Open Access Article
Open Access Articleα-D-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal α-galactosidase A in Fabry disease†
Marta
Artola
 ab,
Christinne
Hedberg
ab,
Christinne
Hedberg
 a,
Rhianna J.
Rowland
a,
Rhianna J.
Rowland
 c,
Lluís
Raich
d,
Kassiani
Kytidou
b,
Liang
Wu
c,
Amanda
Schaaf
a,
Maria Joao
Ferraz
b,
Gijsbert A.
van der Marel
a,
Jeroen D. C.
Codée
c,
Lluís
Raich
d,
Kassiani
Kytidou
b,
Liang
Wu
c,
Amanda
Schaaf
a,
Maria Joao
Ferraz
b,
Gijsbert A.
van der Marel
a,
Jeroen D. C.
Codée
 a,
Carme
Rovira
a,
Carme
Rovira
 de,
Johannes M. F. G.
Aerts
de,
Johannes M. F. G.
Aerts
 b,
Gideon J.
Davies
b,
Gideon J.
Davies
 *c and
Herman S.
Overkleeft
*c and
Herman S.
Overkleeft
 *a
*a
aDepartment of Bio-organic Synthesis, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333 CC Leiden, The Netherlands. E-mail: h.s.overkleeft@lic.leidenuniv.nl
bDepartment of Medical Biochemistry, Leiden Institute of Chemistry, Einsteinweg 55, 2333 CC Leiden, The Netherlands
cDepartment of Chemistry, York Structural Biology Laboratory, University of York, Heslington, York YO10 5DD, UK. E-mail: gideon.davies@york.ac.uk
dDepartament de Química Inorgànica i Orgànica (Secció de Química Orgànica), Institut de Química Teòrica i Computacional (IQTCUB), Universitat de Barcelona, Martí i Franques 1, 08028 Barcelona, Spain
eFundació Catalana de Recerca i Estudis Avancats (ICREA), Passeig Lluís Companys 23, 08010 Barcelona, Spain
First published on 20th August 2019
Abstract
Fabry disease is an inherited lysosomal storage disorder that is characterized by a deficiency in lysosomal α-D-galactosidase activity. One current therapeutic strategy involves enzyme replacement therapy, in which patients are treated with a recombinant enzyme. Co-treatment with enzyme active-site stabilizers is advocated to increase treatment efficacy, a strategy that requires effective and selective enzyme stabilizers. Here, we describe the design and development of an α-D-gal-cyclophellitol cyclosulfamidate as a new class of neutral, conformationally constrained competitive glycosidase inhibitors that act by mimicry of the Michaelis complex conformation. We found that D-galactose-configured α-cyclosulfamidate 4 effectively stabilizes recombinant human α-D-galactosidase (agalsidase beta, Fabrazyme®) both in vitro and in cellulo.
Introduction
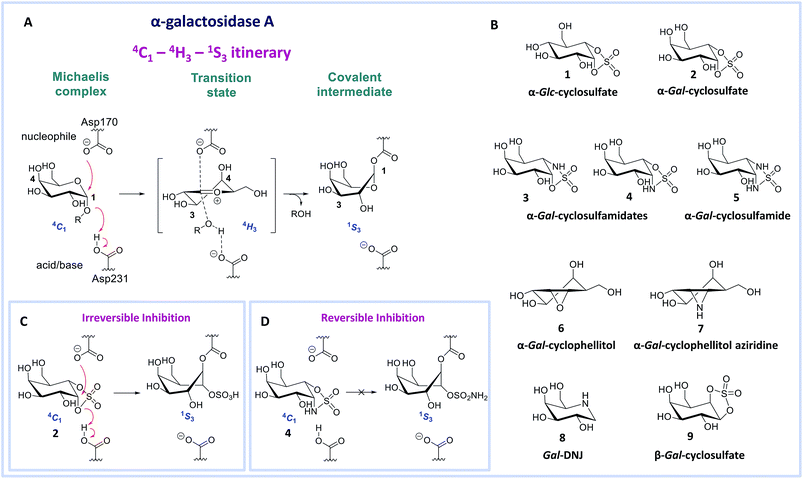
Deficiency of α-galactosidase A (α-gal A, EC 3.2.1.22, a retaining glycosidase of the GH27 glycoside hydrolase family (http://www.cazy.org)1) underlies Fabry disease. This inherited lysosomal storage disorder is characterized by toxic accumulation of glycosphingolipid globotriaosylceramide (Gb3) in lysosomes and its sphingoid base, globotriaosylsphingosine (lyso-Gb3) in plasma and tissues.2,3 Several mutations in the GLA gene encoding α-gal A can result in diminished protein levels and/or enzyme activity, leading to altered metabolite levels and a range of Fabry disease phenotypes. The accumulation of glycosphingolipid metabolites is thought to cause progressive renal and cardiac insufficiency and CNS pathology in Fabry patients.4 Enzyme replacement therapy (ERT) for Fabry disease involves intravenous treatment with recombinant human α-gal A (agalsidase beta, Fabrazyme® or agalsidase alpha, Replagal®), but the clinical efficacy of this therapy is limited.5–7 1-Deoxygalactonojirimycin (Gal-DNJ 8, Migalastat®, Fig. 1B) has recently been approved as a pharmacological chaperone (PC) for the treatment of Fabry disease in patients with amenable mutations.8 Gal-DNJ 8 binds mutant forms of α-gal A, which are catalytically competent but otherwise targeted for degradation due to misfolding. Gal-DNJ 8 stabilizes the protein fold, allowing the mutant α-gal A to be trafficked to lysosomes. However, PC therapy for Fabry disease is limited to specific mutations and its efficacy is hotly debated.9–13 For this reason, an attractive alternative therapeutic intervention strategy, proposed recently, comprises jointly administering a recombinant enzyme and a pharmacological chaperone.14–16 This strategy aims to stabilize the recombinant enzyme in circulation such that larger proportions may reach disease affected tissues, permitting the use of extended intervals between injections and lower enzyme dosages, which should diminish side effects, improve the patient's lifestyle and reduce treatment costs.17,18 For this strategy to become clinical practice, allosteric enzyme stabilizers or orthosteric competitive α-gal A inhibitors that prevent enzyme unfolding and are displaced by the accumulated metabolites in the lysosome recovering the glycosidase activity, with good selectivity and pharmacokinetic/pharmacodynamic properties, are required.16,18,19 We argue that the discovery of such commodities would be greatly facilitated by the design of new inhibitor templates.Human α-gal A hydrolyzes its substrate following a Koshland double displacement mechanism, resulting in net retention of stereochemistry at the anomeric center of the produced galactopyranose.20,21 The reaction coordinates by which α-gal A processes its substrate to form the intermediate covalent adduct are 4C1 → 4H‡3 → 1S3 with respect to the conformation of the galactopyranose moiety in the Michaelis complex → TS → covalent intermediate complex (Fig. 1A).22–24 This same reaction itinerary is also employed by GH31 retaining α-glucosidases, with the difference that a glucopyranose, rather than a galactopyranose, is captured in the enzyme active site.25 We have recently shown that α-glc-cyclosulfate 1 (Fig. 1B) potently, selectively and irreversibly inhibits retaining α-glucosidases. Compound 1 in free solution predominantly resides in a 4C1 chair conformation, thus mimicking the initial Michaelis complex conformation utilized by α-glucosidases25 allowing facile interception by the catalytic nucleophile.
We reasoned that α-gal-cyclosulfate 2 would covalently and irreversibly inhibit α-gal A with equal efficiency and selectivity following the same mode of action (Fig. 1B and C). Building on this concept, we further hypothesized that substitution of one or both of the cyclosulfate ring oxygens for nitrogen, as in compounds 3–5, would lead to competitive α-gal A inhibitors because of the decreased leaving group capacity of cyclosulfamidates/cyclosulfamides, when compared to cyclosulfates (Fig. 1D). Such compounds would then offer competitive enzyme inhibitors to be tested as stabilizers of recombinant α-galactosidase A for Fabry treatment. Here, we show the validity of this reasoning by revealing α-gal-cyclosulfamidate 4 as a first-in-class, effective and selective, competitive α-gal A inhibitor. Structural and computational analysis of the conformational behavior of compound 4 in solution and in the active site of human α-gal A supports our design and provides a molecular rationale why compound 4 is an effective α-gal A inhibitor. We also show compound 4 to be effective in stabilizing recombinant α-gal A in vitro and in cellulo and that sphingolipid levels in Fabry fibroblasts are effectively corrected by co-treatment with α-gal A and 4.
Results
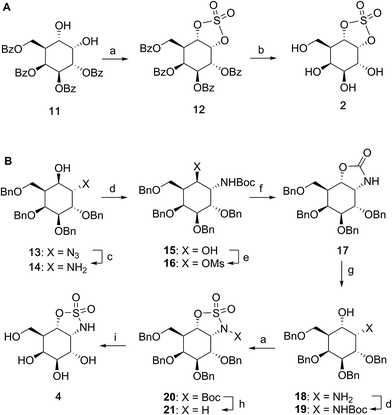
Synthesis of α-D-galactose configured cyclosulfate 2 and cyclosulfamidate 4
Benzoylated diol 11 (see the ESI and Scheme S1† for its synthesis) was treated with thionyl chloride and subsequently oxidized with ruthenium trichloride and sodium periodate to afford protected cyclosulfate 12. α-Gal-cyclosulfate 2 was obtained after benzoyl removal using methanolic ammonia (Scheme 1A). The synthesis of cis-1-amino-6-hydroxy cyclohexane 18, a key intermediate in the synthesis of α-gal-cyclosulfamidate 4 proceeded through oxazolidinone 17, which was obtained from trans-azido alcohol 13 (itself obtained from perbenzylated galacto-cyclophellitol, see the ESI†) as depicted in Scheme 1B. Hydrolysis of the carbamate in 17 and N-bocylation provided 19, which was transformed into fully protected cyclosulfamidate 20. Global deprotection finally yielded the target compound 4. The synthesis of compounds 3, 5 and 9 (Fig. 1B) and intermediates follows related strategies, as is described in the ESI.†α-D-Galactose configured cyclosulfate 2, cyclosulfamidates 3 and 4, and cyclosulfamide 5 are predominantly in the 4C1 conformation
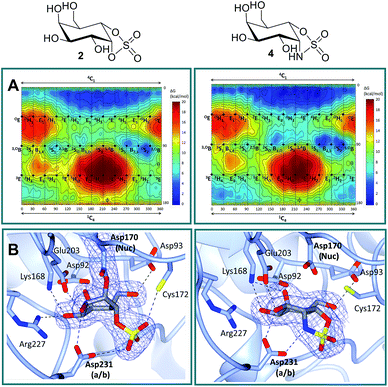
Free energy landscapes (FELs) of inhibitors report the conformational behavior in solution well, and can therefore be used to predict the selectivity for GH active sites.26 We calculated the conformational FELs of compounds 2–5 using ab initio metadynamics (Fig. 2A, S1 and S2†). The FEL of α-gal-cyclophellitol cyclosulfate 2 is strongly biased towards 4C1, with a secondary minimum around B2,5. This B2,5 conformer is unlikely to be enzyme active-site-reactive as it exhibits an equatorial C1–O bond (Fig. S2†). The 4C1 minimum of the substrate extends towards the TS-like 4H3 conformation, indicating that cyclosulfate 2 in a 4H3 conformation could be transiently populated on-enzyme, favoring the nucleophilic attack and formation of a glycosyl-enzyme adduct. The FELs of 3–5 show that substitution of the cyclic sulfate trap by cyclic sulfamidates (3 and 4) or sulfamide (5) does not significantly affect the conformational preferences. The local B2,5 minimum in 4 is more pronounced, probably due to a hydrogen bond between the 2-OH and one cyclosulfamidate oxygen (Fig. S2†).α-D-Galactose configured cyclosulfate 2 and isosteres 4 and 5 inhibit α-gal A in vitro
α-Gal-cyclosulfate 2, α-gal-cyclosulfamidates 3 and 4, and α-gal-cyclosulfamide 5, as well as the known α-galactosidase inhibitors α-gal-cyclophellitol 6,27 α-gal-cyclophellitol aziridine 7,27 Gal-DNJ 828 and β-gal-cyclosulfate 9 were evaluated on their inhibitory potential against recombinant human GH27 α-galactosidase A (α-gal A, Fabrazyme®, agalsidase beta) and their selectivity over human β-galactosidases, galactosidase beta 1 (GLB1, GH35) and galactosylceramidase (GALC, GH59). We first determined the apparent IC50 values by using fluorogenic 4-methylumbelliferyl (4MU)-α- or -β-D-galactopyranose substrates (Table 1). α-Gal-cyclosulfate 2 effectively inhibits α-gal A on a par with α-gal-cyclophellitol 6 (apparent IC50 = 25 vs. 13 μM, respectively), although less potently than α-gal-cyclophellitol aziridine 7 (apparent IC50 = 40 nM). Cyclosulfamidate 4, with the sulfamidate nitrogen taking up the position occupied by the anomeric oxygen in the natural substrate, proved to be a rather good inhibitor (IC50 = 67 μM), whereas isomer 3 is inactive and sulfamide 5 considerably less potent (IC50 = 423 μM).| Compd. | In vitro α-gal A (agalsidase beta) apparent IC50 (μM) | In vitroβ-gal (GLB1) apparent IC50 (μM) | In vitroβ-gal (GALC) apparent IC50 (μM) | Kinetic parameters α-gal A (agalsidase beta) kinact (min−1) and KI (μM) or Ki (μM) | Kinetic parameters α-gal A (agalsidase beta) kinact/KI (min−1 μM−1) |
|---|---|---|---|---|---|
| a Due to weak inhibition. b Due to fast inhibition; N.A., not applicable. Reported values are mean ± standard deviation from 3 technical replicates. | |||||
| α-Gal-cyclosulfate 2 | 25 ± 2.5 | >1000 | >1000 | Irreversible KI = 237 kinact = 0.06 | 0.25 |
| α-Gal-cyclosulfamidate 3 | >1000 | 39 ± 4.6 | 95 ± 14 | N.A. | N.A. |
| α-Gal-cyclosulfamidate 4 | 67 ± 4.7 | >1000 | >1000 | Competitive Ki = 110 | — |
| α-Gal-cyclosulfamide 5 | 423 ± 58 | 38 ± 1.7 | 191 ± 5.5 | N.D.a | N.D.a |
| α-Gal-cyclophellitol 6 | 13 ± 0.95 | 0.84 ± 0.13 | 4.2 ± 0.51 | Irreversible KI = 430 kinact = 0.24 | 0.55 |
| α-Gal-cyclophellitol aziridine 7 | 0.040 ± 0.005 | 0.93 ± 0.06 | 1.1 ± 0.30 | Irreversible N.D.b | 16.4 |
| Gal-DNJ 8 | 0.079 ± 0.004 | 42 ± 0.72 | 433 ± 39 | Competitive Ki = 0.23 | — |
| β-Gal-cyclosulfate 9 | >1000 | >1000 | >1000 | N.A. | N.A. |
We also measured the apparent IC50 values for inhibition against two human β-galactosidases: GLB1 (measured in human fibroblast lysates) and GALC (measured in overexpressing HEK293 cell medium). Cyclosulfate 2 and cyclosulfamidate 4 appear to be more selective than cyclophellitol epoxide 6 and aziridine 7, and we reason that this is due to the 4C1 conformation adopted by 2 and 4, which corresponds to the Michaelis complex conformation in α-galactosidases, but not in β-galactosidases (compound 2 is inactive up to 1 mM whereas 6 and 7 display low micromolar activity towards GLB1 and GALC). β-Gal-cyclosulfate 9, which in principle neither mimics the Michaelis complex nor the transition state conformation of β-galactosidases,29,30 is inactive against β- and α-galactosidases up to 1 mM. Cyclosulfamidate 4 and Gal-DNJ 8 show selectivity over α-glucosidase GAA, but both inhibit human recombinant β-glucosidase (GBA) (Table S1†).
We next explored the reversibility of inhibition by our new cyclic sulfate analogues. Enzymes were pre-incubated for different time periods (30, 60, 120, and 240 min) with inhibitors at concentrations of their corresponding apparent IC50 values, after which residual α-gal A activity was determined (Fig. S3†). Whilst cyclosulfate 2 is an irreversible inhibitor (showing a decrease in α-galactosidase activity with longer incubation time), cyclosulfamidate 4 and cyclosulfamide 5 are reversible inhibitors as revealed by a constant residual activity with extended incubation times. This was confirmed by kinetic studies monitoring the absorbance generated by the hydrolysis of the 2,4-dinitrophenyl-α-D-galactopyranoside substrate (2,4-DNP-α-gal) (Table 1). The irreversible inhibitors 2, 6 and 7 follow pseudo-first order kinetics. Although α-cyclosulfate 2 has a similar kinact/KI ratio to α-cyclophellitol 6 (kinact/KI = 0.25 vs. 0.55 min−1 mM−1, respectively), it has a stronger initial binding constant (KI) and a slower inactivation rate constant (kinact) (2: KI = 237 μM and kinact = 0.06 min−1; 6: KI = 430 μM and kinact = 0.24 min−1). Only a kinact/KI ratio could be measured for α-aziridine 7 due to fast inhibition (kinact/KI = 16.4 min−1 mM−1). Kinetics with increasing 2,4-DNP-α-gal concentrations showed that cyclosulfamidate 4 reversibly inhibits α-galactosidase with a Ki of 110 μM.
Structural analysis of α-gal-cyclosulfate 2 and α-gal-cyclosulfamidate 4 in complex with agalsidase beta
Firstly, in order to confirm the covalent inhibition by the cyclic sulfate, the X-ray structure of agalsidase beta in complex with 2 (PDB:6IBM) was determined to 1.99 Å, revealing a single molecule of 2 covalently bound to the enzyme active site (Fig. 2B). The observed electron density unambiguously shows that α-cyclosulfate 2 has reacted by attack of the catalytic nucleophile Asp170 to form a covalent enzyme–inhibitor complex. This covalent complex adopts a 1S3 conformation, consistent with the conformation of the covalent intermediate in the α-galactosidase reaction itinerary (Fig. 1A). Upon nucleophilic attack to the cyclic sulfate, the sulfate forms hydrogen bonds with Asp231 and Cys172, the latter suffering a shift in position.Armed with the knowledge that 2 indeed forms a covalent adduct to agalsidase beta, we moved on to ascertain if the cyclosulfamidate 4 would, as envisaged, function as a non-covalent, active-centre-directed, inhibitor of the enzyme-replacement enzyme. In contrast to cyclosulfate 2, cyclosulfamidate 4 (PDB:6IBK determined to 2.07 Å) was indeed shown to reversibly bind the catalytic site (Fig. 2B). As expected, the α-galactose configured cyclosulfamidate 4 adopts a 4C1 “Michaelis-like” complex conformation in the active site. Interestingly, the NH from the cyclosulfamidate moiety forms a hydrogen bond with the acid/base Asp231.
Thermostability of agalsidase beta in the presence of α-cyclosulfamidate 4 and Gal-DNJ 8
Competitive α-galactosidase inhibitors, including Gal-DNJ 8, are currently investigated in clinical studies as stabilizers of the recombinant enzyme, where they are employed to enhance enzyme replacement efficacy. In such a treatment regime, the enzyme and active site inhibitor are administered jointly.11,14,16 The stability of a recombinant enzyme relative to the temperature is considered to reflect well its stability in body circulation,31 and can be measured with ease, also in the presence of competitive inhibitors designed to stabilize the protein fold.31,32 Accordingly, we performed thermal stability assays (TSAs) on agalsidase beta alone and in the presence of increasing concentrations of 2, 4 or 8.Thermal melting profiles of lysosomal α-gal A revealed that α-gal A is most thermostable at pH 5.5 (Fig. 3A and S4†), which is consistent with α-gal A being a lysosomal enzyme. α-Gal-cyclosulfamidate 4 stabilizes α-gal A at pH 7.4 with a ΔTmmax of 17.4 °C, compared to a ΔTmmax of 34.3 °C produced by Gal-DNJ 8 (Fig. 3A and S4†). TSA effects at acidic pHs were lower for both 4 and Gal-DNJ 8, with recorded ΔTmmax values of 9.3 °C and 22.3 °C for 4 and Gal-DNJ 8, respectively at pH 5.5, and ΔTmmax values of 9.7 °C and 21.2 °C for the same compounds at pH 4.5. Surprisingly, we observed no thermostabilization effect on α-gal A in the presence of α-gal-cyclosulfate 2, despite this compound being an irreversible α-galactosidase inhibitor. Possibly, the sulfate group does not provide the optimal enzyme–ligand interactions to induce stabilization of α-galactosidase when the ring is in the 1S3 conformation adopted by covalently bound 2, compared to the 4C1 conformations adopted by both 4 and Gal-DNJ 8.
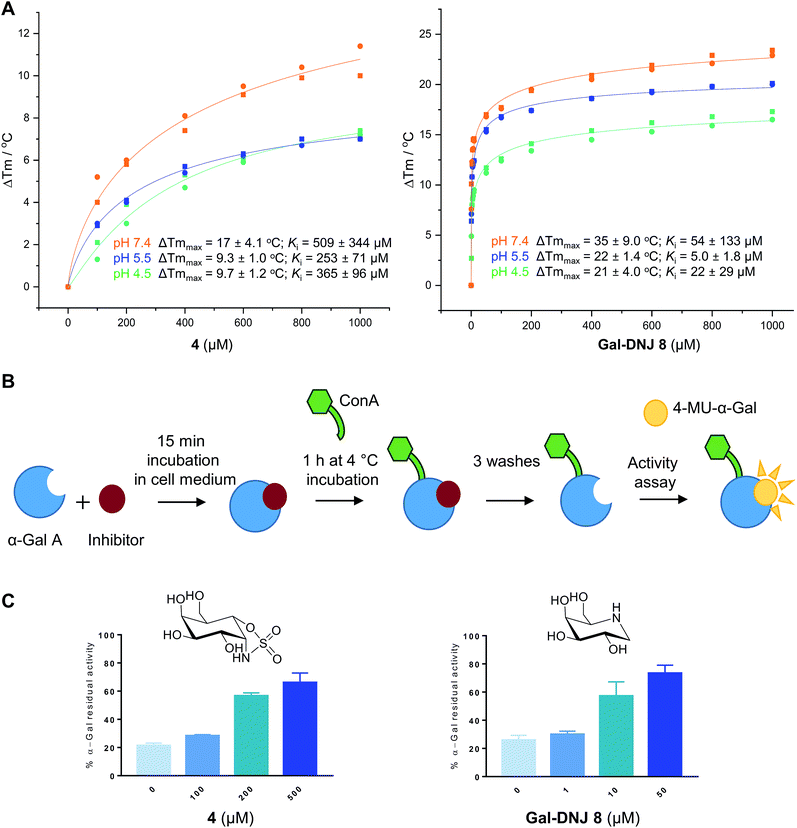 | ||
| Fig. 3 Effect of α-cyclosulfamidate 4 and Gal-DNJ 8 on the thermal stability and cell culture medium stability of agalsidase. (A) Heat-induced melting profiles of lysosomal α-gal A recorded by thermal shift assay, measured at pH 4.5, 5.5 and 7.4 in the presence of α-cyclosulfamidate 4 and Gal-DNJ 8. Melting points (Tm) were determined through thermal shift analyses by monitoring the fluorescence of Sypro Orange dye (λem 585 nm) as a function of temperature (see the ESI†). (B) Schematic representation of stabilization effect assay. Agalsidase beta was incubated with an inhibitor for 15 min in DMEM/F-12 medium and subsequently incubated with ConA sepharose beads for 1 h at 4 °C and washed to remove the inhibitor. Residual α-gal activity was quantified with the 4-MU-α-gal substrate. (C) Percentage of α-gal A residual activity after 15 min of incubation in DMEM/F-12 medium in the presence of inhibitors α-gal-cyclosulfamidate 4 (at 0, 100, 200, and 500 μM) and Gal-DNJ 8 (0, 1, 10 and 50 μM), followed by post final ConA purification. Percentages are calculated considering the 100% activity of α-gal A obtained at 0 min incubation time (n = 2, error bars indicate mean ± standard deviation). | ||
Stabilization of agalsidase beta by α-cyclosulfamidate 4 in cell culture medium
Agalsidase beta shows poor stability in plasma at a pH of 7.3–7.4, with only ∼25% of the hydrolytic activity being retained after incubation of the enzyme at 1 μg mL−1 in human plasma for 30 minutes.33 Given the stabilizing effect observed for 4 in the above-described TSAs, we investigated the ability of this compound to stabilize agalsidase beta in culture medium at physiological pH compared to Gal-DNJ 8.34 We first investigated the stabilization effect of the inhibitors in culture medium alone, as a surrogate measure for plasma stability (Fig. 3B and C). Incubation of agalsidase beta (25 μL of 2.5 μg μL−1) in cell culture medium (Dulbecco's Modified Eagles Medium/Nutrient Mixture F-12 (DMEM/F12), supplemented with 10% fetal calf serum and 1% penicillin/streptomycin) led