Open Access Article
Open Access ArticleA stirring system using suspended magnetically-actuated pillars for controlled cell clustering†
Aref
Saberi‡
a,
Shuaizhong
Zhang‡
 bc,
Carola
van den Bersselaar
a,
Harkamaljot
Kandail
a,
Jaap M. J.
den Toonder
bc and
Nicholas A.
Kurniawan
bc,
Carola
van den Bersselaar
a,
Harkamaljot
Kandail
a,
Jaap M. J.
den Toonder
bc and
Nicholas A.
Kurniawan
 *ac
*ac
aDepartment of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands. E-mail: kurniawan@tue.nl
bDepartment of Mechanical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands
cInstitute for Complex Molecular Systems, Eindhoven, The Netherlands
First published on 15th January 2019
Abstract
Controlled stirring of a solution is a household task in most laboratories. However, most stirring methods are perturbative or require vessels with predefined shapes and sizes. Here we propose a novel stirring system based on suspended magnetically-actuated pillars (SMAPs), inspired by the ability of biological flagella and cilia to generate flow. We fabricated flexible, millimeter-scale magnetic pillars grafted on transparent polydimethylsiloxane (PDMS) substrates and built a simple actuation setup to control the motion of the pillars remotely. We tested the system with a standard 24-well plate routinely used in most research laboratories and demonstrate that the magnetic actuation results in robust bending of the pillars and large-scale fluid flow in the wells. Quantitative analysis using computational fluid dynamics modeling indicates that the flow profile in the well can be tuned by modulating the applied magnetic field and the geometries of the well and the pillar. Finally, we show that, by employing the stirring system in a standard cell culture plate, we were able to obtain controlled clustering of cells. The SMAP stirring system is therefore a promising cost-effective and scalable stirring approach for various types of studies involving colloids as well as soft and biological materials.
Introduction
Stirring a solution or a particle dispersion is a common task performed in everyday life as well as in most industrial and academic research laboratories. Many scientific applications, such as the aggregation and break-up of colloidal suspensions, droplet coalescence, and studies of peptide/protein–particle interactions, require controlled stirring with well-defined speed, flow profile, and intermittence.1 In most research laboratories, controlled stirring is applied using a standard, commercially-available magnetic stirrer plate together with stirrer bars, which are placed at the bottom of the vessel holding the solution to be stirred. This stirring system, while simple, is expensive (hundreds to thousands USD) and necessitates relatively large sample volumes in the range of mL to L. These large required volumes are sometimes prohibitive, especially for studies involving precious materials. Such a stirring system is also much less applicable in the context of cell biological research, as the motion of the stirrer bar at the bottom of the flask would disrupt and damage the culture of adherent cells.Other, more specialized stirring methods have been developed and commercialized for cell research, such as spinner flasks and various types of stirred-tank and wheel reactors (see a recent critical summary by Schnitzler et al.2). These approaches can induce fluid flow in the vessel yet eliminate possible cell damage caused by the stirrer bar, either by effecting whole-vessel movements or by physically separating the cell culture from the impeller. However, all these technologies are costly, involve special equipment and application-specific vessels with predefined shapes and sizes that are often single-use, and still require large sample volumes (mL to L and kL). A scalable, cost-effective solution is required, especially with the growing appreciation and rapid advance of three-dimensional cultures that mimic in vivo environments.3,4
In this study, we introduce a novel stirring system that circumvents the above issues using a suspended magnetically-actuated pillar (SMAP). The approach is inspired by biological flagella, which are ubiquitous structures used by microorganisms to propel themselves forward and ‘swim’.5,6 As the beating motion of the flagella (and, in a similar fashion, the collective beating motion of hairy cilia) is very effective in generating fluid flow, including in low-Reynolds-number environments,7 researchers have over the past decade intensively explored the usefulness of creating synthetic analogs of flagella and cilia for sensing and controlling local flow.8 These artificial cilia and flagella are slender, flexible filaments that are grafted onto the walls of a microchannel and externally actuated, and have been used for pumping9–15 and mixing14–20 purposes in microfluidic setups.
Recently, we developed a micro-molding method to fabricate an array of flexible magnetic artificial cilia made of a composite material consisting of polydimethylsiloxane (PDMS) and magnetic microparticles.21 These artificial cilia were successfully demonstrated to induce versatile flow in microfluidic channels, and have the added advantage of being compatible with biological fluids and cells. Here we ask whether the same principle can be applied at a larger scale (mm–cm), as a stirring system. To ensure that the stirring system is minimally destructive, we built suspended millimeter-sized pillars from the same PDMS-based composite material that can be magnetically actuated, and determined the flow that can be induced experimentally and computationally. We found that SMAP can indeed effectively generate the desired macro-scale flows. As a potential application of this system, we show that SMAP can be used in a typical 24-well cell-culture plate to effectively induce 3D spatial clustering of mammalian cells. The SMAP stirring system therefore offers a scalable, low-cost, and versatile approach that can be easily adopted for a wide range of applications.
