Open Access Article
Open Access ArticleExploring the tolerance of marine yeast to inhibitory compounds for improving bioethanol production†
Darren
Greetham‡
 a,
Abdelrahman Saleh
Zaky‡
a,
Abdelrahman Saleh
Zaky‡
 bcd and
Chenyu
Du
*a
bcd and
Chenyu
Du
*a
aSchool of Applied Science, University of Huddersfield, Queensgate, Huddersfield, HD1 3DH, UK. E-mail: c.du@hud.ac.uk
bDepartment of Microbiology, Faculty of Agriculture, Cairo University, Giza 12613, Egypt
cSchool of Biosciences, University of Nottingham, Nottingham LE12 5RD, UK
dSchool of Biological Sciences, University of Edinburgh, Roger Land Building, King's Buildings, Edinburgh EH9 3FF, UK
First published on 29th April 2019
Abstract
Inhibitor tolerance is one of the key challenges in lignocellulosic bioethanol production. The presence of inhibitors in a lignocellulosic hydrolysate induces stress in microorganisms and thus reduces bioethanol synthesis efficiency. In this study, 166 marine yeasts isolated from different marine environments were compared with 78 terrestrial yeasts for their tolerance to inhibitory compounds such as acetic acid, formic acid, furfural, vanillin and salt, which are commonly found in hydrolysates derived from lignocellulosic materials. Marine yeasts showed higher tolerance to all the inhibitors tested than terrestrial yeasts. The most tolerant marine yeast was Wickerhamomyces anomalus M15 which had IC50 values of 10.7% (w/w) and 83.9 mM for salt and acetic acid, respectively, while those for an industrial terrestrial yeast Saccharomyces cerevisiae NCYC2592 were 6.0% (w/w) and 75.9 mM, respectively. Statistical analysis revealed that marine yeast clustered separately from terrestrial yeast. In fermentation, using simulated wheat straw hydrolysates containing inhibitors and prepared using seawater, three selected marine yeasts Saccharomyces cerevisiae AZ65, W. anomalus M15, and Candida membranifaciens M2 produced 23–24 g L−1 bioethanol from 60 g L−1 glucose, while terrestrial yeast Saccharomyces cerevisiae produced only 12.5 g L−1 bioethanol. Marine yeasts were significantly more tolerant to the presence of inhibitory compounds and are phenotypically distinct from terrestrial yeasts. Significantly, a higher quantity of bioethanol was obtained in fermentation using selected marine yeasts with media containing inhibitory compounds. The study has highlighted the potential of finding microorganisms with industrially favourable characteristics in marine ecosystems and other adverse and challenging environments.
Introduction
Industrial biotechnology and bioenergy has attracted attention in driving the move towards a low-carbon circular economy. Many chemicals, which have relied upon conventional chemical processes, can be replaced by biomass-derived products, including bioethanol production, which has a prime position in terms of commercialisation of bulk biochemicals. Several reports have suggested that demonstration plants have been commissioned for bioethanol production from lignocellulosic raw materials;1 efficient bioethanol production still faces several challenges during pre-treatment such as high energy consumption, inhibitor formation, salt generation, low initial sugar in the media and lack of efficient glucose–xylose fermenting strains.2 In addition, the water footprint (WF) of cellulosic ethanol derived from wheat straw is currently 140 m3 t−1 compared with 205 m3 t−1 for corn or rapeseed.3During cellulosic ethanol production, various compounds are formed, which have the potential to inhibit fermentation.4,5 These compounds include salt,6–8 weak acids (acetic, formic, levulinic, coumaric etc.), furyl compounds (furfural or hydroxymethylfurfural (HMF)) or phenolic compounds (vanillin or vanillic acid).5,9–11 Using a hydrothermal method (130 °C or above) generates between 6 and 90 mM acetic acid depending on the temperature chosen with similar concurrent increases in formic acid, furfural, and HMF.12,13 The presence of inhibitory compounds has been shown to reduce fermentation efficiencies, for example, the presence of 0.5 mM furfural has been shown to reduce ethanol production and at 4 mM it prevented ethanol production.12,14,15 Presence of 20 mM acetic acid has been shown to induce ethanol production,13,14,16 however, higher acetic acid concentrations inhibits ethanol production.17
Pre-treated lignocellulosic materials with diluted NaOH (5%) produced the lowest concentrations of inhibitors but the sample required neutralisation from pH 14, which produced a considerable amount of salt.4 The presence of high salt concentrations in hydrolysates has restricted the conversion of sugars into ethanol using conventional yeast strains; however, procedures using co-saccharification and fermentation have been shown to produce ethanol from hydrolysates under alkaline conditions.18
Marine derived microbes have long adapted their physiology and metabolism to surviving under extreme conditions present in marine environments such as seas and oceans, and closely related environments such as marshes, salt flats and sand dunes. Consequently, this constant exposure to a challenging environment has forced the evolution of protective mechanisms which has led to chemo- and bio-diversity in algae,17 bacteria,19 fungi20 and yeast.21 However, marine yeasts have been poorly investigated for their industrial potential compared to those from terrestrial ecosystems.
