Open Access Article
Open Access ArticleRNA imaging in living mice enabled by an in vivo hybridization chain reaction circuit with a tripartite DNA probe†
Han
Wu
ab,
Ting-Ting
Chen
 a,
Xiang-Nan
Wang
a,
Yonggang
Ke
a,
Xiang-Nan
Wang
a,
Yonggang
Ke
 *b and
Jian-Hui
Jiang
*b and
Jian-Hui
Jiang
 *a
*a
aState Key Laboratory of Chemo/Bio-Sensing and Chemometrics, College of Chemistry and Chemical Engineering, Hunan University, Changsha 410082, P. R. China. E-mail: jianhuijiang@hnu.edu.cn
bWallace H. Coulter Department of Biomedical Engineering, Emory University School of Medicine, Emory University, Atlanta, Georgia 30322, USA. E-mail: yonggang.ke@emory.edu
First published on 21st October 2019
Abstract
RNA imaging in living animals helps decipher biology and creates new theranostics for disease treatment. Due to their low delivery efficiency and high background, however, fluorescence probes for in situ RNA imaging in living mice have not been reported. We develop a new cell-targeting fluorescent probe that enables RNA imaging in living mice via an in vivo hybridization chain reaction (HCR). The minimalistic Y-shaped design of the tripartite DNA probe improves its performance in live animal studies and serves as a modular scaffold for three DNA motifs for cell-targeting and the HCR circuit. The tripartite DNA probe allows facile synthesis with a high yield and demonstrates ultrasensitive RNA detection in vitro. The probe also exhibits selective and efficient internalization into folate (FA) receptor-overexpressed cells via a caveolar-mediated endocytosis mechanism and produces fluorescence signals dynamically correlated with intracellular target expressions. Furthermore, the probe exhibits specific delivery into tumor cells and allows high-contrast imaging of miR-21 in living mice. The tripartite DNA design may open the door for intracellular RNA imaging in living animals using DNA-minimal structures and its design strategy can help future development of DNA-based multi-functional molecular probes.
Introduction
Molecular probes that enable imaging of specific molecular and cellular processes in living animals are indispensable tools in deciphering pathological biology and theranostics for different diseases including cancer.1,2 Current in vivo imaging probes for cancer are mostly developed by targeting tumor-associated enzymes,3–5 protein receptors,6,7 and microenvironments,8–10 which only comprise a considerably small subset of the total hallmarks of cancer.11 Given the crucial biological roles and the increasing number of RNA-based biomarkers identified,12 fluorescent probes targeting RNAs have become a valuable platform to gain insight into tumor biology and theranostic cancer treatment.Fluorescent probes for RNA imaging can be engineered with two different strategies. One utilizes fluorescent protein-tagged RNA-binding proteins for labeling and tracking of RNAs,13 and the other deploys hybridization probes such as molecular beacons,14,15 spherical nucleic acids16,17 and light-up RNA aptamers18,19 for activatable detection of RNAs. However, current probes have only been used for imaging in living cells13–19 or sliced tissues from living animals.20–22 The development of fluorescent probes for imaging of endogenous RNA in living animals remains highly elusive.23 A key challenge for RNA imaging probes is their delivery into specific cells due to their nuclease-mediated degradation within body fluids and low efficiency in cellular uptake, which is also the substantial obstacle to oligonucleotide therapeutics.24 Another major hurdle is the low abundance of RNA targets, which only generate a weak fluorescence response in the absence of signal amplification. To our knowledge, although hybridization chain reaction probes for fluorescence in situ hybridization (FISH) in various model organisms such as zebrafish have been used in recent years,25,26 fluorescent probes that have the ability to achieve in vivo RNA imaging in living mammalian animals have been unexplored. Here we develop a novel fluorescent probe that for the first time enables RNA imaging in living mice via an in vivo hybridization chain reaction (HCR)27 using a cell-targeting tripartite DNA probe. Our previous study had established that a HCR based DNA amplification circuit allowed highly sensitive imaging of mRNA in living cells.28 The advantage of using DNA amplification circuits for RNA imaging is the enzyme-free signal amplification with hundreds of fluorophores activated from a single RNA. We envision that the realization of an in vivo HCR circuit affords the possibility of visualizing low-abundance RNAs in living animals.