Material and methods
Fabrication of suspended magnetic pillars
The magnetic precursor material used to fabricate the SMAPs is composed of thermally curable polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning, Moerdijk, the Netherlands; 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 base to curing agent weight ratio) and carbonyl iron powder (CIP, 5 μm diameter, 99.5%, Sigma-Aldrich, Zwijndrecht, the Netherlands). The weight ratio between PDMS and CIP was kept at 1
1 base to curing agent weight ratio) and carbonyl iron powder (CIP, 5 μm diameter, 99.5%, Sigma-Aldrich, Zwijndrecht, the Netherlands). The weight ratio between PDMS and CIP was kept at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, except for the pillars with concentrated magnetic particles at the tips (see below), for which the weight ratio was 5
1, except for the pillars with concentrated magnetic particles at the tips (see below), for which the weight ratio was 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The magnetic precursor material was prepared by vigorously mixing PDMS and CIP by hand for 10 min.
1. The magnetic precursor material was prepared by vigorously mixing PDMS and CIP by hand for 10 min.
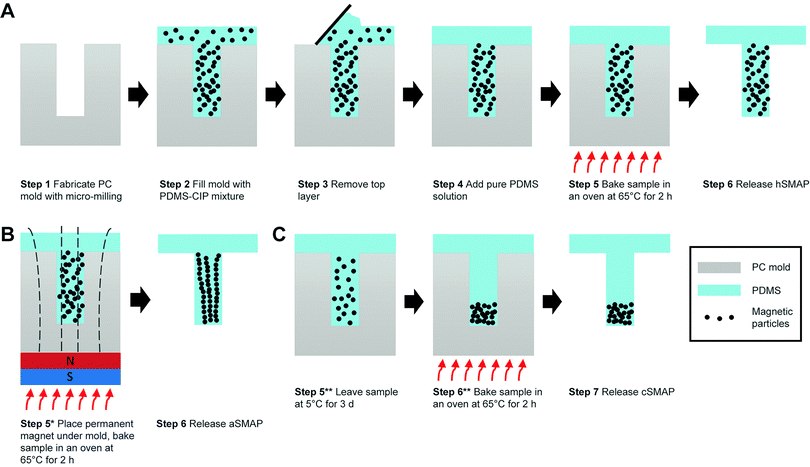
The SMAPs were fabricated using a molding method that was adapted from the micro-molding approach reported earlier for the fabrication of magnetic artificial cilia.21 Briefly, it consisted of 6 main steps (Fig. 1A). Step 1, a polycarbonate (PC) mold was fabricated using micro-milling (Mikron wf 21C), featuring wells with a diameter of 1 mm and a height of 7 mm. Step 2, the prepared magnetic precursor material was poured onto the mold and degassed using a vacuum pump at a vacuum pressure of 2 mbar to remove air bubbles. Step 3, the excess precursor material outside the wells was erased with a scraper and cleanroom tissues (Technicloth TX606, Texwipe). Step 4, pure PDMS solution (i.e., without the CIP) was poured onto the mold. This layer will function as a transparent substrate for suspending the magnetic pillar. Note that, in order to minimize the vertical distance between the SMAP and the actuating magnet in our setup (described in the next section), the thickness of this PDMS substrate was set to 6 mm. Step 5, the mixture was cured in an oven for 2 h at 65 °C. Step 6, the cured PDMS structure was peeled off the PC mold with the help of isopropanol to facilitate the release of the structure. Finally, the magnetic pillars, supported by a transparent PDMS substrate, were obtained. This protocol results in magnetic pillars with a homogeneous distribution of magnetic particles (hSMAP).
Previous studies have indicated that variations in the distribution of magnetic particles can influence the magnetic properties and actuated motion of artificial cilia, due to the anisotropy of the cilia shape and of the particle arrangement.21,22 This provides a possible strategy to tune the magnetic susceptibility of the SMAPs and therefore enhance their actuation properties, by varying the distribution of CIP in the SMAPs. To test this, in addition to the SMAP with random distribution of magnetic particles, we also fabricated two other types of SMAP: pillars with an aligned magnetic particle distribution (aSMAP) and pillars with a concentrated magnetic particle distribution at the tips (cSMAP).
The fabrication processes for aSMAP and cSMAP were based on that of hSMAP. To fabricate the aSMAP, a permanent magnet (10 × 10 × 5 mm3) with a remnant flux density of 1.3 T was placed under the mold during step 5 to align the magnetic particles along the magnetic field (Fig. 1B). The cSMAP was fabricated making use of the relatively high mass density of the magnetic particles compared to that of the PDMS solution (ρCIP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ρPDMS ≈ 8
ρPDMS ≈ 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Utilizing Stokes’ law and the known physical properties of the PDMS solution and the magnetic particles,21 the travel time for the particles to reach the bottom of the mold can be estimated to be 2–3 h under ideal conditions. Directly after step 4, the sample was placed in a fridge at 5 °C for 3 days to slow down the curing of the PDMS, offering enough time for the magnetic particles to settle at the bottom of the mold (Fig. 1C). This is in contrast to the fabrication process of the hSMAP, where the magnetic particles did not have enough time to settle before the PDMS is solidified, resulting in a homogeneous distribution of the particles in the hSMAPs. We also found that when weight ratio between PDMS and CIP was maintained at 1
1). Utilizing Stokes’ law and the known physical properties of the PDMS solution and the magnetic particles,21 the travel time for the particles to reach the bottom of the mold can be estimated to be 2–3 h under ideal conditions. Directly after step 4, the sample was placed in a fridge at 5 °C for 3 days to slow down the curing of the PDMS, offering enough time for the magnetic particles to settle at the bottom of the mold (Fig. 1C). This is in contrast to the fabrication process of the hSMAP, where the magnetic particles did not have enough time to settle before the PDMS is solidified, resulting in a homogeneous distribution of the particles in the hSMAPs. We also found that when weight ratio between PDMS and CIP was maintained at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the interactions between the highly-concentrated particles and the high viscosity of the PDMS solution prevented the particles from settling at the bottom even after 3 days. For this reason, we used a reduced weight ratio of 5
1, the interactions between the highly-concentrated particles and the high viscosity of the PDMS solution prevented the particles from settling at the bottom even after 3 days. For this reason, we used a reduced weight ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for obtaining cSMAPs. After these modified steps, the pillars can be released from the mold to obtain aSMAP and cSMAP.