Yeasts, especially S. cerevisiae, are used for bioethanol production, but wild S. cerevisiae has been described as moderately osmotic and salt tolerant compared with other yeasts.22,23 Marine yeasts have been reported as promising candidates for the production of bioethanol and other chemicals, as they possess high salt tolerance.22 Previous studies have revealed that there is a wide range of phenotypic responses in the presence of these inhibitory compounds in yeasts and yeasts with a tolerant phenotype perform better in fermentation in the presence of these inhibitory compounds.24 Research has revealed that marine yeast strains have the ability of converting monomeric hexose sugars into ethanol more efficiently than terrestrial yeast strains.22,25 A marine yeast, Candida membranifaciens, has been shown to have xylose and arabinose utilising capabilities and whilst ethanol production was not determined using these sugars, this yeast did produce riboflavins from these carbon sources and could ferment hexose sugars such as glucose, sucrose and maltose.26 The ability of yeast to convert sugars present in marine biomass (seaweed) is restricted due to the low or no cellulosic content of this feedstock; however, yeasts with the capability of using sugars derived from seaweed (fucose, mannitol, rhamnose etc.) have been identified.27 However, efficient conversion of these sugars into biofuels or other products has proved difficult, as S. cerevisiae cannot use mannitol, the principal sugar released by certain types of seaweed.28
This study has examined phenotypic responses of yeast isolated from marine environments to inhibitors present in cellulosic fermentation; these yeast populations came from a variety of marine sources and their tolerance and variation of responses were determined in comparison with terrestrial yeast strains. Then, yeasts which possessed higher tolerance in the presence of these inhibitors were selected for an assessment of bioethanol fermentation efficiency using media containing lignocellulosic hydrolysate inhibitors.
Materials and methods
Seawater
Seawater used for fermentation was collected from Skegness, UK, and was taken approximately five meters from the shore and at a depth of one meter. Seawater used for the phenotypic microarray was obtained from the North Sea near Whitby, UK. Seawater was taken at a depth of one meter, near the shore and was allowed to sediment for approximately 24 hours before being filtered through Whatman® glass microfiber filters (pore size 1.2 μm). After filtration, the seawater was autoclaved at 121 °C for 15 min. Sterilized seawater was then stored at 4 °C till required.Samples and sampling
Marine samples used in marine yeast isolation in this study were taken from either Skegness or Aberystwyth, UK. Samples from Skegness (July 2017) came from a wide range of coastal habitats such as dried seaweed, marine organisms, sand, driftwood and mussels. Samples were taken using 50 mL sterile falcon tubes containing 10 mL of Zaky's isolation broth,23 and the samples were kept at room temperature until returned to the lab, where they were incubated at 30 °C. Wet seaweed samples were collected from Aberystwyth bay (October 2017). These samples were couriered to Huddersfield overnight and yeast isolation was performed within 14 days as mentioned above.Terrestrial yeast strains
Terrestrial yeast strains S. cerevisiae NCYC2592 (www.ncyc.co.uk) and 78 Saccharomyces sp. (ESI S1†) were maintained on YPD (2% glucose, 2% peptone, and 1% yeast extract prepared using reverse osmosis water (ROW)).Marine yeast strains and isolates
Yeast isolates (M1–M58) were isolated from different marine sources (dried or wet seaweed, marine organisms, sand, or driftwood). In addition, 116 marine yeast isolates, which have already been isolated and characterized by Zaky et al. (2016)29 were included in this paper and are labelled AZ1–AZ150. All marine yeasts were maintained on YPD–SW (2% glucose, 2% peptone, and 1% yeast extract in seawater (SW)).Isolation method
Yeasts were isolated using an isolation method defined by Zaky et al. (2016)29 with a slight modification. Briefly, the samples were enriched using Zaky's isolation broth media which used either (6%, w/v) glucose, mannitol, rhamnose, arabinose, or xylose as the carbon source respectively and were incubated at 30 °C, with shaking at 180 rpm, for 48 h, in seawater. The cultures obtained were sequentially sub-cultured 4 times, over an approximate two-week period at 0.1% (v/v) inoculation size using identical isolation media and incubated under identical conditions. After enrichment, 10 μL of the yeast culture or subsequent diluted broth (usually 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution) was spread on YPD–SW agar plates (2% glucose, 2% peptone, 1% yeast extract and 2% agar, (w/v) in seawater) and incubated at 30 °C for 48 h. Identification of single colonies indicated a pure culture and these were then stored at −80 °C as 50% glycerol stocks; 58 yeast isolates were obtained and used in this study, and were labelled M1–M58. In addition, 116 marine yeast isolates which had already been isolated and characterized by Zaky et al. (2016)29 were included in this paper and are labelled AZ1–AZ150. All marine yeasts were maintained on YPD–SW (2% glucose, 2% peptone, and 1% yeast extract in seawater (SW)).
100 dilution) was spread on YPD–SW agar plates (2% glucose, 2% peptone, 1% yeast extract and 2% agar, (w/v) in seawater) and incubated at 30 °C for 48 h. Identification of single colonies indicated a pure culture and these were then stored at −80 °C as 50% glycerol stocks; 58 yeast isolates were obtained and used in this study, and were labelled M1–M58. In addition, 116 marine yeast isolates which had already been isolated and characterized by Zaky et al. (2016)29 were included in this paper and are labelled AZ1–AZ150. All marine yeasts were maintained on YPD–SW (2% glucose, 2% peptone, and 1% yeast extract in seawater (SW)).
