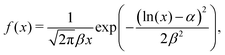Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
In vivo adhesion force measurements of Chlamydomonas on model substrates
Christian Titus
Kreis†
,
Alice
Grangier
and
Oliver
Bäumchen
 *
*
Max Planck Institute for Dynamics and Self-Organization (MPIDS), Am Faßberg 17, D-37077 Göttingen, Germany. E-mail: oliver.baeumchen@ds.mpg.de
First published on 6th March 2019
Abstract
The initial stages of biofilm formation at a surface are triggered by the surface association of individual microorganisms. The biological mechanisms and interfacial interactions underlying microbial adhesion to surfaces have been widely studied for bacteria, while microalgae remained rather unconsidered despite their technological relevance, e.g., in photo-bioreactors. We performed in vivo micropipette force measurements with the model organism Chlamydomonas reinhardtii, a unicellular eukaryotic microalga that dwells in liquid-infused soils and on moist rocks. We characterize the adhesion forces and dissect the influence of intermolecular interactions by probing the adhesion forces of single cells on different model substrates with tailored properties. Our experiments show that the flagella-mediated adhesion of Chlamydomonas to surfaces is largely substrate independent, enabling the cell to adhere to any type of surface. This universal adhesion mechanism allows the microalga to effectively colonize abiotic surfaces in their heterogeneous natural habitats. Our results reveal a dominant contribution of electrostatic interactions governing microalgal adhesion and suggest that flagella membrane processes may cause significant variations of the adhesive properties of the flagella.
Microorganisms can be found on many biotic and abiotic surfaces. Their natural habitats are rather diverse and the surfaces that microbes colonize are typically heterogeneous in their topography as well as in their chemical composition. The chemical compositions of the surface and the underlying substrate determine the interfacial interactions that cells experience in close proximity to a substrate. Indeed, understanding the intermolecular interactions that govern microbial surface colonization is imperative to develop physicochemical pathways for dealing with biofilm-related issues in natural and technological settings.
Microbial adhesion strategies have been extensively studied for bacteria due to their outstanding relevance in biomedical applications.1 Bacterial adhesion is generally mediated by membrane proteins and cellular appendages, like pili and fimbrae, that are often tailored to attach to extracellular material in a host or biofilm, but also enable adhesion to abiotic surfaces.2–6 Atomic force microscopy techniques have been widely employed to study the intermolecular interactions that mediate the adhesion of living bacteria to different substrates.7–9 In contrast to bacteria as well as other representatives of microbial life, e.g. slime molds,10,11 microalgae remained rather unconsidered so far.
Microalgae are photoactive, eukaryotic microorganisms that form the phytoplankton of salt and freshwater ecosystems, which contribute to the global nutrient cycles and represent the basis of food chains.12–15 Although they often live freely suspended in open water bodies, microalgae also colonize light-exposed surfaces in moist habitats, like soil, temporary pools, and streams. On surfaces, microalgae may form biofilms (often in symbiosis with bacteria), which represent a major contribution to biofouling in the aqueous environments of various industrial scenarios, like water cooling systems and ship hulls.16,17 Despite the fact that microalgae inherit profound ecological and technological importance, e.g. for the production of biofuels and drug components in photo-bioreactors, quantitative single-cell adhesion measurements are lacking and microalgal adhesion strategies remain elusive so far. Adhesion studies are limited to force measurements on the adhesive glycoproteins and nanofibres secreted by green algae and diatoms, respectively.18,19
The unicellular, bi-flagellated microalga Chlamydomonas reinhardtii is a model organism to study cellular processes, e.g. flagella biology, cilia-related diseases, and microbial motility.20,21Chlamydomonas shares many common features of flagellated microalgae, e.g. the circadian life cycle, sexual and asexual reproduction, and the fact that it can be found in a free-swimming (planktonic) as well as a surface-associated state. Its surface association is enabled by flagella-surface contacts, which are mediated by adhesive interactions between the flagella membrane glycoprotein (FMG-1B) and the surface.22 Furthermore, the adhesive contact enables the cell to move on the surface, called gliding motility,23 which is widely studied as a model for the dynamics of molecular motors and cellular force transduction.24,25
Chlamydomonas is equipped with a variety of different photoreceptors that may trigger specific biomolecular responses, e.g. its ability to sense the direction of light and adapt its flagella beat accordingly (phototaxis).26,27 In a previous study, we report on our discovery that the flagella-mediated adhesion of Chlamydomonas to surfaces can be reversibly switched on and off by light.28 We showed that the adhesion force of up to several nanonewton in white light is reduced to zero in red light conditions. Although we were able to identify light as a key requirement for the surface colonization, the intermolecular interactions that govern the adhesion of Chlamydomonas to surfaces remained elusive.
In this study, we quantify the adhesion forces of Chlamydomonas reinhardtii (SAG 11-32b) in vivo to a set of ultra-smooth model substrates in controlled environmental conditions. We dissect the fundamental intermolecular interactions underlying microalgal adhesion by tailoring the substrate properties, while the topography of the substrates remains unchanged. We perform single-cell in vivo force spectroscopy experiments29 with the same Chlamydomonas cell on different model substrates and provide a statistical comparison of the measured adhesion forces. Thereby, we systematically probe the effect of hydrophobic interactions, van der Waals interactions, and electrostatic interactions on microalgal adhesion.