1 for obtaining cSMAPs. After these modified steps, the pillars can be released from the mold to obtain aSMAP and cSMAP.
Actuation setup
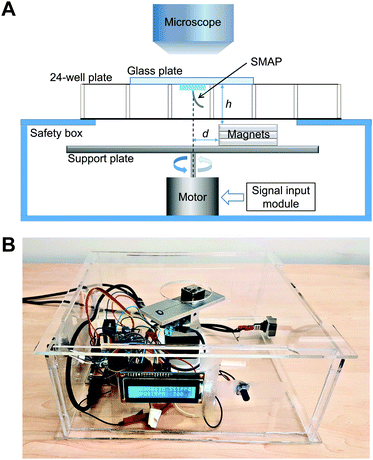
A simple home-built magnetic actuation setup was used to actuate the SMAPs (Fig. 2A). The setup consists of a unipolar stepper motor (Wantai Nema 17) with an aluminium supporting plate for the stacked three pieces of permanent magnets. The setup was fixed in the center of a transparent safety box made from poly (methyl methacrylate) (PMMA) (Fig. 2B). Each magnet has a geometry of 20 × 20 × 5 mm3 with a remnant flux density of 1.3 T. The top surface of the safety box contains a circular opening of 40 mm in diameter, which allows the magnet to rotate freely and minimizes the vertical distance h between the magnets and the SMAP on top of the safety box. In our setup, h was set to be 20 mm, close to the height of a 24-well plate used in the cell experiment. The SMAP, supported by a glass plate which was used as the lid of the well, was aligned with the rotating axis of the motor with the help of a stereo microscope (Olympus SZ61, Leiden, the Netherlands) for capturing the flow in the well. The magnets were placed with an offset distance d of 4 mm with respect to the rotation axis of the motor to induce bending of the pillar towards the stronger magnetic field and thus actuate the SMAP to perform a vertical cone motion. The choice of the parameter values of h and d in this study was based on the constraining dimensions of the well plates and the magnets; other parameter values can be chosen as suited to the specific experimental setup. Note that this actuation scheme, in which the magnetic gradient force actuates the pillar, differs from that used previously for magnetic artificial cilia, where the cilia tend to align with the applied magnetic field due to the magnetic torque acting on them.21 To control the motor, we used a microcontroller board (Arduino Uno), a dual H-bridge drive controller board (L298N), an analog potentiometer to tune the motor speed, and a LED screen to display the speed.Characterization of pillar motion and medium flow
A high-speed camera (Phantom V9, Vision Research, Bedford, UK) mounted on the stereo microscope was used to capture the top-view motion of the SMAPs and the generated liquid flow. The tip deflection of the SMAPs, projected on the x–y (horizontal) plane, was measured from static snapshots and averaged for each type of SMAP. To characterize the flow generated by the SMAP, we filled one well of a 24-well plate with deionized water seeded with 30 μm polylactic acid particles (micromod Partikeltechnologie GmbH, Rostock, Germany) as fiducial markers, and then we placed the SMAP onto this well. Subsequently the SMAP was actuated by the magnetic setup to perform the vertical cone motion. The high-speed camera was used to capture the generated flow at a frame rate of 150 fps. The generated flows close to the SMAP tip were measured by a Manual Tracking analyses in ImageJ.Computational fluid dynamics (CFD) analysis
Flow patterns in the well were quantified by numerically solving the Navier–Stokes equations using ANSYS CFX (Ansys Inc., Canonsburg, PA, USA). An accurate computer-aided design model of the well in a 24-well plate culture chamber, as used in the experimental setup (height = 16.5 mm, bottom diameter = 15.7 mm, top diameter = 16.28 mm), was constructed using SolidWorks (Dassault Systèmes, Vélizy-Villacoublay, France). The fluid domain and the magnetic pillar were meshed using 9633 hexahedral and 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 048 tetrahedral elements, respectively, using ANSYS Meshing (Ansys Inc., Canonsburg, PA, USA). To keep the computational cost affordable, we assumed that the magnetically-actuated bending of the SMAPs and any effect of hydrodynamic forces achieve a steady state, after which there is no further deformation of the pillar. This allows us to model the magnetic pillar as a rigid-body spinning at a constant rotation, thus circumventing the need to couple a finite-element-based structural solver to compute the motion of the magnetic pillar. As a result, ANSYS CFX's build-in immersed solid method was used to resolve the fluid-structure interaction. Furthermore, we made two assumptions: (1) the flow is laminar. This assumption is always satisfied within the range of angular rotations that we experimentally investigated in this study (150–300 rpm; Reynold's number < 1). (2) The fluid is single phase, isothermal, incompressible, and Newtonian. Flow profiles generated by a 7 mm magnetic pillar in water at 37 °C were visualized at four actuation speeds: 150, 200, 250, and 300 rpm. To model stirring using the aSMAPs, the geometry of the deformed pillar during stirring was constructed to capture the experimental tip deflection of the aSMAPs. No-slip boundary conditions were prescribed on the domain walls while the fluid motions was resolved by prescribing a constant rpm to the magnetic pillar.