DNA extraction
A single yeast colony was incubated in 5 mL YPD media at 30 °C, without shaking, and genomic DNA was extracted using an Epicentre DNA extraction kit (MasterPure yeast DNA purification kit); contaminating RNA was removed by digestion of the DNA/RNA samples with RNAse (RNAse from bovine pancreas, Sigma R6513). The extracted DNA was stored at −20 °C. The purity of DNA was analysed by running it on 1% (w/v) agarose gel in 1× TBE (Trizma base, boric acid and EDTA) buffer stained with SYBY Safe (Invitrogen) and visualized under UV light. A 1000 bp DNA ladder (Bioline Hyperladder IV, 100 Lanes, Lot No: H4-107K) marker was used as the size standard.PCR of ITS and D1/D2 regions
Amplification and sequencing of the ITS region: the fungus specific universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3) and ITS4 (5′-TCCTCCGCTTATTGATATG-3′) were used to amplify the ITS region. PCR was performed in a total reaction volume of 50 μL consisting of 50 mM MgCl2, 5× DNA loading buffer (Bioline 5× DNA loading buffer) Taq DNA polymerase (Sigma, US), 0.8 mM deoxynucleoside triphosphates (dNTPs) (0.2 mM each), and 2 μL of DNA template. PCR was carried out on a Techne Prime thermal cycler (Cole-Palmer, UK) under the following conditions: initial denaturation at 94 °C for 5 min; 40 cycles of denaturation (92 °C for 30 s) and annealing (54 °C for 45 s); an extension step (72 °C) and a final extension step (72 °C for 5 min). A negative control was used in each run by replacing DNA with sterile water in the PCR mixture. All amplicons were purified using a PCR clean up system (Qiagen, US). The D1/D2 region of the large-subunit RNA gene was sequenced for species confirmation and clarification. Primers NL1 (5′-GCATATCAATAAGCGGAG GAAAAG-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) were used to amplify this region. The protocol for PCR amplification, PCR product purification and sequencing of the PCR products are the same as described for the ITS region.FASTA sequence identification
Sequence data derived from the ITS and D1/D2 domains were compared using the FASTA nucleotide similarity search function (http://www.ebi.ac.uk/Tools/sss/fasta/nucleotide.html).Phenotypic microarray (PM)
Phenotypic microarray (PM) technology (Biolog, US) assay is based on the detection of metabolic output using a reporter system.30 The reporter system utilises a redox sensitive tetrazolium dye which upon reduction correlates with an increase in metabolic rate as the cell oxidises a carbon source. PM analysis was performed following the procedure of Greetham et al., (2014)13 with further modification for the use of seawater provided by Zaky et al., (2016).29 Stock solutions (1 M) of aliphatic weak acids such as acetic and formic acids were prepared using filtered seawater; furfural and HMF were prepared as 1 M stock solutions in 100% ethanol. These chemicals were prepared as fresh stock solutions for each experiment and added as appropriate to individual microtitre plate wells to make up the relevant concentrations (50 mM for acetic acid or 10 mM for formic acid, furfural, and HMF). In addition, each well (120 μL) contained 6% glucose, 0.67% yeast nitrogen base, 0.2 μL dye D (Biolog, US) and the yeast cells (1 × 106) were buffered in IFY buffer (Biolog, US). The plates were analysed at 30 °C for 96 hours with a reading every 15 minutes under aerobic conditions, with no shaking. Data were converted from the raw PM assay data into Excel compatible data and then analysed.Determination of yeast growth in the presence of different inhibitors
Yeast growth had a starting OD of 1 × 106 was monitored for 24 h with a reading every 15 min using a Spectrostar Nano (BMG, labtech, Germany) plate reader at 30 °C. YPD media with various additional inhibitory compounds were pre-loaded onto 96 well plates, containing either (w/v) NaCl (salt, 3–15%), acetic acid (25–100 mM), formic acid (5–15 mM) or furfural (5–15 mM) The assay was performed in triplicate and an average reading was plotted. The presence of acetic or formic acid reduced the pH of the media, so the media were adjusted to pH 5 using 1 M NaOH, as this has been shown to be the optimum pH for yeast fermentation of cellulosic feedstock.31 Addition of furfural or salt has no effect on the pH of the media so the pH was unchanged from a starting pH of 6.5.Fermentation
Fermentation was conducted in 100 mL (working volume) mini-fermentation vessels (MFV) (Wheaton glass bottles, Sigma-Aldrich, US). Cryopreserved yeast cultures were streaked onto YPD plates and incubated at 30 °C for 48 h. Colonies of yeast strains were used to inoculate 5 mL of YPD–SW broth, and incubated in an orbital shaker at 30 °C and 200 rpm for 24 h. These were then transferred to 200 mL of YPD and grown for 48 h in a 500 mL conical flask with shaking at 200 rpm at 30 °C. Cells were harvested and washed three times with sterile reverse osmosis (RO) water and then re-suspended in 5 mL of RO water. Under control conditions, 1.5 × 107 cells per mL were inoculated in 99.6 mL of medium containing 6% glucose, 2% peptone, and 1% yeast extract with 0.4 mL RO water (w/v).Under inhibitor stress, 1.5 × 107 cells per mL were incubated in 99.6 mL of medium containing 6% glucose, 2% peptone, and 1% yeast extract with 1.04 mM furfural, 62 mM acetic acid, 32.6 mM formic acid, and 0.1 mM vanillin (w/v). Volumes of media were adjusted to account for the addition of the inhibitory compounds (∼400 μL) to ensure that all fermentation processes began with the same glucose content.
Anaerobic conditions were obtained using a sealed butyl plug (Fisher, Loughborough, UK) and aluminium caps (Fisher Scientific). A hypodermic needle attached with a Bunsen valve was pushed through the rubber septum to facilitate the release of CO2. All experiments were performed in triplicate and weight loss was measured at each time point. Fermentations were conducted at 30 °C, on a 15 position magnetic plate (2mag AG, Muenchen, Germany) at 200 rpm. For fermentation using seawater, the samples were prepared as mentioned above except that seawater was used instead of RO water.