1 Materials and methods
Substrate preparation and characterization
As substrates, we used non-functionalized as well as functionalized silicon (Si) wafers and magnesium oxide (MgO) substrates. The silicon wafers with native, thin SiO2-layer of approximately 1.7 nm thickness (Si native) and thermally grown, thick SiO2 layer (Si thick) of 150 nm thickness were obtained from Si-Mat (Kaufering, Germany). Magnesium oxide substrates were obtained from Sigma-Aldrich (Germany). In order to obtain hydrophobic substrates, we functionalized silicon wafers (type Si native) with self-assembling silane molecules featuring a CH3 tail group (octadecyltrichlorosilane, OTS, CAS 112-04-9) by following an established recipe.30In the experiments, we used small substrate pieces of approximately 6 mm × 2 mm that were cut from the bulk substrates. These substrate pieces were glued with polydimethylsiloxane (PDMS; Dow Corning, Midland, Michigan, USA; Sylgard® 184 silicon elastomer kit) to a stainless steel substrate holder. After attaching a pair of substrates to the holder, we immersed the substrate holder for three minutes in an ethanol (purity ≥ 99.9%, ROTISOLV® HPLC grade) ultrasonic bath in preparation for experiments. In order to quantitatively compare the adhesion force of the same cell on different substrates, we always attached two different substrates next to each other on the same substrate holder. In experiments featuring substrates cleaned with so-called ‘piranha solution’, the substrates were cleaned with ‘piranha solution’ and ethanol, respectively, before attaching them to the holder. The ‘piranha solution’ contained sulphuric acid (H2SO4, CAS 7664-93-9, 96.5%) and hydrogen peroxide (H2O2, CAS 7722-84-1, 30%, stabilised) at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The residues of the ‘piranha solution’ were carefully removed over a period of 90 min by rinsing the substrates with ultra-pure water (Millipore, Milli-Q®, 18.2 MΩ cm, <6 ppb total organic carbon content), the water being replaced at least four times in between.
1. The residues of the ‘piranha solution’ were carefully removed over a period of 90 min by rinsing the substrates with ultra-pure water (Millipore, Milli-Q®, 18.2 MΩ cm, <6 ppb total organic carbon content), the water being replaced at least four times in between.
We chose ultra-smooth substrates to dissect the influence of intermolecular interactions on microbial adhesion from the influence of substrate roughness and substrate stiffness. The root-mean-square (rms) surface roughness of the substrates was measured from 1 μm × 1 μm scans using atomic force microscopy (Bruker, Billerica, Massachusetts, USA; DimensionV) in contact mode with a cantilever featuring a nominal tip radius of 7 nm (Olympus Corporation, Tokyo, Japan; OMCL-AC160TS-W2). The Young modulus of the used substrates are on the order of tens of GPa, which is several orders of magnitude stiffer than the extracellular matrix in a biofilm or physiological environments and elastomers.
The surface energy was determined with a three-liquid method31 using ultra-pure water, Glycerol (CAS 56-81-5) and Bromonapthalene (1-bromonapthalene, CAS 90-11-9) as probe liquids. This method allows to determine the surface energy γ from the static contact angles of sessile droplets, which were determined from at least ten independent measurements (dataphysics, OCA).
The isoelectric point pH(I), i.e. the pH-value at which the substrate carries no mean net charge, is inferred from zeta-potential measurements from literature. The relevant substrate properties are summarized in Table 1.
Cell cultivation
Wild-type Chlamydomonas reinhardtii cells, strain SAG 11-32b, were grown axenically in Tris-acetate-phosphate (TAP) medium (Thermo Fisher Scientific) on a 12 h/12 h day-night cycle in a Memmert IPP 100Plus incubator. The daytime temperature was 24 °C with light intensity of 1 to 2 × 1020 photons per m2 per s; the nighttime temperature was 22 °C and the light intensity was reduced to zero. Experiments were performed with vegetative cells taken from the cultures in logarithmic growth phase during the daytime on the second to fourth day after incubation. A small amount of the culture, about 0.1 to 0.2 ml, was injected into the liquid cell to achieve a dilute suspension for force spectroscopy experiments in ambient conditions (24 to 26 °C).In vivo micropipette force spectroscopy
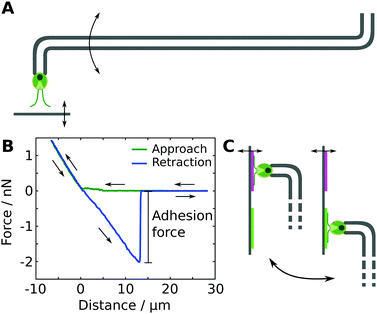
We studied the adhesion of individual living Chlamydomonas cells using micropipette force spectroscopy, following the measurement protocols described in our earlier work28,29 (see Fig. 1). Micropipette force spectroscopy is an experimental technique that employs the deflection of a force-calibrated micropipette cantilever to measure forces of living cells (see Fig. 1A), inspired by the measurement principle of atomic force microscopy techniques.29 We determine the deflection of the cantilever using high-resolution optical microscopy combined with an image auto-correlation analysis that features a sub-pixel resolution of the cantilever's deflection. The force sensors were calibrated using the added weight of a variable mass, such as a water droplet attached to a freely suspended micropipette in air, or a reference cantilever. The spring constants of the force sensors employed in this study varied between 0.2 to 1 nN μm−1, resulting in a force resolution of a few tens of piconewton.Each individual force–distance cycle consists of a substrate–cell approach, a time delay when the cell is in contact with the substrate, and the retraction of the substrate from the cell (see Fig. 1B). The substrate was moved with a constant speed of 1 μm s−1 and the time delay of 15 s resulted in a total cell-surface contact time of approximately 25 s. This contact time enables the cells to establish the gliding configuration on the substrate,28i.e. both flagella are spread out at an angle of approximately 180°, representing the natural flagella configuration of Chlamydomonas in contact with a substrate. After detachment of the cell from the substrate, the regular flagella beating is recovered after 2 to 15 seconds, as obtained from image sequences of the recovery process recorded at 400 fps. The time between two consecutive force–distance curves was about 60 to 90 seconds to ensure that the regular flagella beating was recovered. The loading rate during a force–distance cycle was 0.2 to 1 nN s−1, which is several orders of magnitudes lower than the loading rates in typical bacterial force spectroscopy experiments.35,36 Within this range of loading rates, the adhesion forces of Chlamydomonas were found to be independent of the loading rate.28
All experiments were performed in TAP medium as buffer solution at ambient conditions using white-light illumination. We grasped a Chlamydomonas cell with the micropipette force sensor in an optically controlled orientation and probed the adhesion of the Chlamydomonas flagella to the model substrates (see Fig. 1A). The cell body does not exhibit any adhesiveness.28 Before and after all experiments, we controlled the viability of the microalgae by monitoring the beating of their flagella or the pulsing vacuole at the cell apex.