048 tetrahedral elements, respectively, using ANSYS Meshing (Ansys Inc., Canonsburg, PA, USA). To keep the computational cost affordable, we assumed that the magnetically-actuated bending of the SMAPs and any effect of hydrodynamic forces achieve a steady state, after which there is no further deformation of the pillar. This allows us to model the magnetic pillar as a rigid-body spinning at a constant rotation, thus circumventing the need to couple a finite-element-based structural solver to compute the motion of the magnetic pillar. As a result, ANSYS CFX's build-in immersed solid method was used to resolve the fluid-structure interaction. Furthermore, we made two assumptions: (1) the flow is laminar. This assumption is always satisfied within the range of angular rotations that we experimentally investigated in this study (150–300 rpm; Reynold's number < 1). (2) The fluid is single phase, isothermal, incompressible, and Newtonian. Flow profiles generated by a 7 mm magnetic pillar in water at 37 °C were visualized at four actuation speeds: 150, 200, 250, and 300 rpm. To model stirring using the aSMAPs, the geometry of the deformed pillar during stirring was constructed to capture the experimental tip deflection of the aSMAPs. No-slip boundary conditions were prescribed on the domain walls while the fluid motions was resolved by prescribing a constant rpm to the magnetic pillar.
Cell stirring experiments
MDA-MB-231 cells were cultured in RPMI medium containing 10% FBS and 1% penicillin–streptomycin at 37 °C and 5% CO2. The SMAP, attached to a cover slip, was fixed to the lid of a 24 well-plate with tape and sterilized with 70% ethanol and UV exposure for 5 minutes. For the stirring experiments, 5 × 105 cells were seeded into a 24-well plate culture chamber (Greiner Bio-One CELLSTAR, 662160, Alphen a/d Rijn, the Netherlands). The lid containing the SMAP was placed on top of the 24-well plate, and the sample was transported to the actuation setup in an incubator at 37 °C and 5% CO2. After 20 h of stirring, micrographs of the cells were taken with an EVOS XL Core Cell Imaging System (Thermo Fisher Scientific, Waltham, MA, USA) equipped with a 4× objective lens.Results
Magnetic particle distribution affects pillar deflection
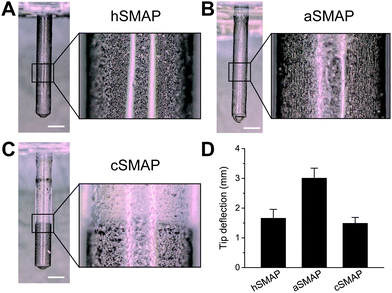
Fig. 3A–C shows optical microscopy images of the 3 types of fabricated SMAPs and the corresponding magnetic particle distributions. The SMAPs are shown to be cylindrically shaped with a pointed tip, reflecting the shape of the mold used in the fabrication process, with a diameter of 1 mm and a length of 7 mm. For the hSMAPs, the magnetic particles are homogeneously distributed as expected, with only a few randomly-located regions of high or low magnetic particle concentrations (Fig. 3A). For the aSMAPs, the application of the magnetic field during the fabrication process results in an ordering of magnetic particles into linear chains oriented along the length of the pillars (Fig. 3B). The slow curing step in the fabrication of the cSMAPs successfully leads to a concentrated region of magnetic particles at the tip of the pillar, with a clear boundary between the CIP-rich section and the pure PDMS section at the root of the pillar (Fig. 3C). With our fabrication protocol, the average length ratio between these two sections is found to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.
2.
To assess how the variations in the magnetic particle distribution influence the effectiveness of SMAPs as a stirring system, we actuated the different types of SMAP with our actuation setup and recorded the motion of the SMAPs. The projected tip deflections of the SMAPs are shown in Fig. 3D. We find that the deflection of the aSMAPs is significantly larger than the deflection of the other two types of SMAPs. This is consistent with the theoretical prediction that aligned magnetic particle distribution enhances the overall magnetic susceptibility.21 The deflection of the cSMAPs is the smallest and is close to that of the hSMAPs. This can be explained by the prediction that the magnetic forces acting on the pillars are proportional to the weight of the magnetic particles,21 as the cSMAPs contain only 20% of the magnetic particles compared to hSMAPs and aSMAPs, resulting in a decreased tip deflection. Since the aSMAPs show the best actuation performance, indicated by the largest tip deflection, we used them for the remainder of the present study.