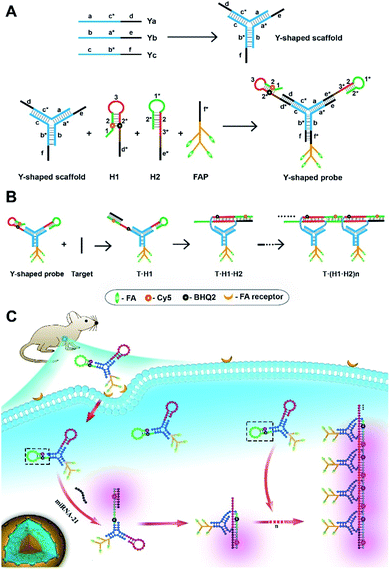
Motivated by this hypothesis, we design a probe using a tripartite Y-shaped DNA for in vivo HCR circuit-based RNA imaging, as illustrated in Scheme 1. The use of a Y-shaped DNA structure has two advantages. One is the tripartite modular scaffold conferring three modules for functionalization with three DNA motifs required for an in vivo HCR circuit. The other is that the Y-shape is a DNA-minimal structure with tripartite functionalities.
The minimal complexity of the probe can enhance its performance in live animal studies compared to alternative DNA structures with more strands. Three DNA motifs are assembled on the Y-shaped probe via sticky-end hybridization (Scheme 1A). One motif is a cell-targeting DNA-peptide (poly-lysine) conjugated folate probe (FAP), in which the poly-lysine domain is grafted with multiple folate (FA) ligands. Since tumor cells are mostly known for overexpression of folate receptors,29 such a multiple ligand presentation in the DNA probe can confer improved affinity and targetability to tumor cells through multivalent interactions. The other two motifs are hairpin probes H1 and H2, which undergo HCR assembly in response to a target RNA (Scheme 1B). First, the target miR-21 hybridizes with the toehold domain 1 and domain 2 of H1 and opens the hairpin. Then the activated domain 3 and domain 2* in H1 hybridize with the domain 3* and domain 2 of H2 and expose the domain 1* and 2* for the next cycle of the HCR reaction. A near-infrared (NIR) fluorescence reporter (Cy5) and a quencher moiety BHQ2 are labeled in the stem region of the H1 probe such that a fluorescence signal is activated to indicate the HCR assembly. When systemically administered into a living mouse via intravenous injection, the tripartite DNA probe can be selectively taken up into tumor cells. The presence of target RNA mediates a cascade of alternating assembly between probes H1 and H2 via the HCR circuit, generating an activated NIR fluorescence response for in vivo RNA imaging in the living mouse (Scheme 1C).
As a proof of principle, we choose microRNA-21 (miR-21) as the target in this work, because of its overexpression in various tumor cells.30 We demonstrate that the tripartite DNA probe allows facile modular synthesis with a high yield and ultrasensitive detection of target RNA in vitro. Live cell assays reveal that the tripartite DNA allows selectively and efficiently internalization in cells with FA receptor overexpression, displaying fluorescence signals dynamically correlated with miR-21 expression. Additionally, its caveolar-mediated endocytosis mechanism affords the potential for efficient escape from lysosomal degradation. Moreover, the tripartite DNA probe affords increased stability of the probe in serum and enables specific delivery into tumor cells for high-contrast imaging of miR-21 in living mice. Because of the broad utility of DNA amplification circuits for RNA detection, the tripartite DNA design may open the door for intracellular RNA imaging in living animals using DNA-minimal structures.
Results and discussion
Design and synthesis of the tripartite DNA probe
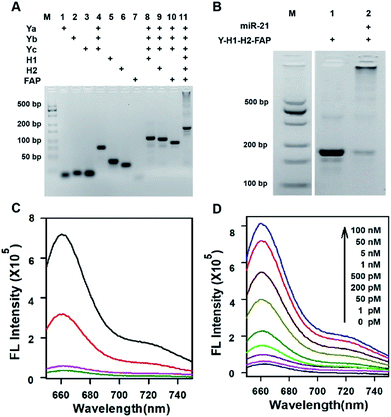
Three motifs are required to realize the in vivo HCR circuit: a cell-targeting unit to deliver the hairpin probes into tumor cells and two hairpin probes responding to target RNA to initiate the HCR assembly. We chose a DNA-minimal Y-shaped scaffold31,32 to construct the modular probe for in vivo realization of the HCR circuit. Such a DNA-minimal structure afforded the advantages of low cost and high efficiency in synthesis and minimized complexity of structural variability for in vivo applications. The Y-shaped DNA was synthesized via self-assembly using three 39-mer single-stranded (ss) DNAs (Ya, Yb and Yc), each containing a 26-mer region to form the double-stranded Y shape and a 13-mer sticky end of different sequences. To synthesize a cell-targeting motif, a poly-lysine peptide with an azido modifier, azido-GGKKKKKKKK, was designed, in which the side chain amino groups were conjugated to multiple FA using the EDC–NHS chemistry (Fig. S1†). The cell-targeting motif, DNA-peptide conjugated FAP, was synthesized by linking the FA conjugated peptide to a ssDNA