Experimental procedure
To probe the influence of substrate properties on the adhesion force of Chlamydomonas, we performed experiments with the exact same cell on a pair of different substrates that were attached next to each other onto the same substrate holder (see Fig. 1C). On each of the two substrates, we carried out two sets of five individual force–distance curves, resulting in a total of ten measurements with the same cell on each substrate. Switching from one substrate to another involves manual repositioning using the micromanipulators, which typically takes between 1 to 5 minutes. The order of the four sets was varied randomly in order to avoid any potential systematic bias in the measured adhesion forces originating from the experimental procedure.2 Characterization of the adhesion force distributions
The results of adhesion force measurements of in total 119 Chlamydomonas cells are shown in Fig. 2. For each cell, we performed 10 individual force–distance experiments on the Si native reference substrate and calculate the mean adhesion force μ and the adhesion force spread, characterized by the standard deviation σ. The mean adhesion forces for different cells vary from almost zero up to 5 nN (see Fig. 2). For each individual cell, the adhesion forces from the 10 individual measurements have a relative standard deviation σr = μ/σ of a few tens of percent of the mean adhesion value (see inset of Fig. 2).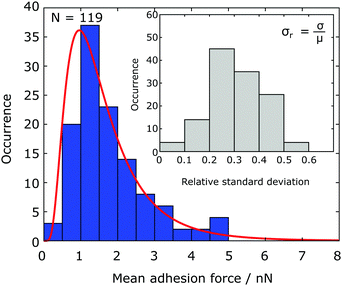 | ||
| Fig. 2 Statistical distribution of the mean adhesion forces for 119 Chlamydomonas cells. The solid red line represents the best fit of a lognormal distribution to the data (see Fig. 6 for a boxplot). Inset: For an individual cell, the relative standard deviation of the adhesion force is a few tens of percent of the mean value. | ||
The mean adhesion forces of 119 cells yields a distribution with mean of 1.77 nN, a median force of 1.48 nN, and a interquartile range of 1.11 nN (25th percentile: 1.10 nN, 75th percentile: 2.21 nN, see Fig. 6). The statistical distribution is in excellent agreement with a logarithmic normal distribution,
In our experiments, about 6% of all cells exhibited mean adhesion forces larger than approximately 5 nN on the Si native reference substrate. We classify these cells as outliers based on a definition using the interquartile range of the data set (differing by more than 2.5× the interquartile range from the 25th/75th quartile). The data obtained from these cells were excluded from any data analysis (see discussion for further details).
3 Force spectroscopy on model substrates with tailored properties
To dissect the intermolecular interactions mediating Chlamydomonas adhesion to abiotic substrates, we performed force–distance curves on four different substrate sets, each consisting of a pair of substrates that differ in their surface or subsurface properties. For each set of model substrates, we quantified and compared the adhesion forces using the exact same cells (see Fig. 1C).Surface energy
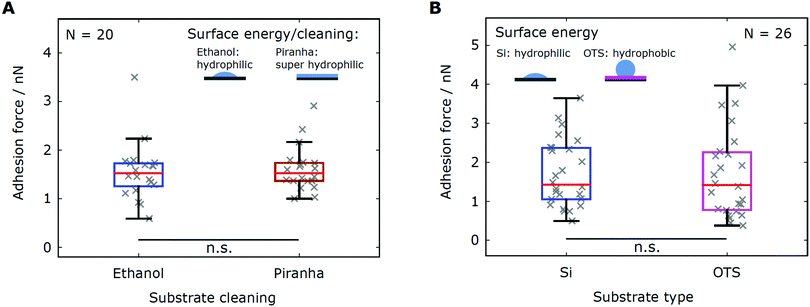
We varied the strength of the short-ranged hydrophobic interactions by performing force spectroscopy experiments on two sets of substrates exhibiting different surface energies. The first substrate set consisted of two pieces of the same silicon wafers that were cleaned in different ways. By cleaning the substrate with a piranha solution, we removed any organic residues from the surface and obtained a hydrophilic substrate (complete wetting, water contact angle θW ≤ 5°) with a high surface energy. The reference substrate was cleaned with ethanol, which yielded a rather moderate surface energy (see Table 1). The second substrate set contained the reference silicon substrate and a hydrophobic substrate with low surface energy, obtained by functionalizing a silicon wafer with a self-assembled silane monolayer (OTS).A direct comparison of the adhesion forces recorded on the first substrate set (N = 20 cells) does not show any influence of the substrate cleaning on cell adhesion (Fig. 3A). The characteristic statistical measures of both adhesion force distributions are found to be in excellent agreement (Table 2). Likewise, force–distance experiments on the second substrate set (N = 26 cells) yield comparable adhesion forces, which is evidenced by the characteristic values of the force distributions (Fig. 3B and Table 2). In summary, the adhesion force distributions obtained from experiments on substrates sets with different surface energies are consistent with each other.