Pillar motion is enough for stirring
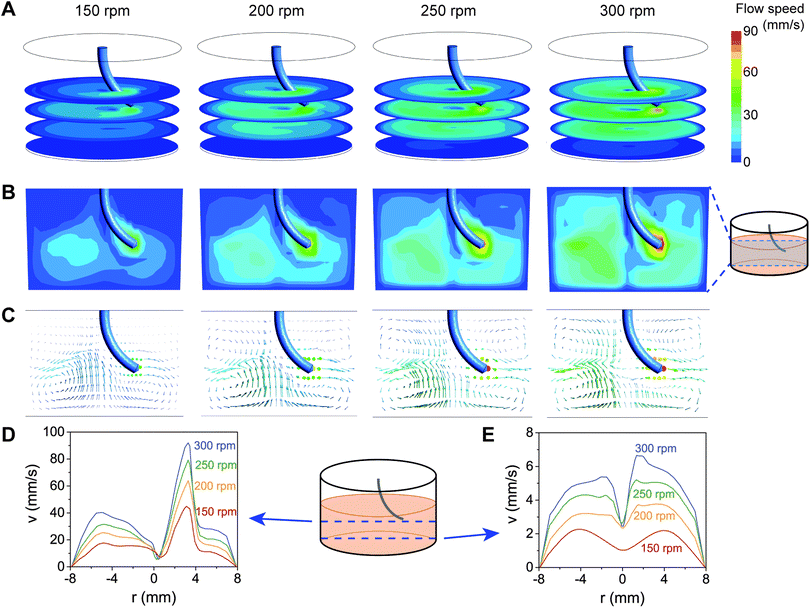
We next asked whether the magnetically-induced bending and rotation of the SMAPs was enough to generate fluid flow in a well. To be able to visualize the fluid flow, we added microparticles as fiducial markers in the fluid and recorded their motion in water resulting from SMAP motion. As shown in Fig. S1 and Video S1 in the ESI,† the SMAP rotating motion is strong enough to generate flow in the whole well. It can be seen that the flow is faster near the tip of the pillars, compared to further away from the pillar tip. However, the flow is not homogeneous, making it difficult to experimentally resolve the flow at different depth and locations in the well.To obtain more insights into the flow induced by the motion of the SMAPs, we performed CFD simulations of the SMAP stirring. Furthermore, we tested the effect of different stirring speeds on the flow by applying angular frequencies in the range of 150–300 rpm. This range of stirring speeds lies within the range accessible for most commercial stirrer plates as well as our magnetic actuation setup. The flow velocity profiles generated by the stirring motion of a 7 mm pillar in a well of the same dimensions as that used in the experiments are shown in Fig. 4. Consistent with the experimental observation, the simulations indicate that the pillar motion can effectively induce large-scale flow in the well, even at heights more than 2 mm below from the tip of the pillars (Fig. 4A). The flow is highest at the trajectory of the pillar, reaching 40–90 mm s−1, and decays with increasing distance from the pillar (Fig. 4B). Increasing the rotation speed results in higher fluid flow at all locations in the well. The vertical cross-sectional flow profiles suggest that circumferential flow is maintained away from the pillar (Fig. 4B and C), reaching ∼40% of the maximum flow at the opposite face of the well (Fig. 4D). Moreover, the stirring motion generated a vortex along the rotation axis of the pillar, characterized by a local drop in the flow velocity. Interestingly, the flow profile near the bottom of the wells show a clear dependence both on the rotation speed and on the radial location (Fig. 4C and E). At the center and near the edge of the well, the flow in the radial direction is close to zero, while at other locations there is a counter-current directed towards the center of the well.
Flow-induced cell clustering
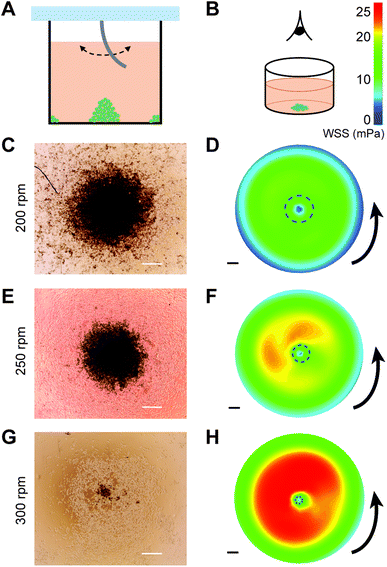
The rotation-speed-dependent radial flow suggests that tuning the stirring speed can be a simple approach to control the shear stresses at the bottom of the wells, which in turn can regulate the spatial deposition of particles. This is particularly interesting for cell culture, as it may offer a way to promote controlled clustering of cells without labeling. As a proof of concept, we experimentally performed the stirring experiment in cell-seeded wells with different stirring speeds (Fig. 5A), and observed the distribution of MDA-MB-231 cells at the bottom of the well after 20 h of culturing (Fig. 5B). The MDA-MB-231 breast cancer cell line, cultured either as single cells, clusters, or spheroids, is commonly used as a tumor model for studying cancer invasion and testing drug efficacy.23–26Stirring using SMAP at 200, 250, and 300 rpm was found to induce clustering of cells in the center of the well (Fig. 5C, E and G) and, to a lower extent, at the edge (i.e., close to the wall) of the well (Fig. S2B–D in the ESI†). Moreover, the stirring speed influenced the size of the cluster: increasing the stirring speed from 200 to 250 and 300 rpm resulted in monotonically decreasing cluster size. The clusters remained stable even after 2 days of further culture and change of medium without stirring. This indicates that the clusters were not only aggregates but the cells actually formed stable cell–cell adhesions that support 3D cell spheroid, as is expected for MDA-MB-231 cells.27 In contrast, a control experiment at 0 rpm did not result in any cluster formation (Fig. S3 in the ESI†), whereas stirring at 150 rpm only generated clusters at the edge, but not in the middle, of the well (Fig. S2A in the ESI†).
