Open Access Article
Open Access ArticleEndoplasmic reticulum targeting fluorescent probes to image mobile Zn2+†
Le
Fang
 a,
Giuseppe
Trigiante
b,
Rachel
Crespo-Otero
a,
Giuseppe
Trigiante
b,
Rachel
Crespo-Otero
 a,
Chris S.
Hawes
a,
Chris S.
Hawes
 c,
Michael P.
Philpott
b,
Christopher R.
Jones
c,
Michael P.
Philpott
b,
Christopher R.
Jones
 a and
Michael
Watkinson
a and
Michael
Watkinson
 *c
*c
aSchool of Biological and Chemical Science, Queen Mary University of London, The Joseph Priestley Building, Mile End Road, London, E1 4NS, UK
bCentre for Cutaneous Research, Institute of Cell and Molecular Science, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London E1 2AT, UK
cThe Lennard-Jones Laboratories, School of Chemical and Physical Science, Keele University, ST5 5BG, UK. E-mail: m.watkinson@keele.ac.uk
First published on 16th October 2019
Abstract
Zn2+ plays an important role in the normal function of the endoplasmic reticulum (ER) and its deficiency can cause ER stress, which is related to a wide range of diseases. In order to provide tools to better understand the role of mobile Zn2+ in ER processes, the first custom designed ER-localised fluorescent Zn2+ probes have been developed through the introduction of a cyclohexyl sulfonylurea as an ER-targeting unit with different Zn2+ receptors. Experiments in vitro and in cellulo show that both probes have a good fluorescence switch on response to Zn2+, high selectivity over other cations, low toxicity, ER-specific targeting ability and are efficacious imaging agents for mobile Zn2+ in four different cell lines. Probe 9 has been used to detect mobile Zn2+ changes under ER stress induced by both tunicamycin or thapsigargin, which indicates that the new probes should allow a better understanding of the mechanisms cells use to respond to dysfunction of zinc homeostasis in the ER and its role in the initiation and progression of diseases to be developed.
Introduction
The endoplasmic reticulum (ER), an organelle in eukaryotic cells, serves a number of important cellular roles, such as protein synthesis and transport, protein folding, carbohydrate metabolism, and lipid and steroid synthesis.1 Proteins synthesized in the ER are normally properly folded and transported to the Golgi apparatus, however, when there are changes to ER function, resulting from factors such as ageing, genetic mutations, or the environment, unfolded or misfolded proteins are synthesized and accumulate in the ER,2 causing ER stress, which activates the unfolded protein response (UPR).3 It has been found that ER stress can result in a wide range of diseases, such as inflammation,4,5 diabetes6,7 and neurodegenerative disorders,8 including Parkinson's and Alzheimer's diseases.9,10 Additionally, the ER acts as an intracellular store for biological mediators, including zinc, which it requires for normal function.11,12 For example, it has been found that zinc can be released from thapsigargin- and inositol 1,4,5-trisphosphate (IP3)-sensitive ER storage in cortical neurons.13 In light of this, it is unsurprising that a large number of reports demonstrate that the depletion of zinc transporters and zinc deficiency can cause ER stress and upregulate the UPR,14–18 however, the role of ‘free’ or ‘mobile’ zinc in this process is little understood due to the lack of suitable molecular tools to image this subcellular region that exist.Small molecule fluorescent probes, which have great advantages of high sensitivity and selectivity, low toxicity, and good photophysical properties, are increasingly important tools for the investigation of biological events19–25 and there have been extensive efforts in the development of a range of probes to detect mobile zinc.26–30 However, a failure to control the probe's subcellular location or indeed their different behaviour across cell lines31 limits their utility somewhat.32,33 Though there is a clear need for ER localised probes to monitor mobile Zn2+ flux, to our knowledge there are only two such reports that have appeared in the last decade and in both cases ER localisation was adventitious.34,35 Thus, there remains a need for effective and reliable methods to localize small molecule Zn2+ probes to target the ER.
The most effective and reliable strategy employed to date to provide organelle targeting capability in small molecule probes has been to incorporate specific organelle targeting vectors into them. For example, the triphenylphosphonium salt (TPP) has been widely demonstrated to be an effective mitochondrial targeting group,36–38 whilst basic ethylenediamine39 or morpholine40–42 groups have been successfully applied to target the lysosome. However, this strategy has not yet been widely explored in ER localized probes.
Previously we have reported a modular ‘click’ synthetic methodology to produce an array of biologically targeted Zn2+ probes for both extracellular and intracellular imaging of zinc through the incorporation of a range of targeting units.43,44 We were therefore hopeful that this approach could be further extended towards the development of ER specific probes, provided that a suitable targeting vector could be identified and incorporated in a modular manner utilising our established chemistry. Herein we present our efforts to this end.
Results and discussion
Probe design and synthesis
Recently a number of reports have appeared in which the methyl sulfonamide group has been used as a targeting unit to visualize hydrogen peroxide,45 methylglyoxal,46 hypochlorite47 and hydrogen sulfide48 in the ER. Inspired by this, we first synthesized fluorescent probe 1 (ref. 49) using our modular synthetic protocol that incorporated the methyl sulfonamide unit (Fig. 1) and whilst the fluorescence response of the probe to Zn2+ was efficacious in vitro and it co-localised to the ER with ER tracker red, it was unfortunately also widely dispersed in other organelles in HeLa cells.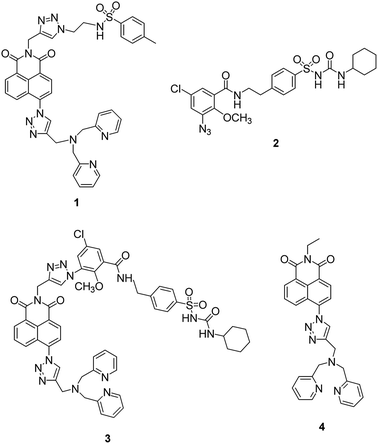 | ||
| Fig. 1 The structure of probe 1, glibenclamide azide 2, unsuccessfully synthesized probe 3 and non-targeting probe 4. | ||
We therefore decided to employ a revised strategy based on the incorporation of glibenclamide as the targeting unit. Glibenclamide, an antidiabetic drug, is employed in the commercial dyes ER tracker red and ER tracker green because it binds and inhibits ATP-sensitive K+ channels that are abundant in the ER membrane.50,51 We were further encouraged by a report of the use of glibenclamide as a targeting group in a small molecule chemical probe which was shown to selectively accumulate in the ER and image glutathione.52 We therefore sought to develop a route to a ‘clickable’ variant of glibenclamide that we could incorporate into our modular synthetic route and prepared ‘click’ glibenclamide 2 through six steps in moderate yield (see Scheme S1, ESI†). Gratifyingly the CuAAC reaction with alkyne S6 proceeded, as expected, in good yield (Scheme S2, ESI†), however, to our bemusement, when we came to install the Zn2+ binding unit 8, the second ‘click’ reaction to give compound 3 failed completely despite a range of Cu(I) sources being tested. Given the success of the first ‘click’ reaction we decided to investigate the development of a ‘reverse’ methodology in which the metal-binding unit was first installed into anhydride S9 before addition of the targeting unit via a second ‘click’ process was attempted (see ESI, Scheme S3†). Although the first ‘click’ reaction again worked well, the second ‘click’ reaction to incorporate the glibenclamide azide completely failed.
At this point we decided to review our synthetic strategy and were attracted by a number of recent reports that reveal the cyclohexyl sulfonylurea moiety of glibenclamide is an important element in binding with ATP-sensitive K+ channels, which are present in the ER membrane.53–55 We therefore designed an alternative route in which this ER targeting unit was directly incorporated into the naphthalimide, which also makes the synthesis route shorter and easier.
To synthesize the probes, we started from 4-(2-aminoethyl)benzenesulfonamide to give ER-targeting group 6 through the route depicted (see Scheme S4, ESI†) and directly introduced this into the fluorophore through reaction with commercially available 4-bromo-1,8-naphthalic anhydride 5 (Scheme 1). This was then conveniently converted to azide 7 using standard conditions for such SNAr reactions, which then underwent smooth and high yielding ‘click’ reactions with alkynes 8 (ref. 56) or 10,44 to yield probes 9 and 12, the latter after deprotection of 11 with TFA. All products were satisfactorily characterized by 1H, 13C NMR and IR spectroscopies as well as high-resolution mass spectrometry.