![[F with combining macron]](https://www.rsc.org/images/entities/i_char_0046_0304.gif) as well as the median F0.5. The spread of a distribution is characterized by the 25th percentile F0.25 and the 75th percentile F0.75. The p-values are obtained from a Mann–Whitney U test; a p-value of p < 0.05 is considered significant
as well as the median F0.5. The spread of a distribution is characterized by the 25th percentile F0.25 and the 75th percentile F0.75. The p-values are obtained from a Mann–Whitney U test; a p-value of p < 0.05 is considered significant
| Substrate | Varied substrate properties | Graphs | # of cells |
![[F with combining macron]](https://www.rsc.org/images/entities/i_char_0046_0304.gif) /nN /nN |
F 0.5/nN | F 0.25/nN | F 0.75/nN | p-Value |
|---|---|---|---|---|---|---|---|---|
| Si native (ethanol) | Surface energy | Fig. 3 | 20 | 1.55 | 1.52 | 1.26 | 1.73 | 0.378 |
| Si native (piranha) | 1.61 | 1.52 | 1.36 | 1.74 | ||||
| Si native (ethanol) | Surface energy | Fig. 3 | 26 | 1.71 | 1.43 | 1.05 | 2.37 | 0.307 |
| OTS (ethanol) | 1.75 | 1.42 | 0.779 | 2.26 | ||||
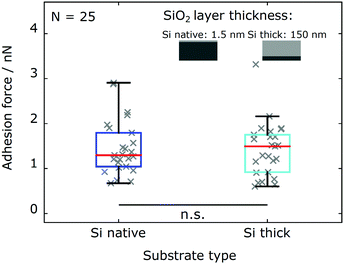
| Si native (ethanol) | van der Waals interactions | Fig. 4 | 25 | 1.46 | 1.30 | 1.04 | 1.79 | 0.454 |
| Si thick (ethanol) | 1.50 | 1.50 | 0.919 | 1.75 | ||||
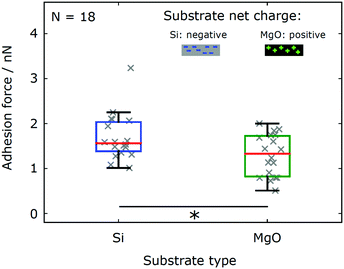
| Si native (ethanol) | Surface charge | Fig. 5 | 18 | 1.70 | 1.56 | 1.38 | 2.03 | 0.0258 |
| MgO (ethanol) | 1.30 | 1.33 | 0.824 | 1.73 | ||||
Van der-Waals interactions
We probe the influence of long-ranged van der Waals (vdW) interactions by comparing silicon wafers with different silicon oxide layer thickness (Si native and Si thick). These model substrates have shown to be well suited to probe the influence of vdW interactions in biological systems, since their surface characteristics are identical within the experimental error while their subsurface contribution is different.33,37–39 As shown in Fig. 4, adhesion force measurements with the same cells (N = 25 cells) on both types of substrates are in excellent agreement. The adhesion force distributions for Chlamydomonas on both types of substrates yield consistent characteristic statistical measures (see Table 2).Electrostatic interactions
In order to determine the effect of electrostatic interactions on the adhesion of Chlamydomonas cells to abiotic surfaces, we probe the adhesion forces on substrates exhibiting different surface charges. The reference silicon substrate (Si native) features an isoelectric point pH(I) ≈ 3, see ref. 32, and carries a net negative charge in the TAP buffer solution with a pH ≈ 7. In contrast, the magnesium oxide substrate (MgO) has an isoelectric point pH(I) = 12.5, see ref. 34, resulting in a positive net charge at pH = 7. As all other relevant surface properties of both substrates are comparable (Table 1), we can dissect the influence of surface charges on the adhesion forces of Chlamydomonas from any other contributions.We find that force–distance experiments with the same set of cells (N = 18 cells) yield adhesion forces that are significantly smaller on the MgO substrate as compared to the Si substrate (see Fig. 5 and Table 2). This difference is evidenced by the data sets recorded on MgO being systematically shifted towards smaller adhesion forces as compared to adhesion forces recorded on the Si substrate (see Fig. 5). From the characteristic statistical measures (see Table 2), we estimate that the adhesion forces of Chlamydomonas on the MgO substrate were approximately 25% smaller than the adhesion forces on the Si substrate.
In aqueous environment, electrostatic interactions can be tuned by varying the ion concentration in the buffer solution. A higher ion concentration leads to a stronger screening of the electrostatic interactions, i.e. smaller Debye length. The standard TAP medium yields a Debye length 1/κ ≈ 1.8 nm, which we calculated from the concentration of ions in the medium (obtained from the website of the supplier). In addition to the adhesion experiments in TAP medium, we performed experiments in a nitrogen-deprived minimal medium (NMM) with lower salt concentration (80 μM MgSO4, 100 μM CaCl2, 3.1 mM K2HPO4, and 3.4 mM KH2PO4, pH 6.8) yielding a larger Debye length 1/κ ≈ 2.6 nm, following the recipe from Berthold et al.40 However, the salt concentration does not only alter the electrostatic interactions, but also directly influences the biology and behavior of Chlamydomonas. In the NMM vegetative cells transform into sexually active cells (gametes) that express additional sexual agglutinins on the flagella surface, which are not reported to be involved in the unspecific adhesion to abiotic substrates. We find that Chlamydomonas gametes‡ in the NMM exhibit consistent adhesion forces with corresponding vegetative cells in TAP medium (see Fig. 6), despite the different ion concentrations in the two buffer media and the resulting alteration of the Debye length by a factor of 1.5.