We hypothesize that the formation of cell clusters in the middle and at the edge of the well is associated with SMAP-induced radial flow of medium (Fig. 4E) and the associated wall shear stress (WSS) at the bottom of the well. To check this, we plot the WSS profiles, which are remarkably consistent with the locations of the cluster formation: areas of low WSS are associated with cluster formation for all stirring speeds (Fig. 5D, F and H), including at the edge of the well (Fig. S2 in the ESI†). This suggests that radial flow near the bottom of the well pushes the cells towards the center and the edge of the well, where flow and WSS are low or even close to zero. When the stirring speed is too low (150 rpm), the WSS is not strong enough to redistribute the cells towards the center of the well and cluster formation only happens at the edge of the well. Comparison between the experimental and simulation results suggests a threshold WSS around 10 mPa (dashed circles in Fig. 5D, F and H), below which local cluster formation occurs. It is worth noting that the magnitudes of WSS experienced by the cells at the bottom of the wells as induced by our stirring system is very low (in the order of mPa), and are thereby not expected to affect cell viability or phenotype.
Discussion
In this paper, we introduce a stirring system using suspended magnetically-actuated pillars (SMAPs) that can be simply constructed with minimal technical requirements. The SMAPs are fabricated by molding a mixture of PDMS and magnetic particles. The dimensions of the mold and therefore the SMAPs can be adapted depending on the required size of the vessel to be stirred. Here we demonstrate the use of SMAPs, together with a home-built magnetic actuator, as a stirring system in a standard 24-well culture plate. While here we focus on the proof-of-concept experiment of stirring in a single well, parallelizing for stirring in multiple wells can be achieved in a straightforward manner by implementing a standard mechanical design of a gear system controlling the rotating motion of multiple magnets. To match the dimensions of the gear system with the well plate, one can either employ magnets and support plates with smaller sizes than those used in the present study or opt for larger wells (e.g., a 6-well plate or even a custom-sized well plate as desired). Scaling down of the stirring system to smaller dimensions (i.e., micrometer scale) has been achieved in our earlier work.21 Scaling up to larger wells (or well plates) is in theory straightforward, by fabricating larger SMAPs. Indeed, the magnetic susceptibility and the resulting deflection of the SMAPs can be tuned in multiple ways: by modifying the distribution of the magnetic particles in the SMAPs (as we showed through the comparison of hSMAP, aSMAP, and cSMAP), by changing the dimensions and/or stiffness of the pillars, by using magnets with different flux densities, or by varying the distance between the SMAP and the actuating magnets. These strategies give flexibility to the experimenter to adjust the design of the stirring system based on the exact experimental setup, and therefore makes the system scalable.The stirring motion of the SMAP is able to generate large-scale fluid flow, which can be characterized numerically using CFD analysis. Here we performed the computational analysis assuming a non-deformable pillar. Although this is sufficient in the case of steady-state and constant stirring rotation, it will not be suited to model experiments with dynamically varying stirring speed. To get an impression on the variation of the flow profiles caused by different geometric modeling of the pillar, we performed an additional set of simulations, assuming that the pillar was a completely rigid (i.e., rather than bendable) rod (Fig. S4 in the ESI†). Although the overall flow profiles are comparable to those of bent pillars (Fig. 4), it can be seen that the magnitude of the flow velocities are slightly larger in the case of the rigid rod. We therefore recommend a careful characterization of flow using realistic pillar geometries and hydrodynamics when dynamic stirring is required.
Our study demonstrates that the flow generated by the SMAP motion can be exploited for controlling spatial distribution of particles. More specifically, qualitative comparison between our experimental and computational data suggests that the radial flow and wall shear stress at the bottom determine the deposition of particles. Indeed, as one potential application of our SMAP stirring system, we showed that the system is able to produce controlled multicellular clustering, which is an important first step to obtain 3D cell culture, in standard 24-well cell culture plates. Comparison between cluster formation at different stirring speeds indicates that the size of the cluster depends on the stirring speed: cluster size decreases with increasing stirring speed. This trend is consistent with previous reports of flow-induced cell clustering obtained using commercial orbital rotary shaker or spinner flasks.28,29 It is worth noting that the magnitude of the angular velocities in these studies (25–75 rpm) is significantly lower than that used in our present study (200–300 rpm), because of the large vessel size necessitated by these methods. Moreover, we find that a 150 rpm stirring speed fails to produce cell clusters, suggesting that there may be an optimal window of the flow speed in the well that enables cell clustering. We emphasize that a quantitative comparison between our experimental and computational results would be speculative at this stage; more detailed flow simulations involving fluid–structure interactions and modeling of the dispersed, deformable particles or cells are required. Further, we note that, in practice, the resulting cell cluster size is dependent not only on the stirring speed, but also on other detailed experimental variables, such as the cell seeding density, fluid volume, well size, and potentially also cell type. A systematic investigation into the effect of modulating these experimental variables on cell clustering can provide an interesting avenue for future studies especially involving 3D cell culture.
3D cell culture is increasingly recognized to better mimic in vivo conditions than conventional 2D cell culture.30,31 To date, various methods to obtain 3D cell culture have been developed, but these methods have several important drawbacks.4 For example, agitation-based approaches such as spinner-flask bioreactors32,33 require specialized equipment and consumables and are commercially available only in predefined shapes and size; the hanging-drop34–36 and forced-floating37 methods often suffer from limitations in the size and variability of cell spheroids and are either labor-intensive (when custom-prepared) or expensive (with commercial products); microwell-based approaches38–40 are suitable for high-throughput production of well-defined cell aggregates but may require extensive and costly optimization process of the fabricated microwell patterns; contactless cell manipulation such as the magnetic levitation method41–43 inherently involves magnetization of cells that raises toxicity and epigenetics questions; whereas microfluidics-based44 and scaffolds-based45 approaches require expertise in microfabrication or polymer chemistry and entail difficult sample retrieval and analysis.