Ethanol detection using GC
Samples were taken after fermentation and centrifuged at 1200g to remove cellular materials. The supernatant was then filtered using a 0.45 μm syringe-filter (Corning, US), diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 10 μL was run on a Bruker CP 3900 gas chromatograph (Agilent, CA, US) and compared with a series of ethanol controls. Conditions for GC were using helium gas as a carrier at 1.2 mL min−1. The temperatures of the injector and interface were maintained at 250 °C and 280 °C, respectively. The temperature program for the column oven was as follows: 70 °C for 2 min, a linear ramp to 250 °C at 10 °C min−1 and a hold time of 5 min. Electron impact (EI)-ionization was performed at 70 eV.32
100 and 10 μL was run on a Bruker CP 3900 gas chromatograph (Agilent, CA, US) and compared with a series of ethanol controls. Conditions for GC were using helium gas as a carrier at 1.2 mL min−1. The temperatures of the injector and interface were maintained at 250 °C and 280 °C, respectively. The temperature program for the column oven was as follows: 70 °C for 2 min, a linear ramp to 250 °C at 10 °C min−1 and a hold time of 5 min. Electron impact (EI)-ionization was performed at 70 eV.32
Statistics
PCA and discrimination analysis (DA) were carried out using XLSTAT, a statistical software package designed for use with Excel (https://www.xlstat.com/en/).Heat maps were determined using ClustVis, a web tool for analysing multivariate data (https://biit.cs.ut.ee/clustvis/).
Results
Marine yeast inhibitor tolerance test using phenotypic microarray (PM)
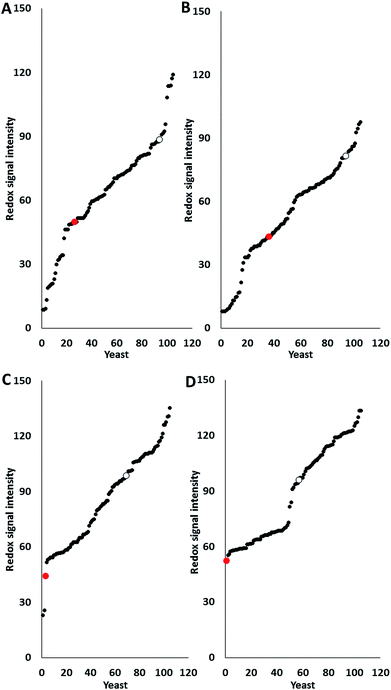
Most pre-treatment methodologies potentially liberate inhibitory compounds of different types such as acetic and formic acids, furfural and HMF. The effect of these compounds on glucose utilisation of 103 marine yeast isolates (AZ1–150) was determined and compared with two terrestrial yeast strains, haploid S. cerevisiae S288C (inhibitor sensitive strain) and industrial polyploid S. cerevisiae NCYC2592 (inhibitor tolerant strain) using phenotypic microarray. Previous research has shown that all yeasts are suitable for analysis using PM technology regardless of their Crabtree status,13 with data expressed as redox signal intensity (Fig. 1). Responses revealed that 11 marine yeasts were more tolerant than S. cerevisiae NCYC2592 to acetic acid (Fig. 1A) and 11 were more tolerant to formic acid (Fig. 1B).Data revealed that 35 marine yeasts were more tolerant in the presence of furfural and 48 marine yeasts were more tolerant in the presence of HMF than the terrestrial S. cerevisiae NCYC2592 (Fig. 1C and D). Analysis revealed that response to furanic compounds was similar for the majority of yeast strains assayed here; for example, strains which were tolerant to furfural also exhibited tolerance to HMF (ESI Fig. S1†). Ranking the yeasts in terms of tolerance to these inhibitory compounds revealed that AZ76, AZ65, AZ101 and AZ116 were the most inhibitor robust strains possessing tolerance in the presence of each of the inhibitory compounds assayed in this study (ESI S2†).
Marine yeast inhibitor tolerance test on a plate reader
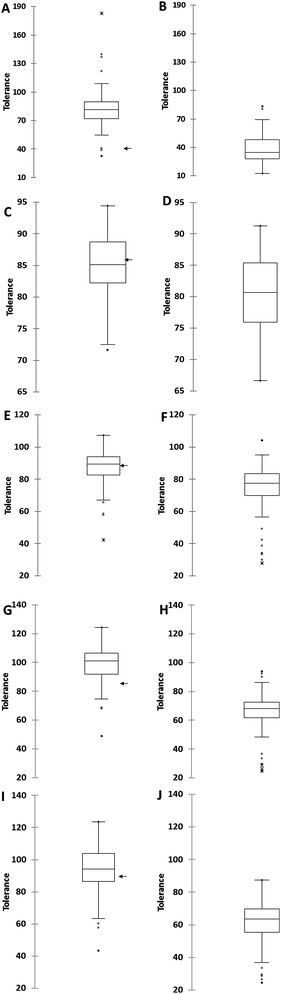
Data derived from the marine yeast inhibitor tolerance test using phenotypic microarray showed that the majority of marine yeasts are more tolerant than S. cerevisiae S288C and some were more tolerant than S. cerevisiae NCYC2592. This result led to more yeasts being isolated from marine environments (seaweed, sand, marine organisms, and driftwood) and their growth curves were determined in the presence of the above inhibitory compounds, which was further extended to include salt (3% NaCl, w/w). The responses of marine yeast strains to these compounds were compared against that of terrestrial yeast strains and the data are expressed as percentage tolerance when compared with control conditions (Fig. 2).We determined that the average tolerance in the presence of salt (NaCl) in marine yeast was 83.8 ± 22.2% compared with 38.43 ± 15.6% for terrestrial yeast (Fig. 2A and B), where 100% tolerance would indicate that there was no difference between the control and the samples in the presence of the inhibitor in terms of growth. Marine yeasts were significantly more tolerant in the presence of salt than terrestrial yeasts (P < 0.0001); marine yeasts were significantly more tolerant in the presence of furfural, acetic and formic acid (Fig. 2C–H) (P < 0.01). The average percentage tolerance of 66 marine yeasts against furfural, acetic acid and formic acid was 99.76 ± 8.0%, 96.23 ± 15.3%, and 106.71 ± 19.49% respectively, while that for the 78 terrestrial yeasts was 79.85 ± 15.56%, 73.98 ± 6.55% and 65.17 ± 14.24%, respectively. There were significant differences in the responses of marine and terrestrial yeasts in the presence of the phenolic compound vanillin (Fig. 2I and J) (P < 0.01), this included several marine yeasts whose growth was stimulated by vanillin (indicated by a response of over 100%). The over 100% relative tolerance of marine yeasts was also observed in experiments in the presence of formic acid. Only one terrestrial yeast S. cerevisiae RM11_1A was stimulated by the presence of acetic acid compared with growth under control conditions (ESI S3†). S. cerevisiae NCYC2592 was taken forward for fermentation studies as this distillery derived strain displayed tolerance above that observed in other yeast strains such as S. cerevisiae Y12 which is derived from sake fermentation or S. cerevisiae YIIc17_E5 which is derived from wine fermentation33 (ESI S3†).