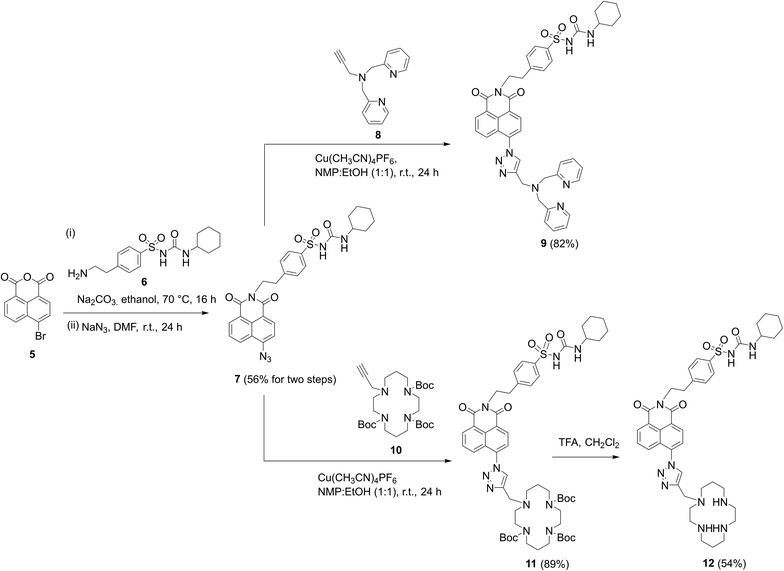 | ||
| Scheme 1 The successful synthetic route to probes 9 and 12 incorporating cyclohexyl sulfonylurea as the ER targeting unit. | ||
Photophysical properties in solution
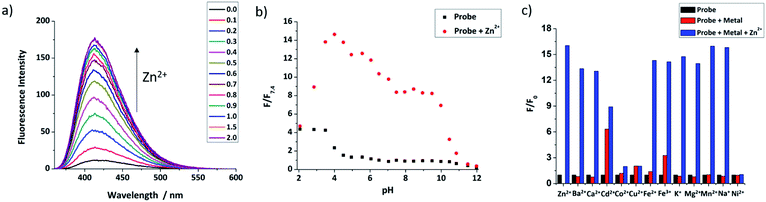
Fluorescence titrations with Zn2+ were first undertaken to show that both probes have a switch-on response to Zn2+. From Fig. 2a and S2 (ESI†), it can clearly be seen that with increasing Zn2+ equivalents, the fluorescence intensity of probes 9 and 12 increased gradually with about 16-fold and 4-fold maximal increase, respectively. Subsequent treatment with TPEN (N,N,N′,N′-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine), a strong Zn2+ chelator, resulted in the expected decrease in fluorescence with the fluorescence of 9 being completely quenched while that of 12 was partly quenched (Fig. S3, ESI†), which is consistent with previous reports.57 The Job's plot (Fig. S4, ESI†) revealed the expected 1:1 binding stoichiometry with Zn2+. The dissociation constants Kd for probes 9 and 12 were obtained through non-linear curve fitting using reported methods58,59 at a number of probe concentrations (Fig. S5 and Table S1, ESI†). However, because of the very low Kd values for these compounds and to mimic the buffered nature of mobile zinc in cellulo, EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid) was used as a competitive chelator27,60,61 and the Kd values again calculated at three different concentrations (Fig. S6, ESI†) with optimal fit being observed at 0.01 μM in all cases (Table S2, ESI†) revealing the dissociation constant of 9 to be 3.5 nM and 4.7 nM for 12. The detection limits of 9 and 12 (Fig. S7, ESI†) were also estimated to be 47 pM and 0.71 nM, respectively.The quantum yields of both probes were measured using anthracene as the standard (Φ = 0.27 in ethanol). It was found that the quantum yield of 9 was 0.041, which increased to 0.25 after binding with 1 eq. of Zn2+. In line with our recent report,44,62 the quantum yield of 12 was much lower, 0.013 and only increased to 0.041 for the complex with 1 equivalent of Zn2+ (Fig. S8, ESI†).
The pH-dependent fluorescence response of both probes was tested to confirm that they have fluorescence responses to Zn2+ in the biologically relevant pH range. Fig. 2b shows that 9 has good switch on fluorescence response to Zn2+ over a wide pH range of 3.0–10.0, and the fluorescence of both probe and complex is increased in an acidic environment. In contrast, 12 (Fig. S11, ESI†) shows a poor response in acid conditions, presumably due to the basic nature of the polyamine ligand, but works well in the pH range 6.0–12.0; given that the reported pH of the ER is the same as the cytoplasm, which is about 7.2,63 both probes should have a response to mobile Zn2+in cellulo. Through non-linear curve fitting (applied with the eqn S(4) and S(5)),† the pKa values of both probes can be obtained, which are pKa1 = 0.81 ± 0.19, pKa2 = 4.34 ± 0.11, pKa3 = 6.83 ± 0.53, pKa4 = 11.63 ± 0.41 for 9, and pKa1 = 3.20 ± 0.14, pKa2 = 8.07 ± 0.11, pKa3 = 11.07 ± 0.58 for 12 (Fig. S10 and S12, ESI†). These values are comparable to those reported for related compounds.44
The selectivity of the two probes over a range of other competing cations was also investigated. As shown in Fig. 2c, beside Zn2+, the fluorescence of 9 was not switched on obviously after addition of 5 equivalents of other cations, except for the stereoelectronic isostere Cd2+, which is a common issue for many Zn2+ probes,64 but is not a problem in biological milieu due to its negligibly low concentration. Subsequent addition of Zn2+ resulted in recovery of a fluorescence response in most cases, however for Co2+, Cu2+ and Ni2+ fluorescence was not recovered, but as they mostly exist in bound forms in biology, rather than the free cations tested here this is unconcerning. For 12, results were similar, as shown in Fig. S13 (see ESI†), with only Cu2+, Fe2+ and Fe3+ continuing to quench fluorescence after the addition of Zn2+. Importantly, the metal ions Na+, K+, Ca2+ and Mg2+, which are abundant in cells, show no effect on either probe. Overall these results indicate that they should show a selective response to ‘mobile’ Zn2+in cellulo.
DFT simulations
DFT and TDDFT calculations were undertaken to understand the binding behaviour of 9 and 12 with Zn2+ and their excited states. The optimised structure of probe 9 with 1 equivalent of Zn2+ (Fig. S14, ESI†) shows the Zn2+ is bound to the nitrogen in ligand N,N-di-(2-picolyl)ethylenediamine (DPEN) and a nitrogen atom of the triazole. For 12 (Fig. S15, ESI†), the triazole nitrogen is involved in binding with Zn2+ in addition to the cyclam nitrogen donors, which is consistent with previous observations in closely related analogues.65,66The calculated vertical excitations and emissions are in relatively good agreement with experimental data (see Table S3 and S4, ESI†). The excited state calculations also show that positions of the absorption maximum and the emission energies are not significantly affected by the formation of the complexes with Zn2+ as observed experimentally. This behaviour can be understood by visualising the S1–S0 electron density plots (Fig. S16 for 9, Fig. S17 for 12, ESI†). The electronic transition is localised on the naphthalimide moiety and the vicinal triazole, and that there is no major difference between probes and their complexes with Zn2+; only a slight decrease of electron density on the nitrogen from the triazole is observed when it is involved in complex formation and the oscillator strength is not significantly affected. These calculations indicate that the enhancement of emissive behaviour of the complexes should be related to a reduced decay through nonradiative pathways after complexation with Zn2+ in contrast with the typical PET mechanism. The role of alternative mechanisms to PET in enhancing the fluorescence of organometallic complexes has been recently highlighted.67,68 The formation of the complex hinders large amplitude vibrations in the vicinity of the fluorophore hampering access to nonradiative mechanisms and increasing the quantum yield of emission. The restriction of intramolecular rotations can also hinder the access to low energy conical intersections associated with ultrafast decay to the ground state. Tang et al. have recently shown how restriction of intramolecular motions in a Zn2+ complex can hamper the access to a dark state enhancing the fluorescence response.69
ER-targeting ability
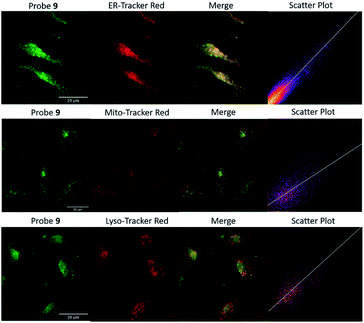
Encouraged by the promising properties of 9 and 12in vitro, we attempted to assess their suitability for imaging Zn2+ in cells. Firstly, the innate toxicity of them was measured by alamarBlue assays with HeLa cells (Fig. S18 and S19, ESI†).22 The cell viabilities were over 90% after 24 h incubation with both probes in a concentration range 10–50 μM compared to the control group, demonstrating both have very low cytotoxicity.70The targeting behaviour of 9 and 12 was then studied using co-staining assays. HeLa cells were co-incubated with 9 and ER-tracker red, and as shown in Fig. 3, the fluorescence of 9 from the green channel overlays well with that of ER-tracker red, which was obtained from the red channel, giving an excellent Pearson's correlation coefficient of 0.92. The negative controls of 9 co-incubated with Mito-tracker red and Lyso-tracker red, did not reveal a good overlay and Pearson's correlation coefficients were 0.52 and 0.58, respectively. Similar results were observed for 12 (see Fig. S22, ESI†) with Pearson's coefficients of 0.85, 0.52, 0.45 being measured for the ER, mitochondria and lysosome respectively. A control probe that did not contain a targeting unit 4 (ref. 49) was observed to be widely distributed in all three organelles (Fig. S23, ESI†) and this is also consistent with our original study.65
To confirm the probes have ER-targeting ability in different cell types, a human breast cancer cell line MCF-7, a sweat gland tissue cell line EC23 and a human liver cancer cell line HepG2 were used to study the ER-targeting behaviour. The results (see Fig. S24–S26 for probe 9, Fig. S27–S29 for probe 12, ESI†) show that probe localisation is similar to that observed in HeLa cells demonstrating the general applicability of the probes. Based on these data, we can conclude that both 9 and 12 show excellent, and general, ER-targeting capability.
Zn2+ fluorescence response in cells
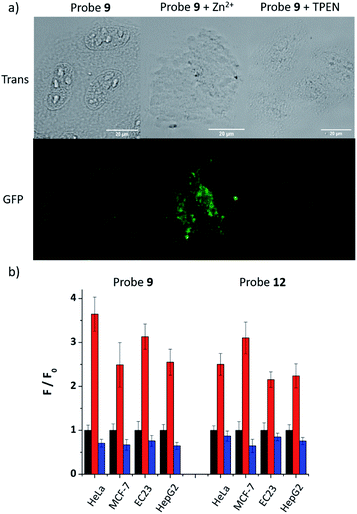
As 9 and 12 can localise to the ER, their fluorescence response to ‘mobile’ Zn2+in cellulo was tested to demonstrate that they have the ability to image Zn2+ in the ER. As shown in Fig. 4a, the fluorescence response of probe 9 in HeLa cells could be clearly observed in the ER. Upon the addition of zinc pyrithione, a membrane permeable zinc source, the fluorescence intensity increased considerably; the addition of TPEN resulted in almost complete quenching of fluorescence, as expected from in vitro control experiments. The fluorescence intensity read from these images (Fig. 4b) clearly reveals these changes. For example, in HeLa cells the intensity of the fluorescence response increases 3.6 times after the addition of zinc pyrithione and decreases by 30% on addition of TPEN compared to that observed for 9 alone. Similar results were observed in other cell lines and also for 12 (Fig. 4b and S30–S36, ESI†) demonstrating that both probes are tractable in their ability to image mobile Zn2+in cellulo.Fluorescence change under ER stress
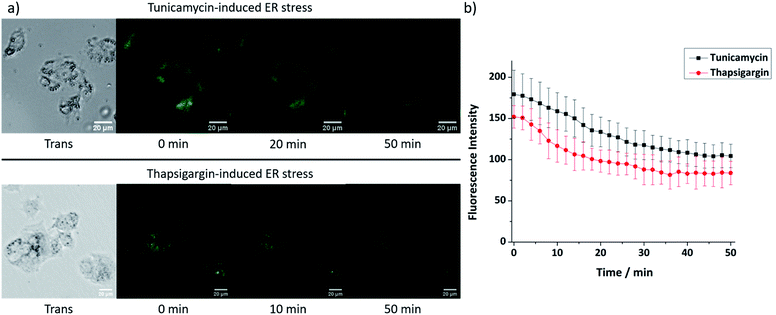
As 9 shows excellent ER-localisation and Zn2+ response, we sought to explore whether it could be applied to monitor changes in mobile Zn2+ levels. It is reported that ER stress can be induced by tunicamycin and thapsigargin with tunicamycin causing unfolded protein accumulation and thapsigargin reducing Ca2+ levels. As shown in Fig. 5, the fluorescence intensity of 9 in HepG2 cells decreased significantly after treatment with both ER stress inducers, indicating that the mobile Zn2+ level in the ER was significantly depleted under ER stress within 1 hour. As a control, no fluorescence intensity change was observed in their absence (Fig. S37, ESI†).Conclusions
In conclusion, the ER localised fluorescent probes 9 and 12 have been developed to image mobile Zn2+ in the ER through the introduction of a simple sulfonylurea targeting group. They show a good fluorescence response to Zn2+, high selectivity over biologically relevant competing cations, and can be used to detect Zn2+ over a wide range of pH in vitro. Their low toxicity, ER-targeting and ability to image mobile Zn2+ in different cell lines should allow them to be applied in imaging Zn2+ related biological processes in the ER. The fluorescence intensity of 9 was also shown to decrease under ER stress inducers, indicating that mobile Zn2+ levels are reduced. The ER localised probes may therefore provide a platform to understand the mechanisms cells use to respond to dysfunction of zinc homeostasis in the ER, and to develop a better understanding of the role of Zn2+ in the initiation and progression of related diseases.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the EPSRC National Mass Spectrometry Service, University of Wales, Swansea for the provision of high-resolution mass spectrometry. DFT calculations were performed on Queen Mary's Apocrita HPC facility, supported by QMUL Research-IT. LF is grateful to the Chinese Scholarship Council for the provision of a PhD studentship.References
- D. S. Schwarz and M. D. Blower, Cell. Mol. Life Sci., 2016, 73, 79–94 CrossRef CAS PubMed.