The SMAP stirring system circumvents some of these drawbacks by allowing the experimenter to steer the organization of cells with any standard culture plate with simple scaling of the SMAP geometry. The stirring procedure is contactless, label-free, and single-step, thereby making the strategy time- and labor-efficient. Another advantage of the SMAP stirring system is that in principle it requires no specialized equipment. In this article we employed a home-built magnetic actuation setup to demonstrate the simplicity and cost-effectiveness of building the complete system (total cost < USD 50). However, the SMAP can also be actuated using common laboratory stirrers with only minimal modification: a magnet should be affixed off-axis on the rotator of the stirrer.
Conclusions
We propose a novel stirring system based on suspended magnetically-actuated pillars (SMAPs). The main strengths of this system are that it is cost-effective, contactless, simple to fabricate, easily scalable to fit the geometry of the vessel of choice, and requires no specialized equipment. Moreover, it is straightforward to modulate the resulting flow by taking into account the geometries of the SMAP and the vessel and to parallelize the system to perform high-throughput experiments. Thus, we anticipate that this system can be a versatile tool for various applications, such as mesofluidic mixing, colloid self-assembly, directed assembly and separation of microparticles and biological cells,46,47 as well as generation of cell spheroids for cell biological and drug screening studies.48,49Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Jurgen Bulsink for help with fabrication of the mold and the supporting plate for the magnets; Erik Homburg, Willie ter Elst, and Andreas Pollet for their support in constructing the actuation setup; Sheen SahebAli for insightful discussions; and Jelle Sleeboom for kindly sharing the cells.References
- D. Li and R. B. Kaner, J. Am. Chem. Soc., 2005, 128, 968–975 CrossRef PubMed.
- A. C. Schnitzler, A. Verma, D. E. Kehoe, D. H. Jing, J. R. Murrell, K. A. Der, M. Aysola, P. J. Rapiejko, S. Punreddy and M. S. Rook, Biochem. Eng. J., 2016, 108, 3–13 CrossRef CAS.
- N. Tandon, D. Marolt, E. Cimetta and G. Vunjak-Novakovic, Biotechnol. Adv., 2013, 31, 1020–1031 CrossRef CAS PubMed.
- S. Breslin and L. O'Driscoll, Drug Discovery Today, 2013, 18, 240–249 CrossRef CAS PubMed.
- S. Vogel, Life in Moving Fluids, Princeton University Press, Princeton, NJ, 1996 Search PubMed.
- C. Brennen and H. Winet, Annu. Rev. Fluid Mech., 1977, 9, 339–398 CrossRef.
- T. Qiu, T. C. Lee, A. G. Mark, K. I. Morozov, R. Munster, O. Mierka, S. Turek, A. M. Leshansky and P. Fischer, Nat. Commun., 2014, 5, 5119 CrossRef CAS PubMed.
- J. den Toonder and P. Onck, Artificial Cilia, RSC Publishing, Cambridge, 2013 Search PubMed.
- F. Fahrni, M. W. J. Prins and L. J. van IJzendoorn, Lab Chip, 2009, 9, 3413–3421 RSC.
- S. Hanasoge, P. J. Hesketh and A. Alexeev, Microsyst. Nanoeng., 2018, 4, 11 CrossRef.
- M. Vilfan, A. Potocnik, B. Kavcic, N. Osterman, I. Poberaj, A. Vilfan and D. Babic, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1844–1847 CrossRef CAS PubMed.
- Y. Wang, J. den Toonder, R. Cardinaels and P. Anderson, Lab Chip, 2016, 16, 2277–2286 RSC.
- Y. Wang, Y. Gao, H. Wyss, P. Anderson and J. den Toonder, Lab Chip, 2013, 13, 3360–3366 RSC.
- J. den Toonder, F. Bos, D. Broer, L. Filippini, M. Gillies, J. de Goede, T. Mol, M. Reijme, W. Talen, H. Wilderbeek, V. Khatavkar and P. Anderson, Lab Chip, 2008, 8, 533–541 RSC.
- A. R. Shields, B. L. Fiser, B. A. Evans, M. R. Falvo, S. Washburn and R. Superfine, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15670–15675 CrossRef CAS PubMed.
- C. Y. Chen, C. Y. Chen, C. Y. Lin and Y. T. Hu, Lab Chip, 2013, 13, 2834–2839 RSC.
- C. Y. Chen, C. C. Hsu, K. Mani and B. Panigrahi, Chem. Eng. Process., 2016, 99, 33–40 CrossRef CAS.
- F. L. Liu, J. Zhang, G. Alici, S. Yan, R. Mutlu, W. H. Li and T. H. Yan, Smart Mater. Struct., 2016, 25, 095049 CrossRef.
- B. P. Zhou, W. Xu, A. A. Syed, Y. Y. Chau, L. Q. Chen, B. Chew, O. Yassine, X. X. Wu, Y. B. Gao, J. X. Zhang, X. Xiao, J. Kosel, X. X. Zhang, Z. H. Yao and W. J. Wen, Lab Chip, 2015, 15, 2125–2132 RSC.
- M. Baltussen, P. Anderson, F. Bos and J. den Toonder, Lab Chip, 2009, 9, 2326–2331 RSC.