linker L complementary to a sticky end with a 5′-alkynyl modifier via copper catalyzed click chemistry33 (Fig. S2 and S3†). The hairpin probes H1 and H2 were carefully designed and optimized according to the miR-21 sequence to minimize the background arising from reaction with non-homologous initiator sequences (Fig. S4†). With the cell-targeting DNA-peptide conjugated FAP and hairpin probes H1 and H2, the tripartite DNA probe Y–H1–H2–FAP was synthesized via assembling the three motifs on the corresponding sticky ends.Gel electrophoresis analysis was used to validate the successful synthesis of the tripartite DNA probe (Fig. 1A). A major band with lower mobility appeared after incubation of the three component ssDNAs (Ya, Yb and Yc), indicating the formation of a Y-shaped scaffold from the three component ssDNAs. With addition of one of the three motifs, major bands were observed with further lowered mobility, verifying the high efficiency in assembly of each of the three motifs on the Y-shaped scaffold. A one-pot synthesis using three component ssDNAs (Ya, Yb and Yc) and three motifs (FAP, H1 and H2) also only yield a major band with substantially lower mobility, evidencing the formation of the complete tripartite DNA probe. The estimated yield of the Y-shaped scaffold was close to 100% and that of the tripartite DNA probe was ∼90%, as determined by densitometry. The high yield, facile one-pot synthesis and minimal design of the tripartite DNA probe afforded unique advantages for its living animal applications that typically require using probes at a relatively high dosage.
In vitro evaluation of the tripartite DNA probe for RNA detection
Because of the minimal structure of the tripartite DNA probe with minimized steric hindrance, we expected that it still allowed an efficient HCR assembly in response to target miR-21. Gel electrophoresis analysis demonstrated that upon incubation with the target miR-21, the tripartite DNA probe Y–H1–H2–FAP gave consecutive bands with decreased mobility, consistent with reported HCR circuits (Fig. 1B). The majority of HCR products were trapped in the well, indicating efficient assembly of the tripartite DNA probes into large HCR polymers. This finding verified the feasibility of using the tripartite DNA probe to realize the HCR circuit in response to miR-21. Further fluorescence assays revealed that the tripartite DNA probe Y–H1–H2–FAP gave substantial enhancement (∼15-fold) in the Cy5 fluorescence signal after incubating with 50 nM target RNA (Fig. 1C). In contrast, a control RNA with the most homogeneity to miR-21 in mammalian cells only generated a very slight increase in the fluorescence peak, suggesting the high specificity of the HCR circuit using the tripartite DNA probe. A control Y-shaped probe Y–H1–FAP, which can also be activated by miR-21 but with no further signal amplification, showed only 40% fluorescence output upon interaction with 50 nM miR-21, which can directly trigger maximally the opening of half of the probe Y–H1–FAP. These results gave direct evidence for selective and signal-amplified fluorescence activation of the tripartite DNA probe in response to target miR-21. Further investigation revealed that the tripartite DNA probe exhibited dynamic responses to miR-21 of varying concentrations from 1 pM to 100 nM (Fig. 1D). The detection limit was estimated to be 0.8 pM according to the linear correlation between peak intensities and target concentrations (Fig. S5†). Moreover, the tripartite DNA probe displayed fast kinetics in the HCR assembly and the reactions almost completed within 40 min (Fig. S6†).Detection of miR-21 in live cells with the tripartite DNA probe
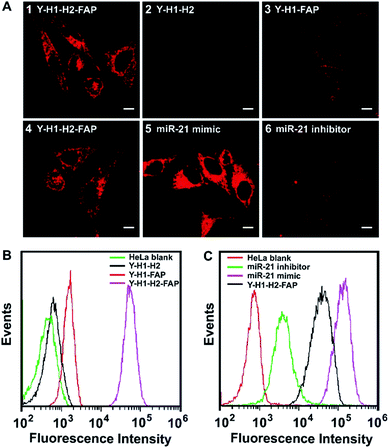
To test the performance of the tripartite DNA probe for imaging of miR-21 in living cells, we chose a model system of HeLa cells, which was known for overexpression of the target miR-21 and FA receptor on the cell membrane.34,35 After HeLa cells were incubated with the tripartite DNA probe Y–H1–H2–FAP (100 nM) for 3 h, we observed bright red fluorescence in the cells (Fig. 2A, image 1). Confocal images at higher resolution showed highly localized fluorescent spots around the nuclei (Fig. S7†), implying that the tripartite DNA probe based HCR circuit had potential for localization detection of target RNA. Time-dependent imaging further revealed that the cells started to show fluorescence after 20 min incubation and the signal became saturated after 60 min (Fig. S8†). This observation suggested that the in cellular HCR kinetics was close to that under in vitro conditions. A control experiment using a Y-shaped probe Y–H1–H2 gave no detectable fluorescence in the cells (Fig. 2A, image 2), indicating that the cell-targeting motif FAP played an essential role in cellular delivery of the DNA probe. An additional control experiment using the non-amplification Y-shaped probe Y–H1–FAP only exhibited weak fluorescence in the cells (Fig. 2A, image 3), suggesting that the tripartite DNA probe based HCR circuit afforded substantial sensitivity enhancement in live cell imaging of miR-21. Flow cytometry assay gave remarkably increased fluorescence signals for the tripartite DNA probe Y–H1–H2–FAP over the non-amplification probe Y–H1–FAP and further over the non-targeting probe Y–H1–H2 (Fig. 2B). The results above demonstrated that the tripartite DNA probe Y–H1–H2–FAP allowed efficient cell uptake and substantial signal amplification.The specific response of the tripartite DNA probe to miR-21 was validated using HeLa cells treated with miR-21 mimic, a synthetic dsRNA enabling the increase of miR-21 expression,36 or miR-21 inhibitor, a synthetic ssRNA complementary to miR-21 and decreasing its expression.36 Confocal images revealed that cells pretreated with the miR-21 mimic displayed stronger fluorescence than untreated HeLa cells, while cells pretreated with the miR-21 inhibitor gave much lower fluorescence (Fig. 2A, image 4–6). Flow cytometry analysis also showed consistent fluorescence responses in a decreasing order of HeLa cells treated with the miR-21 mimic, untreated cells and cells treated with the miR-21 inhibitor (Fig. 2C). These findings revealed a strong correlation between the miR-21 concentration and the fluorescence signals generated by the tripartite DNA probe, indicating the probe's potential for quantitative detection of target RNA in living cells.
FA receptor-specific endocytosis of the tripartite DNA probe
To realize in vivo RNA imaging in tumors, it is critical for the tripartite DNA probe to enable cell-targeting delivery into tumor cells. Because a culture medium containing FA could block the receptors,37 we started to investigate the targeting properties of the tripartite DNA probe to FA receptor-overexpressed tumor cells using HeLa cells incubated in a culture medium with or without FA. As anticipated, after growing in the culture medium containing FA, HeLa cells incubated with the tripartite DNA probe did not show appreciable fluorescence contrast (Fig. S9†), which was also consistent with the data obtained with flow cytometry. This finding suggested that the tripartite DNA probe entered the cells via a FA receptor dependent mechanism. Further evidence was obtained with MCF-7 cells, which were known to have negative expression of the FA receptor but overexpression of the miR-21 target.38,39 After incubation with the tripartite DNA probe, MCF-7 cells displayed a negligible fluorescence signal (Fig. S9†), confirming the FA receptor-specific internalization of the tripartite DNA probe in tumor cells.To further validate the FA receptor-specific cellular uptake of the tripartite DNA probe, a plasmid that induces overexpression of the FA receptor (Scheme S1†) was used for transfection of two cell lines, L-02 and MCF-7, both with low expression of the FA receptor.39 After the transfected cells were incubated with the tripartite DNA probe, we observed fluorescence images as well as flow cytometry profiles of varying fluorescence intensities in these cells (Fig. S10†). MCF-7 cells displayed the brightest fluorescence, indicating the highest expression of miR-21. L-02 cells showed the weakest fluorescence with an indicator for the lowest expression of miR-21. The results were consistent with previous reports38 and our RT-PCR analysis (Fig. S11†). These findings confirmed the FA receptor-specific cellular uptake of the tripartite DNA probe and served as additional evidence for quantitative imaging of miR-21 in living cells with the tripartite DNA probe.
Furthermore, the intracellular uptake mechanism was investigated using inhibitors for different cellular internalization pathways40 with a fluorescent tripartite DNA probe Y–H3–H4–FAP (Fig. S12†). The tripartite DNA probe Y–H3–H4–FAP replaced H1 with a non-responsive hairpin probe H3 and a TARMA labeled version H4 for the hairpin probe H2. When HeLa cells were incubated with the fluorescent tripartite DNA probe in the presence of NaN3, no obvious fluorescence appeared in the cells. As NaN3 was an inhibitor for ATPase involved in all energy-dependent endocytosis,40 this finding confirmed an energy-dependent endocytosis pathway for the tripartite DNA probe. It was also found that cells treated with nystatin (caveolar mediated endocytosis inhibitor40) also displayed weak fluorescence after incubation with the fluorescent tripartite DNA probe. In contrast, chlorpromazine (CPZ, clathrin-mediated endocytosis inhibitor40), amiloride (macropinocytosis mediated endocytosis inhibitor40) and methyl-β-cyclodextrin (lipid-raft mediated endocytosis inhibitor40) had a slight effect on the intracellular fluorescence signals. These results revealed that the tripartite DNA probe entered HeLa cells dominantly via a caveolar-mediated endocytosis mechanism. This finding was also consistent with previous observations that multivalent FA conjugated probes internalized into FA receptor-positive cells via caveolar-mediated endocytosis.41
A further time-dependent study revealed that the tripartite DNA probe Y–H3–H4–FAP did not display substantial colocalization with Lysotracker during the endocytosis (Fig. S13†). This observation was consistent with its caveolar-mediated endocytosis pathway in which caveosomes did not fuse with lysosomes.41 Because this caveolar mediated endocytosis mechanism allowed efficient escape from lysosomal degradation, we expected that the tripartite DNA probe could hold great promise for the delivery of nucleic acid agents for theranostics. Further evidence for the cytoplasm localization of the tripartite DNA probe Y–H1–H2–FAP was obtained using colocalization assay of the HCR products with Lysotracker (Fig. S14†). It was also observed that the HCR products had little colocalization with lysosomes, implying that the tripartite DNA probe entered the cytoplasm and formed a HCR assembly in response to target miR-21. Additionally, a z-axis scan experiment further clearly demonstrated that the HCR products were localized in the cytoplasm (Fig. S15†).
Additionally, the tripartite DNA probe was found to show better stability in 100% fresh fetal bovine serum (FBS) than free hairpin probes H1 and H2 in a fluorescence assay (Fig. S16†). This finding was consistent with previous studies on the stability of other DNA probes.42,43 This result suggested that the tripartite DNA probe improved nuclease resistance of the hairpin probes, implying the potential of the tripartite DNA probe for in vivo imaging applications.
RNA imaging in living mice with the tripartite DNA probe
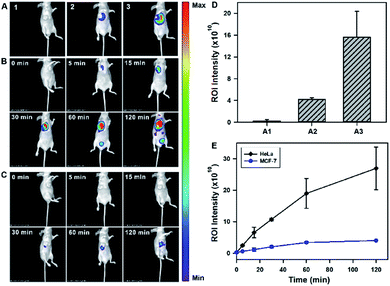
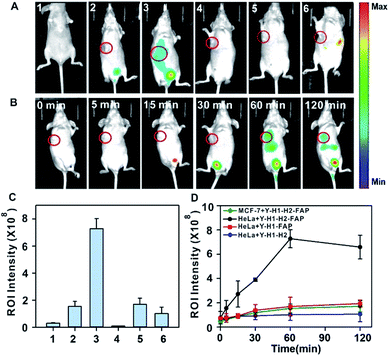
After validating the in vitro and in cellular performance, we then investigated the ability of the tripartite DNA probe for RNA imaging in living mice. We generated tumor xenografts via subcutaneous implantation of MCF-7 cells or HeLa cells (>106 cells) into the right oxter of BALB/c mice. Imaging experiments were performed after the tumors were grown to a 20–25 mm3 size. In the preliminary study, we inspected the feasibility of the tripartite DNA probe for in vivo fluorescence imaging of miR-21 expression in tumors via intratumoral injection. With the injection of the physiological saline, very low background fluorescence was obtained (Fig. 3A, image 1). The intratumoral injection of the tripartite DNA probe in MCF-7 tumor bearing mice exhibited a weak fluorescence signal (Fig. 3A, image 2), attributed to the poor efficiency of the tripartite DNA probe to enter the tumor cells because of the negative expression of the FA receptor on their membranes. An intense fluorescence signal was observed for the HeLa tumor bearing mice (Fig. 3A, image 3), suggesting high expression of miR-21 in HeLa tumors with positive expression of the FA receptor on their membranes. Clearer evidence for the overexpression of miR-21 in the HeLa tumor could be obtained from fluorescence intensities in the tumor regions of interest (Fig. 3D). Time-dependent in vivo imaging also exhibited a rapid increase in the fluorescence responses in the HeLa tumors at 2 h post-injection (Fig. 3B), and only slightly increased fluorescence signals were observed for the MCF-7 tumors (Fig. 3C). The ratio of average fluorescence signals for the HeLa tumors to the MCF-7 tumors at 2 h post-injection was as high as ∼7-fold (Fig. 3E), indicating the high contrast of the tripartite DNA probe for miR-21 imaging in living mice.With the desirable results obtained through intratumoral injection, we started to investigate the capability of the tripartite DNA probe for miR-21 imaging in living mice through tail vein administration. 1 h after the injection of the tripartite DNA probe, mice with no implanted tumor (Fig. 4A, image 1) or with MCF-7 tumors (Fig. 4A, image 2) only displayed very weak fluorescence signals, while the HeLa tumor exhibited strong fluorescence (Fig. 4A, image 3). The average intensities in the region of interest showed that HeLa tumor delivered ∼4-fold enhancement in the fluorescence signals compared to MCF-7 tumors (Fig. 4C). This result evidenced that the tripartite DNA probe enabled rapid cell-targeted delivery of the probe and allowed specific fluorescence activation in response to the miR-21 target. Further experiments using HeLa tumor bearing mice by tail vein administration of physiological saline (Fig. 4A, image 4), the non-amplification control probe Y–H1–FAP (Fig. 4A, image 5) and the non-targeting probe Y–H1–H2 (Fig. 4A, image 6) also did not give an intense fluorescence signal. These controls confirmed that the FAP moiety was essential for cell-targeted delivery of the probe and signal amplification using the HCR circuit was necessary for achieving high contrast imaging of RNA in living animals. Time-dependent imaging of the HeLa tumor bearing mice showed that the fluorescence signals increased gradually after tail vein administration of the tripartite DNA probe and reached the maximum at 1 h followed by a decrease of the response (Fig. 4B). Besides the tumor region, fluorescence signals were also obtained in the bladder region, indicating a rapid clearance of the tripartite DNA probe through the kidney. In contrast, a control experiment of time-dependent imaging of the HeLa tumor bearing mice by injection of the control probe Y–H1–H2 also only displayed weak fluorescence throughout the analysis (Fig. 4D and S17†). This result confirmed the tumor targeting ability of the tripartite DNA probe due to the FAP moiety. Another control with the non-amplification probe Y–H1–FAP also did not display a substantial fluorescence response, implying the significant role of the HCR circuit in in vivo RNA imaging. Furthermore, we merely obtained slightly increased fluorescence signals after administration of the tripartite DNA probe in the MCF-7 tumor-bearing mice (Fig. 4D and S17†). This result verified the specificity of the tripartite DNA probe in targeting cancer cells with folate receptor positive expression.
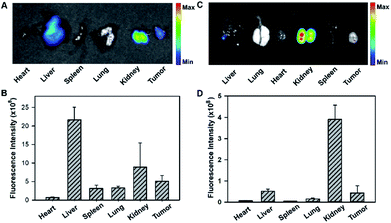
To gain insight into the fluorescence response kinetics, we then performed prolonged time-dependent studies of the HeLa tumor bearing mice via tail vein injection of the tripartite DNA probe. It was observed that the maximum response was achieved at 1 h post-injection and subsequently the fluorescence decreased until approaching the background level at 8 h (Fig. S18†). This finding suggested the rapid activation of the fluorescence response in the tumor area and fast clearance of the probes and the HCR products. Moreover, ex vivo fluorescence imaging of organs dissected at 4 h post-injection, such as the heart, liver, spleen, lung, kidney and tumor, revealed that the activated fluorescence signals were dominantly obtained in the liver, kidney and tumor (Fig. 5A and B). This finding indicated that the probes and the HCR products were mainly metabolized and cleared in the liver and kidney. Ex vivo fluorescence imaging of organs dissected at 8 h post-injection gave different distributions of the probes and the HCR products, and the fluorescence only resided in the kidney at this time (Fig. 5C and D). Presumably, this result can be attributed to the rapid metabolism of the probe in the liver and slow excretion of the HCR product from the tumor into the kidney. Taken together, the live mouse imaging demonstrated that the tripartite DNA probe enabled selective accumulation in tumors with positive folate receptor expression and delivered activated fluorescence signals in response to miR-21 expression in living animals. With the combination of different tumor-targeting ligands, the tripartite DNA probe provides a valuable platform for RNA imaging and related in vivo tumor diagnostics and treatment evaluation.
Conclusions
In summary, we developed a novel tripartite DNA probe that enabled fluorescent RNA imaging in living mice via an in vivo hybridization chain reaction (HCR) circuit. The Y-shaped tripartite DNA probe allowed facile modular synthesis with a high yield. It also demonstrated high sensitivity and specificity in in vitro detection of target RNA with a subpicomolar detection limit. The tripartite DNA probe was demonstrated to be selectively and efficiently internalized into cells with FA receptor overexpression via a caveolar-mediated endocytosis mechanism, affording the potential for efficient escape from lysosomal degradation. Live cell imaging revealed that the probe displayed fluorescence signals dynamically correlated with miR-21 expression. The tripartite DNA probe also showed increased stability in serum and enabled specific delivery into tumor cells for high-contrast imaging of miR-21 in living mice. Because of the broad utility of DNA amplification circuits for RNA detection, the tripartite DNA probe design may open the door for intracellular RNA imaging in living animals using DNA-minimal structures. To our knowledge, it is the first time that DNA probes have been developed for in vivo imaging of RNA in living mice. In virtue of the broad utility of DNA amplification circuits for ultrasensitive RNA detection, the tripartite DNA design may open the door for intracellular RNA imaging in living animals, implying its potential for tumor biology studies and theranostics.Conflicts of interest
There are no conflicts to declare.Acknowledgements
All animal operations were in accord with institutional animal use and care regulations, according to protocol No. SYXK (Xiang) 2008-0001, approved by the Laboratory Animal Center of Hunan. This work was supported by the National Natural Science Foundation of China (Grants 21525522 and 21705039).Notes and references
- B. R. Smith and S. S. Gambhir, Chem. Rev., 2017, 117, 901–986 CrossRef CAS PubMed.
- M. Gao, F. Yu, C. Lv, J. Choo and L. Chen, Chem. Soc. Rev., 2017, 46, 2237–2271 RSC.
- J. Zhang, L. P. Smaga, N. S. R. Satyavolu, J. Chan and Y. Lu, J. Am. Chem. Soc., 2017, 139, 17225–17228 CrossRef CAS PubMed.
- D. Asanuma, M. Sakabe, M. Kamiya, K. Yamamoto, J. Hiratake, M. Ogawa, N. Kosaka, P. L. Choyke, T. Nagano, H. Kobayashi and Y. Urano, Nat. Commun., 2015, 6, 6463 CrossRef CAS PubMed.
- C. E. Callmann, C. V. Barback, M. P. Thompson, D. J. Hall, R. F. Mattrey and N. C. Gianneschi, Adv. Mater., 2015, 27, 4611–4615 CrossRef CAS PubMed.
- Y. Feng, et al. , Chem. Sci., 2017, 8, 3703–3711 RSC.
- J. A. Varela, J. P. Dupuis, L. Etchepare, A. Espana, L. Cognet and L. Groc, Nat. Commun., 2016, 7, 10947 CrossRef CAS PubMed.
- P. Gao, W. Pan, N. Li and B. Tang, Chem. Sci., 2019, 10, 6035–6071 RSC.
- A. Zomer, C. Maynard, F. J. Verweij, A. Kamermans, R. Schäfer, E. Beerling, R. M. Schiffelers, E. de Wit, J. Berenguer, S. I. J. Ellenbroek, T. Wurdinger, D. M. Pegtel and J. van Rheenen, Cell, 2015, 161, 1046–1057 CrossRef CAS.
- X. Zheng, X. Wang, H. Mao, W. Wu, B. Liu and X. Jiang, Nat. Commun., 2015, 6, 5834 CrossRef CAS.
- R. Kole, A. R. Krainer and S. Altman, Nat. Rev. Drug Discovery, 2012, 11, 125–140 CrossRef CAS.
- S. Tyagi, Nat. Methods, 2009, 6, 331–338 CrossRef CAS PubMed.
- N. Daigle and J. Ellenberg, Nat. Methods, 2007, 4, 633–636 CrossRef CAS.
- J. Zheng, R. Yang, M. Shi, C. Wu, X. Fang, Y. Li, J. Li and W. Tan, Chem. Soc. Rev., 2015, 44, 3036–3055 RSC.
- H. Zhang, F. Li, B. Dever, X.-F. Li and X. C. Le, Chem. Rev., 2013, 113, 2812–2841 CrossRef CAS PubMed.
- T. L. Halo, K. M. McMahon, N. L. Angeloni, Y. Xu, W. Wang, A. B. Chinen, D. Malin, E. Strekalova, V. L. Cryns, C. Cheng, C. A. Mirkin and C. S. Thaxton, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 17104–17109 CrossRef CAS PubMed.
- H. Li, B. Zhang, X. Lu, X. Tan, F. Jia, Y. Xiao, Z. Cheng, Y. Li, D. O. Silva, H. S. Schrekker, K. Zhang and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4340–4344 CrossRef CAS PubMed.
- Z.-M. Ying, Z. Wu, B. Tu, W. Tan and J.-H. Jiang, J. Am. Chem. Soc., 2017, 139, 9779–9782 CrossRef CAS.
- R. L. Strack, M. D. Disney and S. R. Jaffrey, Nat. Methods, 2013, 10, 1219–1224 CrossRef CAS.
- T. Lionnet, K. Czaplinski, X. Darzacq, Y. Shav-Tal, A. L. Wells, J. A. Chao, H. Y. Park, V. de Turris, M. Lopez-Jones and R. H. Singer, Nat. Methods, 2011, 8, 165–170 CrossRef CAS.
- H. Y. Park, H. Lim, Y. J. Yoon, A. Follenzi, C. Nwokafor, M. Lopez-Jones, X. Meng and R. H. Singer, Science, 2014, 343, 422–424 CrossRef CAS.
- I. Oomoto, A. Suzuki-Hirano, H. Umeshima, Y.-W. Han, H. Yanagisawa, P. Carlton, Y. Harada, M. Kengaku, A. Okamoto, T. Shimogori and D. O. Wang, Nucleic Acids Res., 2015, 43, e126 CrossRef.
- R. Weissleder, A. Moore, U. Mahmood, R. Bhorade, H. Benveniste, E. A. Chiocca and J. P. Basilion, Nat. Med., 2000, 6, 351–355 CrossRef CAS.
- Y. Xia, R. Zhang, Z. Wang, J. Tian and X. Chen, Chem. Soc. Rev., 2017, 46, 2824–2843 RSC.
- J. Li, S. Liu, L. Sun, W. Li, S.-Y. Zhang, S. Yang, J. Li and H.-H. Yang, J. Am. Chem. Soc., 2018, 140, 16589–16595 CrossRef CAS.
- H. M. T. Choi, C. R. Calvert, N. Husain, D. Huss, J. C. Barsi, B. E. Deverman, R. C. Hunter, M. Kato, S. M. Lee, A. C. T. Abelin, A. Z. Rosenthal, O. S. Akbari, Y. Li, B. A. Hay, P. W. Sternberg, P. H. Patterson, E. H. Davidson, S. K. Mazmanian, D. A. Prober, M. van de Rijn, J. R. Leadbetter, D. K. Newman, C. Readhead, M. E. Bronner, B. Wold, R. Lansford, T. Sauka-Spengler, S. E. Fraser and N. A. Pierce, Development, 2016, 143, 3632–3637 CrossRef CAS.
- H. M. T. Choi, M. Schwarzkopf, M. E. Fornace, A. Acharya, G. Artavanis, J. Stegmaier, A. Cunha and N. A. Pierce, Development, 2018, 145, dev165753 CrossRef PubMed.
- Z. Wu, G.-Q. Liu, X.-L. Yang and J.-H. Jiang, J. Am. Chem. Soc., 2015, 137, 6829–6836 CrossRef CAS.
- H. Elnakat and M. Ratnam, Adv. Drug Delivery Rev., 2004, 56, 1067–1084 CrossRef CAS.
- M. B. Baker, G. Bao and C. D. Searles, Nucleic Acids Res., 2012, 40, e13 CrossRef CAS.
- Y. Li, Y. D. Tseng, S. Y. Kwon, L. d'Espaux, J. S. Bunch, P. L. McEuen and D. Luo, Nat. Mater., 2004, 3, 38–42 CrossRef CAS.
- S. Yue, X. Song, W. Song and S. Bi, Chem. Sci., 2019, 10, 1651–1658 RSC.
- K. Kacprzak, I. Skiera, M. Piasecka and Z. Paryzek, Chem. Rev., 2016, 116, 5689–5743 CrossRef CAS.
- J. A. Chan, A. M. Krichevsky and K. S. Kosik, Cancer Res., 2005, 65, 6029–6033 CrossRef CAS.
- R. Zhao, B. Wang, X. Yang, Y. Xiao, X. Wang, C. Shao and R. Tang, Angew. Chem., Int. Ed., 2016, 55, 5225–5229 CrossRef CAS.
- E. Leone, E. Morelli, M. T. Di Martino, N. Amodio, U. Foresta, A. Gullà, M. Rossi, A. Neri, A. Giordano, N. C. Munshi, K. C. Anderson, P. Tagliaferri and P. Tassone, Clin. Cancer Res., 2013, 19, 2096–2106 CrossRef CAS.
- H. Dong, J. Lei, H. Ju, F. Zhi, H. Wang, W. Guo, Z. Zhu and F. Yan, Angew. Chem., Int. Ed., 2012, 51, 4607–4612 CrossRef CAS.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435, 834–838 CrossRef CAS.
- D. Feng, Y. Song, W. Shi, X. Li and H. Ma, Anal. Chem., 2013, 85, 6530–6535 CrossRef CAS.
- F. Zhao, Y. Zhao, Y. Liu, X. Chang, C. Chen and Y. Zhao, Small, 2011, 7, 1322–1337 CrossRef CAS.
- C. Dalal, A. Saha and N. R. Jana, J. Phys. Chem. C, 2016, 120, 6778–6786 CrossRef CAS.
- J. W. Conway, C. K. McLaughlin, K. J. Castor and H. Sleiman, Chem. Commun., 2013, 49, 1172–1174 RSC.
- A. Lacroix, E. Vengut-Climent, D. de Rochambeau and H. F. Sleiman, ACS Cent. Sci., 2016, 5, 882–891 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc03469b |
| This journal is © The Royal Society of Chemistry 2020 |