Yeasts from different environments are phenotypically different
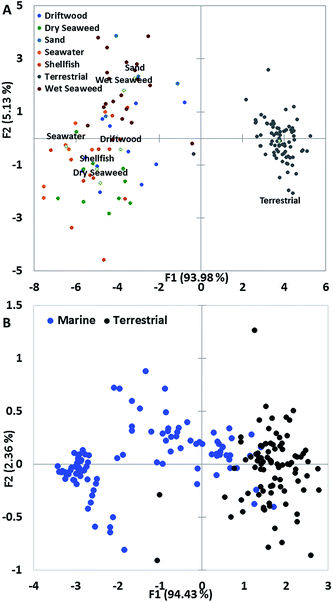
We performed principal component analysis (PCA) on the data generated from populations isolated from marine environments and terrestrial yeasts and this revealed that these populations were very similar in terms of the spread of variance; this was despite the data being derived from different populations of marine yeast and different screening methods. PCA on data derived from the growth of yeast in the presence of inhibitory compounds revealed that yeast isolated from marine environments clustered separately from terrestrial yeast, principally due to their tolerance to salt (Fig. 3A). Only one yeast isolated from wet seaweed and one terrestrial yeast clustered close together (Fig. 3A).Determining a PCA plot on data derived from a PM assay for marine and terrestrial yeasts isolated from seawater revealed that the majority of terrestrial yeasts clustered very tightly but marine yeasts have a disparate range of phenotypes (Fig. 3B). The majority of marine yeast clustered separately from terrestrial yeast with only a few marine yeasts clustering with the terrestrial yeast (Fig. 3B).
Selected marine yeasts exhibit highly reduced potency (IC50) than S. cerevisiae NCYC2592 in the presence of inhibitory compounds
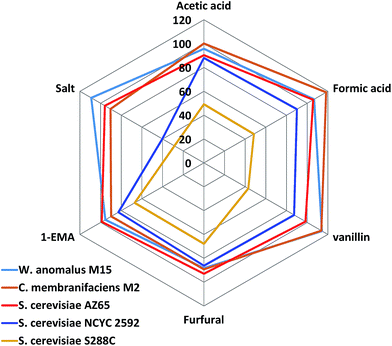
Reviewing the data for marine and terrestrial yeast, it was apparent that marine yeast possessed a more tolerant phenotype than their terrestrial counterparts did; however, the data were compiled using only one concentration of the inhibitors. These concentrations chosen were selected based on expected yeast responses based on published data13,24 or through an initial determination of the effect of the compound on yeast growth (salt). The data could therefore be limited by the concentration chosen, so a dose-dependent curve was plotted and the half-maximal inhibitory concentration (IC50) was determined for selected yeasts against a range of concentrations of salt, furfural, and acetic acid. We determined the IC50 values for the most tolerant marine yeasts (AZ65, M2 and M15), S. cerevisiae NCYC2592 and S. cerevisiae S288C. S. cerevisiae NCYC2592 was the most tolerant terrestrial yeast we tested within this pool of 78 terrestrial strains and has previously been shown to be tolerant in the presence of inhibitory compounds.13S. cerevisiae S288C, has been shown to be sensitive in the presence of inhibitory compounds in this pool of terrestrial yeasts and has been previously24 identified to be inhibitor sensitive (Fig. 4).24 Marine yeasts M2 and M15 were subsequently identified as Candida membranifaciens and Wickerhamomyces anomalus respectively, and AZ65 had already been identified as S. cerevisiae.29IC50 values revealed that AZ65, M2 and M15 had higher (IC50) values in the presence of salt than the IC50 value observed for NCYC2592. W. anomalus M15 was the most tolerant yeast in the presence of salt and had an IC50 value of 10.7% ± 0.2, compared with 6.0% ± 0.3 for NCYC2592, 8.9% ± 1.1 for AZ65, and 10.0% ± 0.2 for C. membranifaciens M2. C. membranifaciens M2 was more tolerant in the presence of acetic acid (95.2 mM ± 1.6) compared with NCYC2592, AZ65 and M15, for which the IC50 values were 75.9 mM ± 1.5, 81.3 mM ± 0.9 and 83.9 mM ± 1.0 respectively. There was little difference in tolerance to furfural except for the S288C strain, which was clearly sensitive in the presence of this compound (Table 1).
| Yeast | Inhibitory compound | ||
|---|---|---|---|
| Furfural (mM) | Salt (%) | Acetic acid (mM) | |
| NCYC2592 | 26.3 ± 0.3 | 6.0 ± 0.3 | 75.9 ± 1.5 |
| AZ65 | 25.9 ± 0.5 | 8.9 ± 1.1 | 81.3 ± 0.9 |
| AZ101 | 26.9 ± 0.8 | 8.2 ± 0.5 | 93.9 ± 0.2 |
| M15 | 24.8 ± 0.2 | 10.7 ± 0.2 | 83.9 ± 1.0 |
| S288C | 15.9 ± 0.2 | 3.9 ± 0.3 | 45.0 ± 1.8 |
Marine yeasts produced more ethanol in the presence of inhibitory compounds than terrestrial yeasts
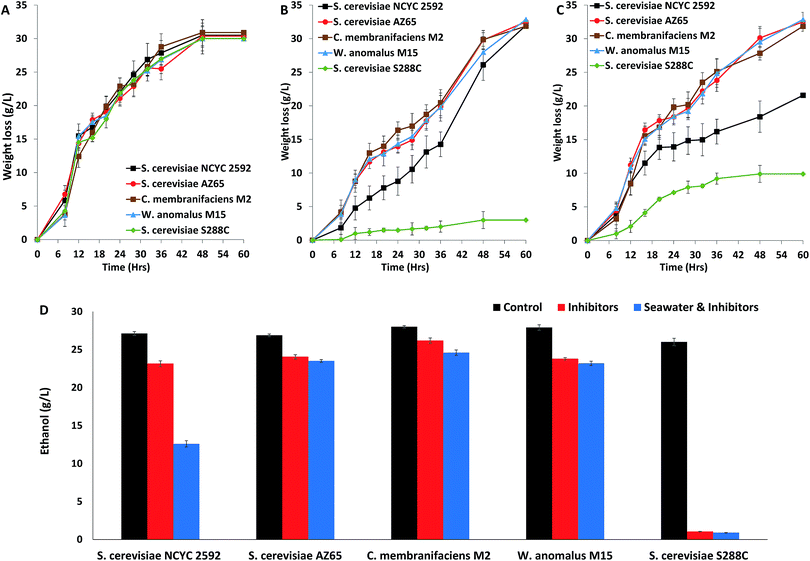
Marine yeast strains W. anomalus M15, C. membranifaciens M2 and S. cerevisiae AZ65 were compared with terrestrial S. cerevisiae strains NCYC2592 and S288C in terms of performance in fermentation. These fermentations were under control conditions and in the presence of inhibitory compounds, with the concentrations of inhibitors based on those produced following hydrothermal (190 °C) pre-treatment of straw.13 Fermentations were monitored by weight loss (g L−1), which is analogous to production of CO2 during the fermentation. Under control conditions, there were no differences in weight loss (g L−1) observed between any of the yeast strains (Fig. 5A). Fermentations in the presence of inhibitory compounds showed that fermentation using S. cerevisiae S228C was very slow when compared with that using the other yeast strains (Fig. 5B); this result was expected as this yeast strain has been shown to be sensitive in the presence of inhibitory compounds.24 Analysing rates of fermentation revealed that all marine yeasts had faster rates of fermentation than S. cerevisiae NCYC2592 (Fig. 5B). Measuring the ethanol content at the end of the fermentation revealed that there was no significant difference between marine yeast and S. cerevisiae NCYC2592 under control conditions (26.8–28.0 g L−1). S. cerevisiae S288C produced slightly less ethanol (25.0 ± 0.19 g L−1) (Fig. 5D). C. membranifaciens M2 produced 26.2 ± 0.3 g L−1 ethanol in the presence of inhibitory compounds compared to 23.2 ± 0.3 g L−1 by S. cerevisiae NCYC2592 (Fig. 5D). The performance of these yeasts when using seawater (salt inhibition) was assessed and compared with fermentations using RO water. Marine yeasts S. cerevisiae AZ65, W. anomalus M15, and C. membranifaciens M2 had faster rates of fermentation than terrestrial yeast S. cerevisiae NCYC2592 (Fig. 5C). Measuring the ethanol content in the fermentation using seawater revealed that marine-derived yeasts produced more ethanol (23.1–24.5 g L−1) than S. cerevisiae NCYC2592 (12.5 ± 0.42 g L−1) (Fig. 5D).Discussion
Isolating strains from natural and industrial habitats is a suitable technique for searching novel strains with specific properties.23,34 Marine environments are an underinvestigated source that could potentially contain strains with various industrially desired phenotypes.23 Zaky et al. (2016) screened marine yeasts for their capability of utilizing sugar monomers, leading to the discovery and identification of several high fermentation capacity marine yeasts.29 This study further explored the inhibitor tolerance ability of marine yeasts. Several marine yeasts performed better than an industrially tolerant terrestrial yeast S. cerevisiae NCYC2592 under acetic acid, formic acid, furfural and HMF stress. We isolated more marine yeasts from a variety of marine resources and habitats and explored the tolerance of these strains in comparison with terrestrial yeasts. Comparing 66 marine yeasts as a clade with a clade of 78 terrestrial yeasts demonstrated that statistically, marine yeasts had better tolerance and therefore a better growth profile than terrestrial yeasts in the presence of inhibitory compounds (Fig. 2). Tolerance of marine yeasts to salt was expected but was confirmed in this study; however, these yeasts also displayed improved tolerance to a range of organic acids and phenolic compounds. The results provided another example that yeasts have global stress response (GSR) that developed via an evolutionary adaptation to stressful environments.35 This suggested that the probability of obtaining a strain with the desired stress tolerance ability is higher from a stressful environment than a non-stressful environment. Constructing a tolerance web graph confirmed that several marine yeasts resisted all five inhibitors better than the most tolerant terrestrial yeast S. cerevisiae NCYC2592 within a pool of 78. A similar strategy has also been successful in identifying xylose utilizing yeast strains from forest floors and manure heaps.36Exploiting the genetic diversity of natural ecosystems could lead to the generation of novel yeast strains with desirable phenotypes for improved bioethanol production.37 Identifying yeasts with innate tolerance capabilities could potentially identify genes that could confer resistance to the chemical constituents of lignocellulosic biomass. Previous studies have indicated that wild-type yeasts respond in a different manner to ethanol stress when compared to laboratory strains.38 PCA analysis has correlated differences in colony morphology with distinct populations in terms of osmotic and ethanol tolerance39 and has been used to identify the key parameters in the brewing process40 and this approach suggests that marine yeasts contain a rich pool of novel genes for improving yeast inhibitor tolerance; for the production of biofuels from lignocellulosic material, raw materials require a pre-treatment to liberate sugars from the plant cell walls, generating inhibitory compounds such as acetic acid, furfural, vanillin and long-chain weak acids.4,13 The presence of these compounds limits the conversion of the sugars into desirable products. Research has helped to identify inhibitor tolerant strains;24,41,42 however, most of these strains could only tolerate individual inhibitory compounds and there is a need for strains that can tolerate a combination of inhibitors. Although there were no differences in bioethanol production in fermentations using inhibitor free media, marine yeast C. membranifaciens M2 produced significantly higher bioethanol than the terrestrial counterpart S. cerevisiae NCYC2592 in the presence of inhibitors. When seawater was replaced with freshwater in the medium, C. membranifaciens M2 produced nearly twice as much bioethanol as S. cerevisiae NCYC2592. The results indicated the feasibility of highly efficient bioethanol fermentation using un-treated lignocellulosic hydrolysates, possibly together with the replacement of freshwater by seawater. Marine yeast clearly possesses important physiological properties; these properties make the use of marine yeast in an industrial setting more likely.43
Conclusions
Marine yeast possesses improved tolerance to a wide range of inhibitory compounds and demonstrates excellent fermentative bioethanol synthesis capacity in a stressful environment. Clustering revealed that marine yeasts are phenotypically distinct from terrestrial yeasts and produce significantly higher concentrations of ethanol in the presence of inhibitory compounds. Future work will focus on the biotechnological use of marine yeasts in industrial biotechnology including the use of seawater as a water source in fermentation; future research will focus on how these yeasts tolerate the presence of inhibitory compounds, their performance in large-scale fermentations and suitability in other industrial processes such as brewing, baking and production of high-value chemicals.Declarations
This manuscript did not involve the use of human participants, data or human tissue; therefore ethical approval is not applicable. There are no data of individual people used in this manuscript so consent for publication is not applicable.The datasets supporting the conclusions of this article are included within the article (and its additional file(s)). Raw data such as growth curves are available upon request.
All authors consent for this manuscript to be submitted for publication purposes.
Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
The authors would also like to acknowledge the help of Dr Jessica Adams (Aberystwyth University) for supplying the seaweed used to isolate yeast strains. The authors acknowledge the funding from the University of Huddersfield URF (URF/408) for supporting this research.References
- A. K. Chandel, V. K. Garlapati, A. K. Singh, F. A. F. Antunes and S. S. da Silva, Bioresour. Technol., 2018, 264, 370–381 CrossRef CAS PubMed.
- D. N. Moyses, V. C. Reis, J. R. de Almeida, L. M. de Moraes and F. A. Torres, Int. J. Mol. Sci., 2016, 17, 207 CrossRef PubMed.
- V. Mathioudakis, P. W. Gerbens-Leenes, T. H. Van der meer and A. Y. Hoekstra, J. Cleaner Prod., 2017, 571–582 CrossRef.
- S. Wilkinson, D. Greetham and G. A. Tucker, Biofuel Res. J., 2016, 5, 11 Search PubMed.
- M. J. Taherzadeh and K. Karimi, Int. J. Mol. Sci., 2008, 9, 1621–1651 CrossRef CAS PubMed.
- A. Zaky, N. Pensupa, Á. Andrade-Eiroa, G. A. Tucker and C. Du, J. Food Compos. Anal., 2017, 56, 8 CrossRef.
- H. B. Klinke, A. B. Thomson and B. K. Ahring, Appl. Microbiol. Biotechnol., 2004, 66, 16 CrossRef PubMed.
- O. J. Sanchez and C. A. Cardona, Bioresour. Technol., 2008, 99, 5270–5295 CrossRef CAS PubMed.
- O. Mafe, N. Pensupa, E. Roberts and C. Du, Advanced generation of bioenergy, Royal Society of Chemistry, 2014 Search PubMed.
- J. Zhang, W. X. Zhang, Z. Y. Wu, J. Yang, Y. H. Liu, X. Zhong and Y. Deng, Prep. Biochem. Biotechnol., 2013, 43, 1–21 CrossRef PubMed.
- D. Steinbach, A. Kruse and J. Sauer, Biomass Convers. Biorefin., 2017, 7, 27 Search PubMed.
- D. Greetham, J. Bioprocess. Biotech., 2014, 5, 192 Search PubMed.
- D. Greetham, T. Wimalasena, D. W. Kerruish, S. Brindley, R. N. Ibbett, R. L. Linforth, G. Tucker, T. G. Phister and K. A. Smart, J. Ind. Microbiol. Biotechnol., 2014, 41, 931–945 CrossRef CAS PubMed.
- S. Helle, D. Cameron, J. Lam, B. White and S. Duff, Enzyme Microb. Technol., 2003, 33, 7 CrossRef.
- N. Banergee, R. Bhatnagar and L. Vismanathan, Eur. J. Appl. Microbiol. Biotechnol., 1981, 11, 3 Search PubMed.
- D. Greetham, A. J. Hart and G. A. Tucker, Biomass Bioenergy, 2016, 85, 53 CrossRef CAS.
- D. B. Stengel, S. Connan and Z. A. Popper, Biotechnol. Adv., 2011, 29, 483–501 CrossRef CAS PubMed.
- Z. Yuan, Y. Wen and G. Li, Bioresour. Technol., 2018, 259, 228–236 CrossRef CAS PubMed.
- H. L. Woo and T. C. Hazen, Front. Microbiol., 2018, 9, 922 CrossRef PubMed.
- L. C. P. Wentzel, F. J. Inforsato, Q. V. Montoya, B. G. Rossin, N. R. Nascimento, A. Rodrigues and L. D. Sette, Microb. Ecol., 2019, 77, 12–24 CrossRef CAS PubMed.
- A. S. Jones EBG and K. L. Pang, Malays. J. Sci., 2013, 32, 6 Search PubMed.
- A. S. Zaky, D. Greetham, G. A. Tucker and C. Du, Sci. Rep., 2018, 8, 12127 CrossRef PubMed.
- A. S. Zaky, G. A. Tucker, Z. Y. Daw and C. Du, FEMS Yeast Res., 2014, 14, 813–825 CrossRef CAS PubMed.
- T. T. Wimalasena, D. Greetham, M. E. Marvin, G. Liti, Y. Chandelia, A. Hart, E. J. Louis, T. G. Phister, G. A. Tucker and K. A. Smart, Microb. Cell Fact., 2014, 13, 47 CrossRef PubMed.
- D. Greetham, A. Zaky, O. Makanjoula and C. Du, Current Opinion in Green and Sustainable Chemistry, 2018, 14, 7 CrossRef.
- L. Wang, Z. Chi, X. Wang, L. Ju, Z. Chi and N. Guo, Microbiol. Res., 2008, 163, 255–266 CrossRef CAS PubMed.
- E. T. Kostas, D. A. White, C. Du and D. J. Cook, J. Appl. Phycol., 2016, 28, 1427–1441 CrossRef CAS PubMed.
- N. Wei, J. Quarterman and Y. S. Jin, Trends Biotechnol., 2013, 31, 70–77 CrossRef CAS PubMed.
- A. S. Zaky, D. Greetham, E. J. Louis, G. A. Tucker and C. Du, J. Microbiol. Biotechnol., 2016, 26, 1891–1907 CrossRef CAS PubMed.
- B. R. Bochner, Nature, 1989, 339, 157–158 CrossRef CAS PubMed.
- Y. Lin and S. Tanaka, Appl. Microbiol. Biotechnol., 2006, 69, 627–642 CrossRef CAS PubMed.
- T. Somboon and S. Sansuk, Food Chem., 2018, 253, 300–304 CrossRef CAS PubMed.
- G. Liti, D. M. Carter, A. M. Moses, J. Warringer, L. Parts, S. A. James, R. P. Davey, I. N. Roberts, A. Burt, V. Koufopanou, I. J. Tsai, C. M. Bergman, D. Bensasson, M. J. O'Kelly, A. van Oudenaarden, D. B. Barton, E. Bailes, A. N. Nguyen, M. Jones, M. A. Quail, I. Goodhead, S. Sims, F. Smith, A. Blomberg, R. Durbin and E. J. Louis, Nature, 2009, 458, 337–341 CrossRef CAS PubMed.
- A. D. Ferreira, S. I. Mussatto, R. M. Cadete, C. A. Rosa and S. S. Silva, Yeast, 2011, 28, 547–554 CrossRef CAS PubMed.
- B. R. Gibson, S. J. Lawrence, J. P. R. Leclaire, C. D. Powell and K. A. Smart, FEMS Microbiol. Rev., 2007, 31, 25 CrossRef PubMed.
- A. Farooq, D. Greetham, A. Somani, M. E. Marvin, E. J. Louis and C. Du, Journal of Applied Microbiology and Biochemistry, 2018, 2(3), 9 Search PubMed.
- Y. L. Zheng and S. A. Wang, PLoS One, 2015, 10, e0133889 CrossRef PubMed.
- J. A. Lewis, I. M. Elkon, M. A. McGee, A. J. Higbee and A. P. Gasch, Genetics, 2010, 186, 1197–1205 CrossRef CAS PubMed.
- V. R. Reis, A. Antonangelo, A. P. G. Bassi, D. Colombi and S. R. Ceccato-Antonini, Braz. J. Microbiol., 2017, 48, 268–274 CrossRef CAS PubMed.
- S. Ohnuki, K. Enomoto, H. Yoshimoto and Y. Ohya, J. Biosci. Bioeng., 2014, 117, 278–284 CrossRef CAS PubMed.
- T. Jansen, J. W. Hoff, N. Jolly and W. H. van Zyl, Folia Microbiol., 2018, 63, 155–168 CrossRef CAS PubMed.
- D. Gonzalez-Ramos, A. R. Gorter de Vries, S. S. Grijseels, M. C. van Berkum, S. Swinnen, M. van den Broek, E. Nevoigt, J. M. Daran, J. T. Pronk and A. J. van Maris, Biotechnol. Biofuels, 2016, 9, 173 CrossRef PubMed.
- V. Mukherjee, D. Radecka, G. Aerts, K. J. Verstrepen, B. Lievens and J. M. Thevelein, Biotechnol. Biofuels, 2017, 10, 216 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: S1 – origins of terrestrial Saccharomyces spp. used in this study, S2 – ranking of phenotypic response and S3 – comparison of growth curves of terrestrial and marine-derived yeast. See DOI: 10.1039/c9se00029a |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2019 |