- S. Zhang, Y. Wang, R. Lavrijsen, P. R. Onck and J. M. J. den Toonder, Sens. Actuators, B, 2018, 263, 614–624 CrossRef CAS.
- F. Khademolhosseini and M. Chiao, J. Microelectromech. Syst., 2012, 22, 131–139 Search PubMed.
- K. M. Charoen, B. Fallica, Y. L. Colson, M. H. Zaman and M. W. Grinstaff, Biomaterials, 2014, 35, 2264–2271 CrossRef CAS PubMed.
- W. Sun, N. A. Kurniawan, A. P. Kumar, R. Rajagopalan and C. T. Lim, Cell. Mol. Bioeng., 2014, 7, 205–217 CrossRef CAS.
- W. Sun, C. T. Lim and N. A. Kurniawan, J. R. Soc., Interface, 2014, 11, 20140638 CrossRef PubMed.
- H. H. Truong, J. de Sonneville, V. P. S. Ghotra, J. L. Xiong, L. Price, P. C. W. Hogendoorn, H. H. Spaink, B. van de Water and E. H. J. Danen, Biomaterials, 2012, 33, 181–188 CrossRef CAS PubMed.
- A. Ivascu and M. Kubbies, Int. J. Oncol., 2007, 31, 1403–1413 CAS.
- Y. Wang, B. K. Chou, S. Dowey, C. He, S. Gerecht and L. Cheng, Stem Cell Res., 2013, 11, 1103–1116 CrossRef CAS PubMed.
- R. L. Carpenedo, C. Y. Sargent and T. C. McDevitt, Stem Cells, 2007, 25, 2224–2234 CrossRef PubMed.
- F. Pampaloni, E. G. Reynaud and E. H. K. Stelzer, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS PubMed.
- N. A. Kurniawan and C. V. C. Bouten, Extreme Mech. Lett., 2018, 20, 59–64 CrossRef.
- H. Kurosawa, J. Biosci. Bioeng., 2007, 103, 389–398 CrossRef CAS PubMed.
- M. Z. Ismadi, P. Gupta, A. Fouras, P. Verma, S. Jadhav, J. Bellare and K. Hourigan, PLoS One, 2014, 9, e106493 CrossRef PubMed.
- N. E. Timmins and L. K. Nielsen, Methods Mol. Med., 2007, 140, 141–151 CrossRef CAS PubMed.
- R. Foty, J. Vis. Exp., 2011, 51, e2720 Search PubMed.
- Y. C. Tung, A. Y. Hsiao, S. G. Allen, Y. S. Torisawa, M. Ho and S. Takayama, Analyst, 2011, 136, 473–478 RSC.
- J. Friedrich, C. Seidel, R. Ebner and L. A. Kunz-Schughart, Nat. Protoc., 2009, 4, 309–324 CrossRef CAS PubMed.
- M. D. Ungrin, C. Joshi, A. Nica, C. Bauwens and P. W. Zandstra, PLoS One, 2008, 3, e1565 CrossRef PubMed.
- C. C. Miranda, T. G. Fernandes, J. F. Pascoal, S. Haupt, O. Brustle, J. M. Cabral and M. M. Diogo, Biotechnol. J., 2015, 10, 1612–1624 CrossRef CAS PubMed.
- S. Selimovic, F. Piraino, H. Bae, M. Rasponi, A. Redaelli and A. Khademhosseini, Lab Chip, 2011, 11, 2325–2332 RSC.
- H. Jaganathan, J. Gage, F. Leonard, S. Srinivasan, G. R. Souza, B. Dave and B. Godin, Sci. Rep., 2014, 4, 6468 CrossRef CAS PubMed.
- D. Fayol, G. Frasca, C. Le Visage, F. Gazeau, N. Luciani and C. Wilhelm, Adv. Mater., 2013, 25, 2611–2616 CrossRef CAS PubMed.
- G. R. Souza, J. R. Molina, R. M. Raphael, M. G. Ozawa, D. J. Stark, C. S. Levin, L. F. Bronk, J. S. Ananta, J. Mandelin, M. M. Georgescu, J. A. Bankson, J. G. Gelovani, T. C. Killian, W. Arap and R. Pasqualini, Nat. Nanotechnol., 2010, 5, 291–296 CrossRef CAS PubMed.
- D. Huh, G. A. Hamilton and D. E. Ingber, Trends Cell Biol., 2011, 21, 745–754 CrossRef CAS PubMed.
- B. A. Justice, N. A. Badr and R. A. Felder, Drug Discovery Today, 2009, 14, 102–107 CrossRef CAS PubMed.
- J. Branscomb and A. Alexeev, Soft Matter, 2010, 6, 4066–4069 RSC.
- S. Sohrabi, J. Tan, D. E. Yunus, R. He and Y. Liu, Biomicrofluidics, 2018, 12, 042206 CrossRef PubMed.
- G. Mehta, A. Y. Hsiao, M. Ingram, G. D. Luker and S. Takayama, J. Controlled Release, 2012, 164, 192–204 CrossRef CAS PubMed.
- M. F. Gencoglu, L. E. Barney, C. L. Hall, E. A. Brooks, A. D. Schwartz, D. C. Corbett, K. R. Stevens and S. R. Peyton, ACS Biomater. Sci. Eng., 2018, 4, 410–420 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S4 and Movie S1, reporting additional experiments and sample analysis. See DOI: 10.1039/c8sm01957f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |