivated by a simple degassing method. In addition, both photocatalytic activity and lifetime were optimized by tuning the Pdots size.
Results and discussion
Pdots preparation and characterization
In this study, we selected conducting polymer PFODTBT as a model polymer since it showed a high photocatalytic activity in our previous work.26 Moreover, a modified method was used to prepare Pdots, as reported in previous publications.24,26,31 The molecular structure of the conducting polymer PFODTBT and the surface stabilizer PEG–COOH–grafted polystyrene (PS–PEG–COOH) are shown in Scheme 1 (left bottom). Both PFODTBT and PS–PEG–COOH were dissolved in tetrahydrofuran (THF). This polymer mixture was then added into pure water under ultrasonication. After a few minutes, a clear mixture solution was observed, which formed well-dispersed organic droplets (Fig. S1b†). THF was then slowly removed with continuous argon (Ar) stripping under ultra-sonication (Experimental detail in method).The morphology of the Pdots was analysed by scanning transmission electron microscopy (STEM), as shown in Fig. 1. Interestingly, hollow nanostructures with a fine controlled size of the PFODTBT Pdots were observed in this study. The nanostructure of Pdots was formed by self-assembled small domains (or primary particles), which were composed of particles of diameters 1–3 nm (Fig. 1b), resulting in a nanoscale porous polymer shell with a number of hydrophilic channels, which may allow efficient proton diffusion. The average thickness of the vesicle shell was 6 nm, which was surprisingly close to the lipid bilayer thickness for cell membranes.32 Fig. S1† shows the possible mechanism for hollow Pdots formation.
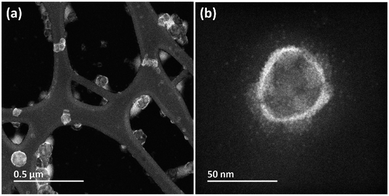 | ||
| Fig. 1 STEM images of polymer nanostructures: (a) HAADF-STEM image with low magnification; (b) HAADF-STEM image with high magnification. | ||
Physiochemical environment of hollow polymer vesicles
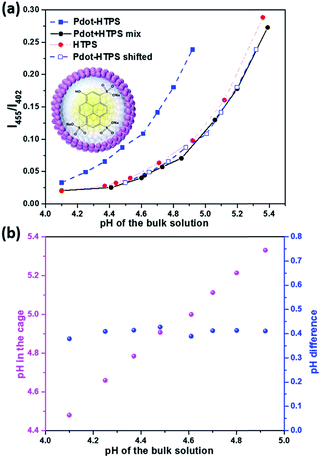
Hollow nanostructures are highly favourable for catalytic reactions due to the nature of the structures allowing much more reactive sites and efficient mass transport for both reactant agents and products in and out of the catalytic system.33,34 In addition, the confinement effect of the hollow sphere highly benefits catalytic performance, which has been recognized and therefore has attracted considerable interests in recent studies.35–39 A confined catalytic reaction environment could differ from the bulk solution since the confinement effect could change properties of aqueous phase in nanospace. In particular, cavities in Pdots could have different local environment, such as pH, which can be key for catalytic reactions.35,40–42 H+/OH− flux and pH gradient in block-co-polymer self-assembled vesicles have been studied recently.43,44 Thus, in order to understand the role of hollow structure of Pdots for photocatalytic processes for the first time, sufficient understanding of the physiochemical environment inside Pdots is necessary. To detect pH condition inside Pdots as well as to study proton diffusion across a Pdots' membrane, a well-known fluorometrical pH probe, water soluble 8-hydroxypyrene-1,3,6-trisulfonic acid trisodium salt (HTPS) was used and encapsulated in polymer vesicles (denoted as Pdot-HTPS).45,46 HTPS has a highly sensitive pH-dependent absorption shift, allowing ratiometric measurements by taking a ratio of fluorescence intensity at emission of λ = 515 nm with excitation at λ = 455 nm and λ = 402 nm.47 Moreover, HTPS does not permeate through membranes,43 which results in stable Pdots-HTPS hybrid nanoparticles (NPs) suitable for analysis. Encapsulation of HTPS inside the Pdots was performed by a method similar to the preparation of Pdots. Instead of pure water, 0.5 mM of HTPS aqueous solution was used. After the complete removal of THF, HTPS-encapsulated polymer vesicles were formed. In order to get rid of the non-encapsulated HTPS, a dialysis membrane with molecular weight cut-off (MWCO) of 20 kDa was used to purify the Pdot-HTPS colloids (Fig. S2†). Dynamic light scattering (DLS) analyses show that the average size of Pdot-HTPS NPs around 90 nm (see Fig. S3†). We were unable to get PFODTBT Pdots of sizes smaller than 90 nm in the presence of HTPS probably because the self-assembly HTPS in Pdots increases the volume of the nano-particles.In order to study if the pH condition within the Pdots is different from that in the bulk environment, different pH values of bulk solution with 0.2 M of acetic acid were adjusted by adding a certain amount of NaOH. Acetic acid used in the system had the same concentration as that of ascorbic acid used in the photocatalytic reactions to avoid side effects, such as hydrogen production and diffusion, during the analysis. Since photocatalytic reaction was performed at the optimal pH of 4,48 pH environment inside the Pdots in the range of pH 4 to pH 5.5 was studied. A standard calibration curve was generated with free HTPS and Pdots (avg. 90 nm) mixture (denoted as Pdot + HTPS mix), as shown in Fig. 2a (black dots), in which X axis represents the pH values and Y axis is the ratio of fluorescence intensities of maximum emission with excitation at 455 nm (I455) and 402 nm (I402). The curve of Pdot + HTPS mix almost overlaps with the curve of bare HTPS, proving that there was no significant interaction between free HTPS and Pdots. However, the pH curve of Pdot-HPTS NPs (Fig. 2a, blue square) is located differently from the previous two samples, indicating that the pH inside the Pdots is indeed different from the bulk solution. Interestingly, a similar phenomenon was found for the nanosized viral protein cage in a certain pH range, which contains pores on the protein shell and allows ions to freely diffusing in and out.47 By shifting the Pdot-HTPS pH response, it can be noted that it follows the same trend as free probes (see empty blue square in Fig. 2a). The internal pH was then calculated accordingly: pH inside polymer vesicle = pH of bulk solution + shift (ΔpH). The internal pH of Pdots was a bit higher than the pH of the bulk solution (Fig. 2b, purple dots). Interestingly, ΔpH remained constant at 0.4, as shown in Fig. 2b (blue dots).
Unlike block-co-polymer assembled polymer vesicles or liposomes with a hydrophobic interlayer, which are able to create large pH gradient across the (polymer) membrane,41 both slow and quick proton diffusions were observed depending on the polymer structure assembly.46 In this study, PEG–COOH–grafted PS polymer and PFODTBT conducting polymer-integrated Pdots showed a small pH gradient and rapid proton diffusion for the whole Pdots' shell. pH inside Pdots response along with the bulk solution and a rapid proton diffusion could be directly observed by the visible eye depending on the color change of Pdot-HTPS. In order to give a clear insight of the fast proton diffusion, fluorescence signals of HTPS were kinetically recorded and presented in Fig. 3. Both encapsulated HTPS and free HTPS were studied and compared, as shown in Fig. 3a and b (Detail information see ESI Section 2c†), and both initial and final pH of bulk solution measured by a pH meter. Again, pH probe within the vesicle (Fig. 3a) shows a phenomenon similar to that of a free probe (Fig. 3b), a sudden increase in proton inside vesicles was observed immediately after adding a solution of HCl (see enlarged plots in Fig. 3a). This behaviour was almost same as that observed for free HTPS (see enlarged plots in Fig. 3b). Note that porous shells with hydrophilic channels contribute to this fast proton diffusion, which also benefits photocatalytic proton reduction in our study.
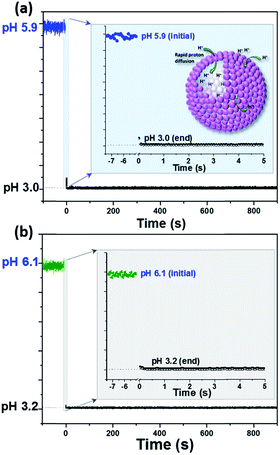 | ||
| Fig. 3 Kinetic monitoring of pH change in (a) Pdot-HTPS and (b) free HTPS, respectively, under experimental condition of 1 mM phosphate buffer, 3 mL in total, add 2.5 μL of 2 M HCl at t = 0. | ||
To exclude the fast pH probe signal change for Pdot-HTPS is caused by leakage in HTPS during the experiment, we first measured the particle stability of Pdot-HTPS hybrid NPs under acidic conditions (pH 3.0), nearly neutral conditions (pH 6.8) and alkaline conditions (pH 9.2) with DLS measurement. As shown in Fig. S7,† particles are stable over a large pH range. Pdot-HTPS hybrid NPs after proton diffusion experiment were then purified using a dialysis cassette with MWCO 20 kDa. UV-vis analysis indicates no leakage of HTPS molecules from Pdots by comparing absorption of Pdot-HTPS before and after experiments (as shown in Fig. S8†). No significant change was found from the absorption spectra of Pdot-HTPS solution before and after proton diffusion experiment. This proves that the Pdot-HTPS NPs are stable. Smaller primary particles self-assembled vesicles with porous shell may offer more mass transport channels, which contributes to rapid proton transport observed in our study.
Photocatalytic experiments of hollow structures
Hollow structures play an important role for high hydrogen generation performance. However, photocatalytic activity of the studied system dramatically slowed down after 2 h of photocatalytic reaction. Hydrogen bubbles may be generated during the photocatalytic process and might be formed both inside and outside the polymer vesicles that blocked photocatalytic active sides; therefore, a photocatalytic process was terminated when the amount of hydrogen in or around the hollow structure reached to a certain point. Detailed hydrogen bubble formation in the hollow polymer vesicles is unclear since there are only few theoretical studies about nanobubble nucleation and formation for nanochannels or nanospaces.49–51 Although there is a high possibility that nanobubbles might be generated on Pdots with the assistance of remaining free amphiphilic surfactant in solution,52 Pdot photocatalysts can be reactivated for hydrogen generation by degassing the system with Ar. This proves that the deactivation process is actually caused by generated hydrogen. Thus, this recycling process can be repeated many times, although the amount of hydrogen generation in solution shows a decrease with repeating cycles. The first cycle is maintained at 80% of the initial run and it drops to 47% after the fifth run (Fig. 4). The decrease in hydrogen generation is primarily caused by nanoparticle aggregation during the degassing procedure and degradation of PFODTBT polymer over reaction time.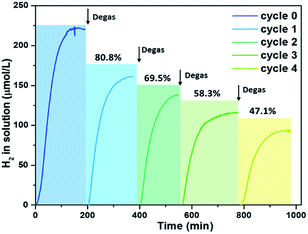 | ||
| Fig. 4 Recycling experiment to prove channels could be a problem for hydrogen diffusion, Pdots (70 nm) at PFODTBT concentration of 23 μg mL−1, 0.2 M ascorbic acid, and pH 4. | ||
Effect of size and morphology on photocatalysis
Although a production limited reaction can be reactivated using a facile degassing method, this procedure is tedious. As shown in Fig. S12a,† in order to improve the photocatalytic performance of photocatalysts in the first run (cycle 0) and enhance the production diffusion from polymer vesicles, Pdots with sizes of 50 nm, 70 nm and 90 nm were prepared and studied. Here, as shown in Fig. S12b,† both HER rates and total amount of hydrogen generation in the first photocatalysis run were compared and studied to investigate the effect of particles size on photocatalytic activity. HER rates of Pdots with various sizes were monitored using hydrogen sensors. Fig. 5 presents the kinetic study of the hydrogen evolution of Pdots of various sizes. The photocatalytic reaction time lasted for 100 min for 90 nm Pdots with eventual H2 concentration in a solution of 92.5 μmol L−1 (Fig. 5, blue line) and for 120 min for 70 nm Pdots with a final H2 concentration in a solution of 222.4 μmol L−1 (Fig. 5, red line). For 50 nm Pdots, H2 reached a level close to that for maximum H2 solubility in aqueous conditions (Fig. 5, black line), which is difficult to be measured accurately using hydrogen sensors. Therefore, as shown in Fig. S13,† H2 in headspace was measured using gas chromatography (GC), and continuous H2 production was observed in this system, with reactions lasting up to 10 h. Notably, for 50 nm Pdots, visible hydrogen bubbles were observed after 2 h of photocatalytic reaction and recorded on video, which is provided separately in the ESI.† The initial HER rates are 6.3 ± 0.1 mmol g−1 h−1, 8.1 ± 1.4 mmol g−1 h−1 and 22.6 ± 0.3 mmol g−1 h−1; and the eventual hydrogen production are 26.5 ± 0.2 mmol g−1 (2.6 mmol g−1 h−1), 46.4 ± 0.3 mmol g−1 (4.6 mmol g−1 h−1) and 181.6 ± 1.2 mmol g−1 (18.1 mmol g−1 h−1) for Pdots with 90 nm, 70 nm and 50 nm, respectively. Decreasing the particle size highly increased the total surface area, which highly enhanced mass transport and light capture by light scattering in the hollow structure. This further improved the photocatalytic activity and photocatalyst lifetime in one photocatalytic run. To highlight the importance of Pdots' hollow structure in photocatalysis, solid Pdots were prepared (Fig. S14†), which showed a much lower photocatalytic activity (Fig. 5, purple dash line), e.g., 0.4 mmol g−1 h−1 were observed (enlarged image shown in Fig. S15†), showing about 50 times decrease in the photocatalytic activity as compared to hollow Pdots with 50 nm. Note that hollow Pdots as photocatalysts have advantages, such as large light harvesting, high specific surface area and spatial anisotropic charge separation,53–57 which significantly enhances photocatalytic activity of the hollow structure.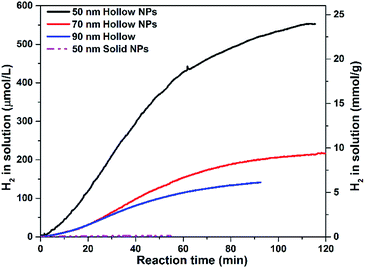 | ||
| Fig. 5 Kinetic record of hydrogen generation of hollow Pdots with sizes of 50 nm, 70 nm and 90 nm and solid NPs with 50 nm (purple dashed line). | ||
Conclusions
In conclusion, a nature-mimicking artificial photocatalyst, light-harvesting conducting polymer dots based on PFODTBT polymer with hollow nanovesicles was obtained. In order to have better understanding of the properties of hollow Pdots, we studied pH inside the polymer vesicle as well as proton diffusion across vesicle shells. The results show that there is a pH gradient between the inner space of the Pdots and bulk solution, and a slightly higher pH was found inside the vesicles. The pH difference was always remained at 0.4. Most importantly, a rapid proton diffusion through Pdots shell was detected, which should benefit its outstanding photocatalytic activity. Furthermore, as compared to solid NPs, 50 times enhancement in initial HER rate for hollow structures was observed. Excitingly, both hydrogen evolution rate and total hydrogen production were dramatically improved when particle size decreased to 50 nm. Moreover, visible hydrogen bubbles released from the solution could be observed by naked eyes. We believe that this work could give a new insight into conjugated polymer-based photocatalysts in the application of solar energy conversion and storage. In particular, the hollow polymer vesicle structures with some interesting properties found will aid in design and preparation of efficient polymer-based nano-photocatalyst for solar fuel production.Experimental
Materials
Semiconducting polymer PFODTBT polymer was purchased from Solaris. The co-polymer, polystyrene grafted with ethylene oxide and carboxyl groups (PS–PEG–COOH, back bone chain MW 8500, graft chain MW 4600, total chain MW 36500), was purchased from Polymer Source Inc., Canada. 8-Hydroxypyrene-1,3,6-trisulfonic acid trisodium salt (HPTS), ascorbic acid (AA) and other chemical reagents were purchased from Sigma-Aldrich and used as-received unless indicated otherwise. All experiments and measurements were performed at room temperature unless indicated otherwise.Preparation of hollow polymer vesicles
For the preparation of well-dispersed hollow polymer vesicles, we followed a modified method according to literature.26 Because of the low solubility of PFODTBT, PFODTBT was dissolved in tetrahydrofuran (THF) at a concentration of 50 μg mL−1, and PS–PEG–COOH was dissolved in THF at a concentration of 1.0 mg mL−1. Then, 100 μL of PS–PEG–COOH solution was added into 10 mL of PFODTBT solution under sonication (or magnetic stirring), and the mixture was stirred for 0.5 h. Then, this mixture was add into 20 mL of distilled water dropwise in an ultrasonic bath, yielding a clear and dark red solution after 5 min of sonication. Argon stripping was then performed in order to remove THF; moreover, ultrasonication was performed for 10 min every 10 min, 4 h in total (2 h of sonication in total; ultra-sonication: 20 kHz, Fisherbrand™ S-Series). Since the temperature might affect the structure and size of polymer particles, temperature of the sonication bath was maintained at room temperature during preparation. After completely removing THF, the colloidal solution was filtered through a 0.45 μm syringe filter before proceeding with other analysis. All samples were prepared using the same method otherwise noted. Pdots with different sizes were prepared by varying the initial concentration of PFODTBT. Fig. S6† shows concentration-dependent particle sizes. Final PFODTBT concentration in Pdots colloid were determined as follows. An aqueous solution of Pdots was freeze-dried first, THF was added to completely dissolve PFODTBT, and then analyzed using UV-vis spectroscopy to determine the concentration.Preparation of HTPS encapsulated Pdots
HTPS encapsulated Pdots (denote as Pdot-HTPS) were prepared using a similar method as that for Pdots. Rather than distilled water, 0.5 mM of HPTS aqueous solution was used to mix with the polymer solution. To remove the non-encapsulated dye molecules, the colloidal solution was dialyzed using a dialysis membrane with a molecular weight cutoff (MWCO) of 20 kDa against distilled water for 2 days. Note that the distilled water was changed four times a day. The remaining HPTS outside the dialyzed membrane was analyzed through UV-vis spectroscopy. Result shows that less than 0.8% of HPTS molecules remained in the colloidal solution, which is a negligible amount for further study (Fig. S2b†). For long-term storage (longer than a week), samples were purified again with a dialyze cassette with MWCO (20 kDa) before use.Dynamic light scattering (DLS) measurements
Hydrodynamic diameter was measured by Zetasizer Nano-S from Malvern Instruments Nordic AB. Average data was obtained from at least five runs of measurements.Hydrogen generation
Hydrogen in solution was detected by using a Unisense microsensor, while hydrogen in headspace was measured using Gas Chromatography (PerkinElmer Clarus 500, using argon (Ar) as carrier gas). A typical measurement was performed as follows. The reaction cuvette was filled with 3 mL of polymer vesicle solution containing 0.2 M of ascorbic acid and sealed with a septum. As studied in other literatures, pH of the mixture was adjusted to pH 4 with 1.0 M NaOH in order to give the maximum hydrogen output.24,26,48 The resultant reaction mixture was degassed with Ar bubbling before illumination. A LED PAR38 lamp (17 W, 5000 K, Zenaro Lighting GmbH, λ > 420 nm) used as the light source. The light intensity illuminated on the active area of the sample was 50 mW cm−2, which was measured by a pyranometer (CM11, KIPP&ZONEN, DELET/HOLLAND). The LED light source basically had a similar intensity as that for standard 1 sun condition between 420 nm and 750 nm.(Scanning) Transmission Electron Microscope (STEM)
A JEOL JEM 2100 transmission electron microscope (TEM) operated at 200 kV and equipped with a Schottky field emission gun and a JEOL energy dispersive X-ray spectrometer was used in this study. High-angle annular dark-field (HAADF-STEM), also known as Z-contrast imaging, and bright-field (BF-STEM) were simultaneously acquired by a JEOL ADF and Gatan BF detector, respectively. A drop (10 μL) of samples was casted on a copper grid with holey carbon supporting films and left for 10 min before the excess solvent was removed with filter paper.Gas Chromatography (GC) Measurements
Gas analysis was performed using a HPR-20 benchtop gas analysis system (HIDEN Analytical) using Ar as a carrier gas.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Wenner-Gren Foundation (UPD2917-0209) and Swedish Energy Agency (44641-1). Yocefu Hattori, Lei Tian, Dr Bo Xu and Dr Jing Huang were highly acknowledged for helpful discussion.Notes and references
- J. Rongé, T. Bosserez, D. Martel, C. Nervi, L. Boarino, F. Taulelle, G. Decher, S. Bordiga and J. A. Martens, Chem. Soc. Rev., 2014, 43, 7963 RSC
.
- W. Vijselaar, P. Westerik, J. Veerbeek, R. M. Tiggelaar, E. Berenschot, N. R. Tas, H. Gardeniers and J. Huskens, Nat. Energy, 2018, 3, 185 CrossRef CAS
.
- E. Kovács-Bogdán, J. Soll and B. Bölter, Biochim. Biophys. Acta Mol. Cell Res., 2010, 1803, 740 CrossRef PubMed
.
- K. Y. Lee, S.-J. Park, K. A. Lee, S.-H. Kim, H. Kim, Y. Meroz, L. Mahadevan, K.-H. Jung, T. K. Ahn, K. K. Parker and K. Shin, Nat. Biotechnol., 2018, 36, 530 CrossRef CAS PubMed
.
- D. Gust, T. A. Moore and A. L. Moore, Acc. Chem. Res., 2009, 42, 1890 CrossRef CAS PubMed
.
- D. R. Whang and D. H. Apaydin, ChemPhotoChem, 2018, 2, 148 CrossRef CAS
.
- S. Ye, R. Chen, Y. Xu, F. Fan, P. Du, F. Zhang, X. Zong, T. Chen, Y. Qi, P. Chen, Z. Chen and C. Li, J. Catal., 2016, 338, 168 CrossRef CAS
.
- Y. Imanaka, T. Anazawa, T. Manabe, H. Amada, S. Ido, F. Kumasaka, N. Awaji, G. Sánchez-Santolino, R. Ishikawa and Y. Ikuhara, Sci. Rep., 2016, 6, 35593 CrossRef CAS PubMed
.
- M. Watanabe, Sci. Technol. Adv. Mater., 2017, 18, 705 CrossRef CAS PubMed
.
- Y. Tachibana, L. Vayssieres and J. R. Durrant, Nat. Photonics, 2012, 6, 511 CrossRef CAS
.
- A. M. Kalsin, M. Fialkowski, M. Paszewski, S. K. Smoukov, K. J. M. Bishop and B. A. Grzybowski, Science, 2006, 312, 420 CrossRef CAS PubMed
.
- D. J. Martin, P. J. T. Reardon, S. J. A. Moniz and J. Tang, J. Am. Chem. Soc., 2014, 136, 12568 CrossRef CAS PubMed
.
- A. S. Weingarten, R. V. Kazantsev, L. C. Palmer, D. J. Fairfield, A. R. Koltonow and S. I. Stupp, J. Am. Chem. Soc., 2015, 137, 15241 CrossRef CAS PubMed
.
- X. Wang, K. Maeda, A. Thomas, K. Takanabe, G. Xin, J. M. Carlsson, K. Domen and M. Antonietti, Nat. Mater., 2008, 8, 76 CrossRef PubMed
.
- M. G. Schwab, M. Hamburger, X. Feng, J. Shu, H. W. Spiess, X. Wang, M. Antonietti and K. Müllen, Chem. Commun., 2010, 46, 8932 RSC
.
- D. J. Woods, R. S. Sprick, C. L. Smith, A. J. Cowan and A. I. Cooper, Adv. Energy Mater., 2017, 7, 1700479 CrossRef
.
- T. Banerjee, K. Gottschling, G. Savasci, C. Ochsenfeld and B. V. Lotsch, ACS Energy Lett., 2018, 3, 400 CrossRef CAS PubMed
.
- X. Jiang, P. Wang and J. Zhao, J. Mater. Chem. A, 2015, 3, 7750 RSC
.
- V. S. Vyas, F. Haase, L. Stegbauer, G. Savasci, F. Podjaski, C. Ochsenfeld and B. V. Lotsch, Nat. Commun., 2015, 6, 8508 CrossRef CAS PubMed
.
- L. Stegbauer, K. Schwinghammer and B. V. Lotsch, Chem. Sci., 2014, 5, 2789 RSC
.
- R. S. Sprick, J.-X. Jiang, B. Bonillo, S. Ren, T. Ratvijitvech, P. Guiglion, M. A. Zwijnenburg, D. J. Adams and A. I. Cooper, J. Am. Chem. Soc., 2015, 137, 3265 CrossRef CAS PubMed
.
- L. Li, Z. Cai, Q. Wu, W.-Y. Lo, N. Zhang, L. X. Chen and L. Yu, J. Am. Chem. Soc., 2016, 138, 7681 CrossRef CAS PubMed
.
- X. Wang, L. Chen, S. Y. Chong, M. A. Little, Y. Wu, W.-H. Zhu, R. Clowes, Y. Yan, M. A. Zwijnenburg, R. S. Sprick and A. I. Cooper, Nat. Chem., 2018, 10, 1180 CrossRef CAS PubMed
.
- L. Wang, R. Fernández-Terán, L. Zhang, D. L. A. Fernandes, L. Tian, H. Chen and H. Tian, Angew. Chem., Int. Ed., 2016, 55, 12306 CrossRef CAS PubMed
.
- S. Admassie, O. Inganäs, W. Mammo, E. Perzon and M. R. Andersson, Synth. Met., 2006, 156, 614 CrossRef CAS
.
- P. B. Pati, G. Damas, L. Tian, D. L. A. Fernandes, L. Zhang, I. B. Pehlivan, T. Edvinsson, C. M. Araujo and H. Tian, Energy Environ. Sci., 2017, 10, 1372 RSC
.
- P.-J. Tseng, C.-L. Chang, Y.-H. Chan, L.-Y. Ting, P.-Y. Chen, C.-H. Liao, M.-L. Tsai and H.-H. Chou, ACS Catal., 2018, 8, 7766 CrossRef CAS
.
- J. Kosco, M. Sachs, R. Godin, M. Kirkus, L. Francas, M. Bidwell, M. Qureshi, D. Anjum, J. R. Durrant and I. McCulloch, Adv. Energy Mater., 2018, 1802181 CrossRef
.
- M. Xiao, Z. Wang, M. Lyu, B. Luo, S. Wang, G. Liu, H.-M. Cheng and L. Wang, Adv. Mater., 2018, 1801369 CrossRef PubMed
.
-
B. Lytle and M. H. Gerardi, in The Biology and Troubleshooting of Facultative Lagoons, 2015, ch. 9, p. 73, DOI:10.1002/9781118981771.ch9
.
- C. Wu, T. Schneider, M. Zeigler, J. Yu, P. G. Schiro, D. R. Burnham, J. D. McNeill and D. T. Chiu, J. Am. Chem. Soc., 2010, 132, 15410 CrossRef CAS PubMed
.
- K. Mitra, I. Ubarretxena-Belandia, T. Taguchi, G. Warren and D. M. Engelman, Proc. Natl. Acad. Sci. U.S.A., 2004, 101, 4083 CrossRef CAS PubMed
.
- X. Xia, Y. Wang, A. Ruditskiy and Y. Xia, Adv. Mater., 2013, 25, 6313 CrossRef CAS PubMed
.
- L. Yu, H. B. Wu and X. W. D. Lou, Acc. Chem. Res., 2017, 50, 293 CrossRef CAS PubMed
.
- H. Li, J. Xiao, Q. Fu and X. Bao, Proc. Natl. Acad. Sci. U.S.A., 2017, 114, 5930 CrossRef CAS PubMed
.
- A. Liu, C. H. H. Traulsen and J. J. L. M. Cornelissen, ACS Catal., 2016, 6, 3084 CrossRef CAS
.
- H. Chen, K. Shen, Q. Mao, J. Chen and Y. Li, ACS Catal., 2018, 8, 1417 CrossRef CAS
.
- Q. Fu, W.-X. Li, Y. Yao, H. Liu, H.-Y. Su, D. Ma, X.-K. Gu, L. Chen, Z. Wang, H. Zhang, B. Wang and X. Bao, Science, 2010, 328, 1141 CrossRef CAS PubMed
.
- Y. Liu, J. Du, M. Yan, M. Y. Lau, J. Hu, H. Han, O. O. Yang, S. Liang, W. Wei, H. Wang, J. Li, X. Zhu, L. Shi, W. Chen, C. Ji and Y. Lu, Nat. Nanotechnol., 2013, 8, 187 CrossRef CAS PubMed
.
- F. Yang, D. Deng, X. Pan, Q. Fu and X. Bao, Natl. Polit. Sci. Rev., 2015, 2, 183 CrossRef CAS
.
- N. Bertrand, C. Bouvet, P. Moreau and J.-C. Leroux, ACS Nano, 2010, 4, 7552 CrossRef CAS PubMed
.
- C. Bonfio, E. Godino, M. Corsini, F. Fabrizi de Biani, G. Guella and S. S. Mansy, Nature Catalysis, 2018, 1, 616 CrossRef
.
- W. F. Paxton, D. Price and N. J. Richardson, Soft Matter, 2013, 9, 11295 RSC
.
- I. A. Chen and J. W. Szostak, Proc. Natl. Acad. Sci. U.S.A., 2004, 101, 7965 CrossRef CAS PubMed
.
- H.-J. Choi and C. D. Montemagno, Nano Lett., 2005, 5, 2538 CrossRef CAS PubMed
.
- S. Yu, T. Azzam, I. Rouiller and A. Eisenberg, J. Am. Chem. Soc., 2009, 131, 10557 CrossRef CAS PubMed
.
- S. J. Maassen, P. van der Schoot and J. J. L. M. Cornelissen, Small, 2018, 14, 1802081 CrossRef PubMed
.
- A. S. Weingarten, R. V. Kazantsev, L. C. Palmer, M. McClendon, A. R. Koltonow, A. P. S. Samuel, D. J. Kiebala, M. R. Wasielewski and S. I. Stupp, Nat. Chem., 2014, 6, 964 CrossRef CAS PubMed
.
- N. Arai, T. Koishi and T. Ebisuzaki, Phys. Rev. E, 2017, 96, 042802 CrossRef
.
- T. Koishi, S. Yoo, K. Yasuoka, X. C. Zeng, T. Narumi, R. Susukita, A. Kawai, H. Furusawa, A. Suenaga, N. Okimoto, N. Futatsugi and T. Ebisuzaki, Phys. Rev. Lett., 2004, 93, 185701 CrossRef CAS
.
- K. S. a. P. Keblinski, J. Chem. Phys., 2014, 141, 234508 CrossRef PubMed
.
- S. Li, Y. Du, T. He, Y. Shen, C. Bai, F. Ning, X. Hu, W. Wang, S. Xi and X. Zhou, J. Am. Chem. Soc., 2017, 139, 14277 CrossRef CAS PubMed
.
- C. C. Nguyen, N. N. Vu and T.-O. Do, J. Mater. Chem. A, 2015, 3, 18345 RSC
.
- S. Zhao, Y. Zhang, J. Fang, H. Zhang, Y. Wang, Y. Zhou, W. Chen and C. Zhang, ACS Sustain. Chem. Eng., 2018, 6, 8291 CrossRef CAS
.
- J. Sun, J. Zhang, M. Zhang, M. Antonietti, X. Fu and X. Wang, Nat. Commun., 2012, 3, 1139 CrossRef
.
- Z. Zhao, X. Wang, Z. Shu, J. Zhou, T. Li, W. Wang and Y. Tan, Appl. Surf. Sci., 2018, 455, 591 CrossRef CAS
.
- S. Ghosh, N. A. Kouamé, L. Ramos, S. Remita, A. Dazzi, A. Deniset-Besseau, P. Beaunier, F. Goubard, P.-H. Aubert and H. Remita, Nat. Mater., 2015, 14, 505 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ta12146j |
| This journal is © The Royal Society of Chemistry 2019 |