- H. Yoshida, FEBS J., 2007, 274, 630–658 CrossRef CAS PubMed.
- K. Mori, Cell, 2000, 101, 451–454 CrossRef CAS PubMed.
- K. Zhang and R. J. Kaufman, Nature, 2008, 454, 455–462 CrossRef CAS PubMed.
- A. D. Garg, A. Kaczmarek, O. Krysko, P. Vandenabeele, D. V. Krysko and P. Agostinis, Trends Mol. Med., 2012, 18, 589–598 CrossRef CAS PubMed.
- D. L. Eizirik, A. K. Cardozo and M. Cnop, Endocr. Rev., 2008, 29, 42–61 CrossRef CAS PubMed.
- S. H. Back and R. J. Kaufman, Annu. Rev. Biochem., 2012, 81, 767–793 CrossRef CAS PubMed.
- C. Hetz and S. Saxena, Nat. Rev. Neurol., 2017, 13, 477–491 CrossRef CAS PubMed.
- J. H. Lin, P. Walter and T. Benedict Yen, Annu. Rev. Phytopathol., 2008, 3, 399–425 CrossRef CAS PubMed.
- L. Ozcan and I. Tabas, Annu. Rev. Med., 2012, 63, 317–328 CrossRef CAS PubMed.
- Y. Hattori, H. Koseki, T. Sato, H. Ohno, T. Watanabe, H. Izumi, T. Hara, W. Ohashi, T. Fukada, K. Hase, A. Hijikata, T. Iwanaga, K. Mishima, Y. Furusawa, O. Ohara, T. Irié, H. Mori, S. Robine, H. Watarai and S. Kimura, PLoS Genet., 2016, 12, e1006349 CrossRef PubMed.
- C. D. Ellis, F. Wang, C. W. MacDiarmid, S. Clark, T. Lyons and D. J. Eide, J. Cell Biol., 2004, 166, 325–335 CrossRef CAS PubMed.
- C. J. Stork and Y. V. Li, J. Mol. Signaling, 2010, 5, 1–6 CrossRef PubMed.
- M.-H. Kim, T. B. Aydemir, J. Kim and R. J. Cousins, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E5805–E5814 CrossRef CAS PubMed.
- M. Wang, Q. Xu and M. Yuan, Plant Signaling Behav., 2011, 6, 77–79 CrossRef CAS PubMed.
- C. Giunta, J. M. Walker, A. E. Palmer, J. Jeong, B. Steinmann, M. Rohrbach, D. J. Eide, J. G. Park and F. Wang, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E3530–E3538 CrossRef PubMed.
- T. S. Le Nguyen, K. Kohno and Y. Kimata, Biosci., Biotechnol., Biochem., 2013, 77, 1337–1339 CrossRef CAS PubMed.
- Z. Zhou, D. Dong, W. Zhang, Q. Li, Q. Sun, X. Sun, X. Sun, W. Zhong and X. Tan, Am. J. Physiol. Gastrointest. Liver Physiol., 2015, 308, G757–G766 CrossRef PubMed.
- D. Wu, A. C. Sedgwick, T. Gunnlaugsson, E. U. Akkaya, J. Yoon and T. D. James, Chem. Soc. Rev., 2017, 46, 7105–7123 RSC.
- Y. Chen, Y. Bai, Z. Han, W. He and Z. Guo, Chem. Soc. Rev., 2015, 44, 4517–4546 RSC.
- H. M. Kim and B. R. Cho, Chem. Rev., 2015, 115, 5014–5055 CrossRef CAS PubMed.
- D. Ding, K. Li, B. Liu and B. Z. Tang, Acc. Chem. Res., 2013, 46, 2441–2453 CrossRef CAS PubMed.
- H. M. Kim and B. R. Cho, Acc. Chem. Res., 2009, 42, 863–872 CrossRef CAS PubMed.