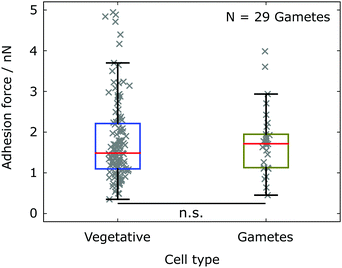 | ||
| Fig. 6 Mean adhesion forces of Chlamydomonas measured in different buffer media yielding different salt compositions and concentrations. The different buffer medium results in a transition from vegetative cells (TAP medium) to gametes (NMM). The adhesion force statistics for gametes (olive, N = 29 cells) is compared to the statistics for vegetative cells (blue, see also Fig. 2). The statistical significance test yields p = 0.2618. | ||
4 Discussion
Statistics of adhesion forces
Micropipette force measurements with Chlamydomonas cells yielded adhesion forces up to a few nanonewtons. These adhesion forces are comparable to adhesion forces reported for single-cell bacterial adhesion studies.35,41,42 In these bacterial adhesion studies, a distinct cell-to-cell variability is rather common in force–distance experiments with different cells of the same species. Such adhesion force variations are probably based on spatially inhomogeneous adhesion protein distributions in the bacterial cell wall and heterogeneities in the bacterial population. Ramified signatures in the force–distance curves of bacteria are usually attributed to nanomechanical properties and rupture events of individual adhesive bonds formed by the bacteria's adhesins.41,43 In contrast to the multitude of different types of adhesins on the bacterial cell wall, adhesion of Chlamydomonas to abiotic surfaces is exclusively attributed to one type of adhesion protein, the flagella membrane glycoprotein FMG-1B, which uniformly covers both flagella.21 A protein-unspecific pronase treatment of the flagella reduced the adhesion force to zero, which confirms that the adhesion of Chlamydomonas is mediated by proteins.28On each flagellum there are about 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies of the adhesion protein FMG-1B, as estimated from polyacrylamide gel electrophoresis.44 The flagella surface area is about 7.5 μm2 (flagella length: 12 μm, diameter: 200 nm), which is conserved for all cells independent of the cell body size. Hence, the average protein density in the flagella membrane would be about 12
000 copies of the adhesion protein FMG-1B, as estimated from polyacrylamide gel electrophoresis.44 The flagella surface area is about 7.5 μm2 (flagella length: 12 μm, diameter: 200 nm), which is conserved for all cells independent of the cell body size. Hence, the average protein density in the flagella membrane would be about 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 proteins per μm2, based on the estimation by Adair et al.44 The glycosylated ectodomains of these glycoproteins presumably represent the prominent glycocalyx seen in transmission electron micrographs of the flagellum.45 Although the average number of FMG-1B copies is known for a Chlamydomonas population, the amount of flagella membrane proteins of an individual cell depends on the expression of the gene encoding the protein. Protein expression is a stochastic process that leads to variations in the amount of protein in cells, including differences in the protein density in the flagella membrane.46–49 Thus, we hypothesise that the variations in the measured mean adhesion forces of different cells (see Fig. 2) might be due to a cell-to-cell variability of the FMG-1B density on the flagella.
000 proteins per μm2, based on the estimation by Adair et al.44 The glycosylated ectodomains of these glycoproteins presumably represent the prominent glycocalyx seen in transmission electron micrographs of the flagellum.45 Although the average number of FMG-1B copies is known for a Chlamydomonas population, the amount of flagella membrane proteins of an individual cell depends on the expression of the gene encoding the protein. Protein expression is a stochastic process that leads to variations in the amount of protein in cells, including differences in the protein density in the flagella membrane.46–49 Thus, we hypothesise that the variations in the measured mean adhesion forces of different cells (see Fig. 2) might be due to a cell-to-cell variability of the FMG-1B density on the flagella.
For a very small subset of cells, we recorded adhesion forces that were up to several times larger than the mean adhesion force of all other cells. The expression of the adhesion-mediating FMG-1B is reported to increase about five-fold upon deflagellation.50 Such large differences in the protein expression could potentially explain exceptionally high adhesion forces between 5 to 10 nN, assuming that deflagellation and flagella regrowth occurred just before selecting the cell. To avoid any statistical bias from cells that might have experienced flagella regrowth in their history, we exclude the data obtained from these cells.
The relative error of the adhesion force of an individual Chlamydomonas cell is in the range of a few tens of percent (see inset of Fig. 2), which might originate from dynamic flagella membrane processes. First, the adhesion-promoting protein FMG-1B is known to redistribute inside the flagellum, as seen by labelling FMG-1B with a fluorescent dye: any labelled FMG-1B is replaced from the flagella within tens of minutes.51 This phenomenon in the flagella membrane is called ‘protein turnover’ and could, indeed, result in temporal variations in the protein density during adhesion force measurements from consecutive force–distance curves. Second, the variation of the adhesion force of an individual cell might be affected by the dynamics of adhesive sites. Interference reflection microscopy data of Chlamydomonas cells in contact with a substrate suggests that the area of the adhesive contact between flagellum and the substrate is variable.23 That is, for the same cell, multiple contact sites, which may be different in size, on both flagella may be simultaneously involved in flagella-substrate adhesion. The position of these contact sites changes during gliding, which indicates that these adhesive sites are essential for the mechanical force transduction between flagellum and surface.
Universal adhesion mechanism
Our experiments revealed that Chlamydomonas cells adhere to all tested model substrates with adhesion forces of a few nN. Our results suggest that Chlamydomonas features a substrate unspecific and universal mechanism for adhering to abiotic surfaces. A statistical analysis of the adhesion force distributions on complementary model substrates yields three main results: (1) Experiments on substrates with different surface energies display consistent adhesion forces. (2) Substrates that differ in van der Waals interactions do not show any significant differences in the adhesion force distributions. (3) Variation of the electrostatic interactions shows a significant influence on the adhesion forces.In contrast to our findings for microalgal adhesion, protein-mediated bacterial adhesion to surfaces does usually depend on the physicochemical properties of the substrates.33,41,42,52,53 For example, single-cell force spectroscopy experiments with Staphylococcus yield adhesion forces of several nN on hydrophobic OTS substrates and only tens of pN on hydrophilic Si substrates.54 These results might be linked to the fact that the adhesins involved in bacterial adhesion are often tailored for attaching to certain biotic substances, e.g. the extracellular material in a host or biofilm.2,3,5,6,41
The adhesion of Chlamydomonas is mediated by the major flagella membrane glycoprotein FMG-1B, which is uniformly distributed on the flagella's surface55 with an average protein density of about 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 proteins per μm2 flagella surface (see above). Besides FMG-1B, so far no other protein has been identified to contribute to substrate adhesion, in particular the mastigonemes (flagella appendages of 0.9 to 1.0 μm length and 16 nm diameter) appear not to be involved.22,56,57 FMG-1B consists of 4149 amino acids (predicted) with an ectodomain of more than 4100 amino acids; the amino acid composition suggests a similar amount of positively and negatively charged amino acids (at pH = 7.4), as well as a similar amount of polar and apolar amino acids. The structure of the protein and its individual domains is unknown, thus, there are no information about hydrophilic or hydrophobic patches, as well as, parts of the protein carrying a significant net charge. Yet, the iodination essay, which identified FMG-1B as the adhesion-mediating protein,22 qualitatively showed that FMG-1B can bind to polar (glass beads) and apolar (polystyrene microspheres) materials.
000 proteins per μm2 flagella surface (see above). Besides FMG-1B, so far no other protein has been identified to contribute to substrate adhesion, in particular the mastigonemes (flagella appendages of 0.9 to 1.0 μm length and 16 nm diameter) appear not to be involved.22,56,57 FMG-1B consists of 4149 amino acids (predicted) with an ectodomain of more than 4100 amino acids; the amino acid composition suggests a similar amount of positively and negatively charged amino acids (at pH = 7.4), as well as a similar amount of polar and apolar amino acids. The structure of the protein and its individual domains is unknown, thus, there are no information about hydrophilic or hydrophobic patches, as well as, parts of the protein carrying a significant net charge. Yet, the iodination essay, which identified FMG-1B as the adhesion-mediating protein,22 qualitatively showed that FMG-1B can bind to polar (glass beads) and apolar (polystyrene microspheres) materials.
The ectodomain of FMG-1B is heavily N-glycosylated,51 and there is experimental evidence that this glycosylation is (indirectly or directly) responsible for the flagella surface adhesiveness. A treatment with tunicamycin blocks protein glycosylation, which lead to a loss of flagellar adhesiveness, as judged by the ability to bind microspheres, while the flagella length or morphology remained unaffected.58 A possible indirect influence of protein glycosylation on the adhesive capability is that protein glycosylation leads, among others, to a proper protein folding during the protein synthesis. While the lack of glycosylation does not inhibit protein transport to the surface and protein exposure on the surface of the flagella, the protein stability might be heavily influenced.59 Consequently, the absence of FMG-1B glycosylation in the flagella membrane would result in a misfolded protein and adhesion-mediating protein domains might not be properly exposed to the substrate. Another possibility is that the tunicamycin treatment resulted in the loss of the carbohydrates and, thus, the protein glycosylation would be directly responsible for the adhesion. The latter functionality of the glycosylation is supported by force spectroscopy studies of the mannose-rich glycosylation of yeast that mediates adhesion forces of individual carbohydrates of tens of piconewton.60 There is evidence that the carbohydrates of FMG-1B are exposed at the protein surface: anti-protein monoclonal antibodies cannot access the protein epitopes of FMG-1B51,55 and the glycans normally extend as flexible, hydrated branches by 3 nm and more from the protein surface.61 The monosaccharide composition of the glycosylation in two Chlamydomonas strains was determined,23 which suggests that the carbohydrates do not carry a net charge, as there are no negatively charged sialic acid residues attached to the carbohydrate.23,62 Although the general backbone structure of the N-linked glycosylation is known, the structure of the glycosylation in Chlamydomonas remains unclear.
In the context of glycoprotein-mediated adhesion, the results of our adhesion force study suggest that the surface-exposed parts of FMG-1B are not predominantly hydrophobic, since otherwise an increased adhesion on the hydrophobized substrates should have been observed. This is supported by the observation that glycan oligosaccharides are predominantly polar61 and that the protein glycosylation potentially prevents a direct protein/surface interaction so that short-ranged hydrophobic interactions are negligible.
Our results for substrates that carry different net charges suggest that electrostatic interactions play an important role in the adhesion of Chlamydomonas, as changing the sign of the substrate's net charge resulted in a systematic shift of the measured adhesion forces. We hypothesise that there might be protein domains (or individual amino acids) carrying opposite charges that both can be exposed to the surface by changing the protein alignment with respect to the surface or conformational changes of the proteins due to electrostatic interactions. Another possibility is that individual side-chains of the protein or individual amino acids undergo an oxidation or, respectively, reduction in contact with a substrate. Such chemical modifications could locally alter the charge of the protein and, thus, influence the electrostatic interactions. Potential candidates are the amino acids aspargine and glutamine, which can transform to aspartic acid and glutamic acid, respectively. In both forms, these amino acids account for approximately 8% of the total amino acid content in FMG-1B.23
5 Conclusions
In summary, we find that the protein-mediated adhesion of Chlamydomonas microalgae to abiotic surfaces is largely independent of the type of substrate. In conjunction with phototaxis and light-switchable adhesion,28 the ability to adhere to any kind of surface appears highly beneficial for accomplishing optimal conditions for photosynthesis and might have evolved as an evolutionary advantage for Chlamydomonas, which dwells in moist habitats exhibiting heterogeneous surface properties and variable light conditions. We present potential mechanisms underlying unspecific protein-mediated adhesion, for which the N-linked glycosylation of the flagella membrane glycoproteins might play a decisive role. As a result of the ability of microalgae to colonize any substrate in aqueous environments, it might be extraordinarily challenging to develop physicochemical pathways, e.g. by applying non-toxic surface coatings, to inhibit microalgal adhesion to surfaces in technological settings.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors thank Maike Lorenz from the Göttingen Algae Culture Collection (SAG) for providing the Chlamydomonas reinhardtii strains SAG11-32a and SAG11-32b and for fruitful discussions. Open Access funding provided by the Max Planck Society.References
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Nat. Rev. Microbiol., 2004, 2, 95–108 CrossRef CAS PubMed.
- S. N. Abraham, D. Sun, J. B. Dale and E. H. Beachey, Nature, 1988, 336, 682–684 CrossRef CAS PubMed.
- S. J. Hultgren, S. Abraham, M. Caparon, P. Falk, J. W. Geme and S. Normark, Cell, 1993, 73, 887–901 CrossRef CAS PubMed.
- J. Pizarro-Cerdá and P. Cossart, Cell, 2006, 124, 715–727 CrossRef PubMed.
- C. Beloin, A. Roux and J. M. Ghigo, in Escherichia coli Biofilms, ed. T. Romeo, Springer Berlin Heidelberg, Berlin, Heidelberg, 2008, pp. 249–289 Search PubMed.
- T. Proft and E. N. Baker, Cell. Mol. Life Sci., 2008, 66, 613 CrossRef PubMed.
- R. Bos, H. C. van der Mei and H. J. Busscher, FEMS Microbiol. Rev., 1999, 23, 179 CrossRef CAS PubMed.
- H. J. Busscher, W. Norde and H. C. van der Mei, Appl. Environ. Microbiol., 2008, 74, 2559–2564 CrossRef CAS PubMed.
- Y. F. Dufrêne, Trends Microbiol., 2015, 23, 376–382 CrossRef PubMed.
- M. Tarantola, A. Bae, D. Fuller, E. Bodenschatz, W.-J. Rappel and W. F. Loomis, PLoS One, 2014, 9, e106574 CrossRef PubMed.
- N. Kamprad, H. Witt, M. Schröder, C. Kreis, O. Bäumchen, A. Janshoff and M. Tarantola, Nanoscale, 2018, 10, 22504–22519 RSC.
- K. G. Porter, Am. Sci., 1977, 65, 159–170 Search PubMed.
- C. B. Field, M. J. Behrenfeld, J. T. Randerson and P. Falkowski, Science, 1998, 281, 237–240 CrossRef CAS PubMed.
- B. Finlay and G. Esteban, Biodiversity & Conservation, 1998, 7, 1163–1186 Search PubMed.
- K. R. Arrigo, Nature, 2005, 437, 349–355 CrossRef CAS PubMed.
- M. P. Schultz, Biofouling, 2007, 23, 331–341 CrossRef PubMed.
- M. P. Schultz, J. A. Bendick, E. R. Holm and W. M. Hertel, Biofouling, 2011, 27, 87–98 CrossRef CAS PubMed.
- J. A. Callow, S. A. Crawford, M. J. Higgins, P. Mulvaney and R. Wetherbee, Planta, 2000, 211, 641–647 CrossRef CAS PubMed.
- T. M. Dugdale, R. Dagastine, A. Chiovitti, P. Mulvaney and R. Wetherbee, Biophys. J., 2005, 89, 4252–4260 CrossRef CAS PubMed.
- E. H. Harris, Annu. Rev. Plant Physiol. Plant Mol. Biol., 2001, 52, 363–406 CrossRef CAS PubMed.
- E. H. Harris, D. B. Stern and G. B. Witman, The Chlamydomonas Sourcebook, Academic Press, London, 2nd edn, 2009 Search PubMed.
- R. A. Bloodgood and L. J. Workman, Cell Motil., 1984, 4, 77–87 CrossRef CAS PubMed.
- R. A. Bloodgood, in Ciliary and Flagellar Membranes, ed. R. A. Bloodgood, Springer US, Boston, MA, 1990, ch. Gliding Motility and Flagellar Glycoprotein Dynamics in Chlamydomonas, pp. 91–128 Search PubMed.
- J. A. Laib, J. A. Marin, R. A. Bloodgood and W. H. Guilford, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3190–3195 CrossRef CAS PubMed.
- S. M. Shih, B. D. Engel, F. Kocabas, T. Bilyard, A. Gennerich, W. F. Marshall and A. Yildiz, eLife, 2013, 2, e00744 CrossRef PubMed.
- K. W. Foster and R. D. Smyth, Microbiol. Rev., 1980, 44, 572–630 CAS.
- K. W. Foster, J. Saranak, N. Patel, G. Zarilli, M. Okabe, T. Kline and K. Nakanishi, Nature, 1984, 311, 756–759 CrossRef CAS PubMed.
- C. T. Kreis, M. Le Blay, C. Linne, M. M. Makowski and O. Bäumchen, Nat. Phys., 2018, 14, 45–49 Search PubMed.
- M. Backholm and O. Bäumchen, Nat. Protoc., 2019, 14, 594–615 Search PubMed.
- M. Lessel, O. Bäumchen, M. Klos, H. Hähl, R. Fetzer, M. Paulus, R. Seemann and K. Jacobs, Surf. Interface Anal., 2015, 47, 557–564 CrossRef CAS.
- C. van Oss, Colloids Surf., A, 1993, 78, 1–49 CrossRef CAS.
- L. Bousse, S. Mostarshed, B. V. D. Shoot, N. de Rooij, P. Gimmel and W. Göpel, J. Colloid Interface Sci., 1991, 147, 22–32 CrossRef CAS.
- P. Loskill, H. Hähl, N. Thewes, C. T. Kreis, M. Bischoff, M. Herrmann and K. Jacobs, Langmuir, 2012, 28, 7242–7248 CrossRef CAS PubMed.
- M. Robinson, J. A. Pask and D. W. Fuerstenau, J. Am. Ceram. Soc., 1964, 47, 516–520 CrossRef CAS.
- N. Thewes, A. Thewes, P. Loskill, H. Peisker, M. Bischoff, M. Herrmann, L. Santen and K. Jacobs, Soft Matter, 2015, 11, 8913–8919 RSC.
- N. Thewes, P. Loskill, C. Spengler, S. Hümbert, M. Bischoff and K. Jacobs, Eur. Phys. J. E: Soft Matter Biol. Phys., 2015, 38, 140 CrossRef PubMed.
- K. Autumn, M. Sitti, Y. A. Liang, A. M. Peattie, W. R. Hansen, S. Sponberg, T. W. Kenny, R. Fearing, J. N. Israelachvili and R. J. Full, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 12252–12256 CrossRef CAS PubMed.
- P. Loskill, J. Puthoff, M. Wilkinson, K. Mecke, K. Jacobs and K. Autumn, J. R. Soc., Interface, 2012, 10, 20120587 CrossRef PubMed.
- P. Loskill, H. Hähl, T. Faidt, S. Grandthyll, F. Müller and K. Jacobs, Adv. Colloid Interface Sci., 2012, 179, 107–113 CrossRef PubMed.
- P. Berthold, S. P. Tsunoda, O. P. Ernst, W. Mages, D. Gradmann and P. Hegemann, Plant Cell, 2008, 20, 1665–1677 CrossRef CAS PubMed.
- R. M. A. Sullan, A. Beaussart, P. Tripathi, S. Derclaye, S. El-Kirat-Chatel, J. K. Li, Y.-J. Schneider, J. Vanderleyden, S. Lebeer and Y. F. Dufrene, Nanoscale, 2014, 6, 1134–1143 RSC.
- G. Zeng, T. Müller and R. L. Meyer, Langmuir, 2014, 30, 4019–4025 CrossRef CAS PubMed.
- R. M. A. Sullan, J. K. Li, P. J. Crowley, L. J. Brady and Y. F. Dufrêne, ACS Nano, 2015, 9, 1448–1460 CrossRef CAS PubMed.
- W. Adair, C. Hwang and U. W. Goodenough, Cell, 1983, 33, 183–193 CrossRef CAS PubMed.
- R. A. Bloodgood and G. S. May, J. Cell Biol., 1982, 93, 88–96 CrossRef CAS PubMed.
- H. McAdams and A. Arkin, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 814–819 CrossRef CAS.
- M. B. Elowitz, A. J. Levine, E. D. Siggia and P. S. Swain, Science, 2002, 297, 1183–1186 CrossRef CAS PubMed.
- J. R. S. Newman, S. Ghaemmaghami, J. Ihmels, D. K. Breslow, M. Noble, J. L. DeRisi and J. S. Weissman, Nature, 2006, 441, 840–846 CrossRef CAS PubMed.
- A. Sigal, R. Milo, A. Cohen, N. Geva-Zatorsky, Y. Klein, Y. Liron, N. Rosenfeld, T. Danon, N. Perzov and U. Alon, Nature, 2006, 444, 643–646 CrossRef CAS PubMed.
- G. J. Pazour, N. Agrin, J. Leszyk and G. B. Witman, J. Cell Biol., 2005, 170, 103–113 CrossRef CAS PubMed.
- R. A. Bloodgood, M. P. Woodward and N. L. Salomonsky, J. Cell Biol., 1986, 102, 1797–1812 CrossRef CAS PubMed.
- R. Yongsunthon and S. K. Lower, J. Electron Spectrosc. Relat. Phenom., 2006, 150, 228–234 CrossRef CAS.
- A. Beaussart, S. El-Kirat-Chatel, R. M. A. Sullan, D. Alsteens, P. Herman, S. Derclaye and Y. F. Dufrêne, Nat. Protoc., 2014, 9, 1049–1055 CrossRef CAS PubMed.
- N. Thewes, P. Loskill, P. Jung, H. Peisker, M. Bischoff, M. Herrmann and K. Jacobs, Beilstein J. Nanotechnol., 2014, 5, 1501–1512 CrossRef PubMed.
- R. A. Bloodgood, in The Chlamydomonas Sourcebook, ed. E. H. Harris, D. B. Stern and G. B. Witman, Academic Press, London, 2nd edn, 2009, pp. 309–368 Search PubMed.
- R. A. Bloodgood, J. Cell Biol., 1977, 75, 983–989 CrossRef CAS PubMed.
- S. Nakamura, G. Tanaka, T. Maeda, R. Kamiya, T. Matsunaga and O. Nikaido, J. Cell Sci., 1996, 109, 57–62 CAS.
- R. A. Bloodgood, Adv. Mol. Cell Biol., 1987, 1, 97–130 CrossRef.
- H. Lodish, A. Berk, P. Matsudaira, C. A. Kaiser, M. Krieger, M. P. Scott, S. L. Zipursky and J. Darnell, Molecular Cell Biology, W. H. Freeman and Company, New York, 2004 Search PubMed.
- D. Alsteens, V. Dupres, K. M. Evoy, L. Wildling, H. J. Gruber and Y. F. Dufrêne, Nanotechnology, 2008, 19, 384005 CrossRef PubMed.
- A. Helenius and M. Aebi, Annu. Rev. Biochem., 2004, 73, 1019–1049 CrossRef CAS PubMed.
- P. Hermentin, R. Witzel, E.-J. Kanzy, G. Diderrich, D. Hoffmann, H. Metzner, J. Vorlop and H. Haupt, Glycobiology, 1996, 6, 217–230 CrossRef CAS PubMed.
Footnotes |
| † Present address: Department of Physical and Environmental Sciences, University of Toronto-Scarborough, 1065 Military Trail, Toronto, Ontario, Canada. |
| ‡ Gametes of both mating types (strain SAG 11-32a and SAG 11-32b) were mixed to observe the sexual mating, as a control for the successful formation of gametes. |
| This journal is © The Royal Society of Chemistry 2019 |