- L. Yuan, W. Lin, K. Zheng and S. Zhu, Acc. Chem. Res., 2013, 46, 1462–1473 CrossRef CAS PubMed.
- M. H. Lee, J. S. Kim and J. L. Sessler, Chem. Soc. Rev., 2015, 44, 4185–4191 RSC.
- G. K. Walkup, S. C. Burdette, S. J. Lippard and R. Y. Tsien, J. Am. Chem. Soc., 2000, 122, 5644–5645 CrossRef CAS.
- T. Hirano, K. Kikuchi, Y. Urano, T. Higuchi and T. Nagano, J. Am. Chem. Soc., 2000, 122, 12399–12400 CrossRef CAS.
- F. Qian, C. Zhang, Y. Zhang, W. He, X. Gao, P. Hu and Z. Guo, J. Am. Chem. Soc., 2009, 131, 1460–1468 CrossRef CAS PubMed.
- S. Chen, R. Bleher, S. A. Garwin, T. V. O'Halloran, T. K. Woodruff, A. R. Bayer, B. Y. Kong, E. L. Que, F. E. Duncan, S. C. Gleber, V. P. Dravid and S. Vogt, Nat. Chem., 2014, 7, 130–139 Search PubMed.
- J. M. Goldberg, F. Wang, C. D. Sessler, N. W. Vogler, D. Y. Zhang, W. H. Loucks, T. Tzounopoulos and S. J. Lippard, J. Am. Chem. Soc., 2018, 140, 2020–2023 CrossRef CAS PubMed.
- W. Maret, Metallomics, 2015, 7, 202–211 RSC.
- H. Zhu, J. Fan, J. Du and X. Peng, Acc. Chem. Res., 2016, 49, 2115–2126 CrossRef CAS PubMed.
- W. Xu, Z. Zeng, J. H. Jiang, Y. T. Chang and L. Yuan, Angew. Chem., Int. Ed., 2016, 55, 13658–13699 CrossRef CAS PubMed.
- W. Lin, D. Buccella and S. J. Lippard, J. Am. Chem. Soc., 2013, 135, 13512–13520 CrossRef CAS PubMed.
- X. Gan, P. Sun, H. Li, X. Tian, B. Zhang, J. Wu, Y. Tian and H. Zhou, Biosens. Bioelectron., 2016, 86, 393–397 CrossRef CAS PubMed.
- H. M. Kim, C. S. Lim, G. Masanta, H. J. Kim, B. R. Cho and J. H. Han, J. Am. Chem. Soc., 2011, 133, 5698–5700 CrossRef PubMed.
- H. M. Kim, B. R. Cho, G. Masanta, C. S. Lim, C. H. Heo and N. Y. Baek, Chem. Commun., 2012, 48, 4546 RSC.
- Z. Guo, Z. Liu, W. He, C. Zhang and Y. Chen, Chem. Commun., 2012, 48, 8365 RSC.
- Q. Liu, D. Zhu, H. Jiang, L. Xue and G. Li, Inorg. Chem., 2012, 51, 10842–10849 CrossRef PubMed.
- H. Zhu, J. Fan, S. Zhang, J. Cao, K. Song, D. Ge, H. Dong, J. Wang and X. Peng, Biomater. Sci., 2014, 2, 89–97 RSC.
- H. J. Lee, C. W. Cho, H. Seo, S. Singha, Y. W. Jun, K. H. Lee, Y. Jung, K. T. Kim, S. Park, S. C. Bae and K. H. Ahn, Chem. Commun., 2016, 52, 124–127 RSC.
- C. Du, S. Fu, X. Wang, A. C. Sedgwick, W. Zhen, M. Li, X. Li, J. Zhou, Z. Wang, H. Wang and J. L. Sessler, Chem. Sci., 2019, 10, 5699–5704 RSC.
- J. Pancholi, D. J. Hodson, K. Jobe, G. A. Rutter, S. M. Goldup and M. Watkinson, Chem. Sci., 2014, 5, 3528–3535 RSC.
- L. Fang, G. Trigiante, C. J. Kousseff, R. Crespo-Otero, M. P. Philpott and M. Watkinson, Chem. Commun., 2018, 54, 9619–9622 RSC.
- H. Xiao, P. Li, X. Hu, X. Shi, W. Zhang and B. Tang, Chem. Sci., 2016, 7, 6153–6159 RSC.
- M. Yang, J. Fan, J. Zhang, J. Du and X. Peng, Chem. Sci., 2018, 9, 6758–6764 RSC.
- J.-T. Hou, H. S. Kim, C. Duan, M. S. Ji, S. Wang, L. Zeng, W. Ren and J. S. Kim, Chem. Commun., 2019, 2533–2536 RSC.
- W. Lin, G. Xu, S. Gao, Y. Ma, Y. Tang and A. Xu, Sci. Rep., 2017, 7, 1–9 CrossRef PubMed.
- L. Fang, G. Trigiante, R. Crespo-Otero, M. P. Philpott, C. R. Jones and M. Watkinson, Org. Biomol. Chem. 10.1039/c9ob01855g , in press.
- S. Phaniraj, Z. Gao, D. Rane and B. R. Peterson, Dyes Pigm., 2016, 135, 127–133 CrossRef CAS PubMed.
- D. Greentree, É. Rousseau, J. Teijeira, K. Côté and L. Picard, J. Mol. Cell. Cardiol., 2004, 34, 1163–1172 Search PubMed.
- S. J. Kim, Y. Wi, J. S. Kim, J. E. Song, K. Sunwoo, H. Y. Yoon, G. Han, H. T. Le, P. Verwilst, C. Kang and T. W. Kim, Chem. Commun., 2018, 54, 8897–8900 RSC.
- J. L. Challinor-Rogers, D. C. Kong, M. N. Iskander and G. A. McPherson, J. Pharmacol. Exp. Ther., 1995, 273, 778–786 CAS.
- E. Yuriev, D. C. M. Kong and M. N. Iskander, Eur. J. Med. Chem., 2004, 39, 835–847 CrossRef CAS PubMed.
- N. R. Johnston, R. K. Mitchell, E. Haythorne, M. P. Pessoa, F. Semplici, J. Ferrer, L. Piemonti, P. Marchetti, M. Bugliani, D. Bosco, E. Berishvili, P. Duncanson, M. Watkinson, J. Broichhagen, D. Trauner, G. A. Rutter and D. J. Hodson, Cell Metab., 2016, 24, 389–401 CrossRef CAS PubMed.
- J. Chaignon, S. E. Stiriba, F. Lloret, C. Yuste, G. Pilet, L. Bonneviot, B. Albela and I. Castro, Dalton Trans., 2014, 43, 9704–9713 RSC.
- L. E. Mcquade and S. J. Lippard, Inorg. Chem., 2010, 49, 9535–9545 CrossRef CAS PubMed.
- P. Thordarson, Chem. Soc. Rev., 2011, 6, 1305–1323 RSC.
- D. B. Hibbert and P. Thordarson, Chem. Commun., 2016, 52, 12792–12805 RSC.
- C. J. Fahrni and T. V. O. Halloran, J. Am. Chem. Soc., 1999, 121, 11448–11458 CrossRef CAS.
- M. Taki, J. L. Wolford and T. V. O. Halloran, J. Am. Chem. Soc., 2004, 126, 712–713 CrossRef CAS PubMed.
- K. Jobe, C. H. Brennan, M. Motevalli, S. M. Goldup and M. Watkinson, Chem. Commun., 2011, 47, 6036–6038 RSC.
- M. M. Wu, J. Llopis, S. Adams, J. M. McCaffery, M. S. Kulomaa, T. E. Machen, H. P. H. Moore and R. Y. Tsien, Chem. Biol., 2000, 7, 197–209 CrossRef CAS PubMed.
- J. Wang, Y. Xiao, Z. Zhang, X. Qian, Y. Yang and Q. Xu, J. Mater. Chem., 2005, 15, 2836–2839 RSC.
- E. Tamanini, A. Katewa, L. M. Sedger, M. H. Todd and M. Watkinson, Inorg. Chem., 2009, 48, 319–324 CrossRef CAS PubMed.
- E. Tamanini, K. Flavin, M. Motevalli, S. Piperno, L. A. Gheber, M. H. Todd and M. Watkinson, Inorg. Chem., 2010, 49, 3789–3800 CrossRef CAS PubMed.
- H. Su, X. Chen and W. Fang, Anal. Chem., 2014, 86, 891–899 CrossRef CAS PubMed.
- D. Escudero, Acc. Chem. Res., 2016, 49, 1816–1824 CrossRef CAS PubMed.
- Y. Tu, J. Liu, H. Zhang, Q. Peng, J. W. Y. Lam and B. Z. Tang, Angew. Chem., 2019, 131, 15053–15056 Search PubMed.
- A third probe S16 (see ESI†) proved to be cytotoxic and was not taken forward to studies in cellulo.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc04300d |
| This journal is © The Royal Society of Chemistry 2019 |