to 80% loss of activity within 15 min, in line with the poor stability of this enzyme in blood plasma. To assess the stabilizing effects of 2, 4 and Gal-DNJ 8 in cell culture media, agalsidase beta was incubated with increasing concentrations of these compounds, followed by capture of the enzyme on concanavalin A (ConA) sepharose beads, washing to remove the bound inhibitor, and quantification of residual α-galactosidase activity with the 4MU-α-gal substrate. Media stabilization of agalsidase beta followed the same trend as observed in TSAs, with 2 failing to stabilize the enzyme and instead irreversibly inhibiting agalsidase beta. In contrast, α-gal-cyclosulfamidate 4 and Gal-DNJ 8 both prevented inactivation of agalsidase beta in cell culture media (pH 7.2) and ∼75% residual α-gal-A activity was retained after incubation with 4 (at 500 μM) or Gal-DNJ 8 (at 50 μM) (Fig. 3C).Competitive activity-based protein profile (ABPP) on recombinant α-galactosidases
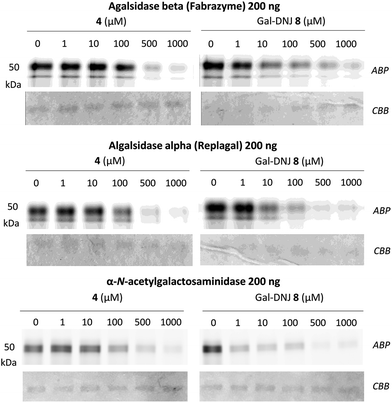
We studied the binding of α-cyclosulfamidate 4 and Gal-DNJ 8 to the commercial α-galactosidases: agalsidase beta (Fabrazyme®), agalsidase alpha (Replagal®) and α-galactosidase B (N-acetylgalactosaminidase, NAGA) by competitive activity-based protein profiling (ABPP, Fig. 4). Enzymes were incubated with increasing concentrations (ranging from 0 to 1000 μM) of both α-cyclosulfamidate 4 and Gal-DNJ 8 for 30 min at 37 °C, followed by incubation with 0.2 μM of an α-galactosidase Cy5 activity-based probe (ABP 10, Fig. S5†) for 30 min at 37 °C. After incubation, the samples were analyzed using sodium dodecyl sulfate–polyacrylamide gels (SDS–PAGE), followed by a fluorescent scan of the gels as previously described.27,33 Competitive ABPP revealed that α-cyclosulfamidate 4 (100–500 μM) and Gal-DNJ 8 (1–10 μM) inhibit both recombinant human α-galactosidases and N-acetylgalactosaminidase.In situ treatment of cultured fibroblasts from patients with Fabry disease
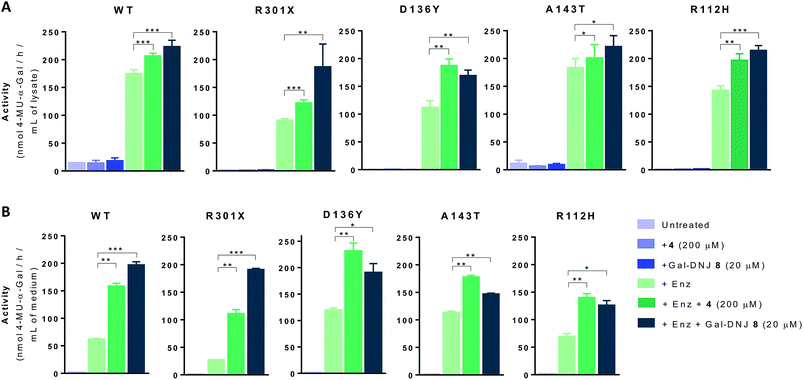
We next investigated whether the stabilizing effect of α-cyclosulfamidate 4 towards agalsidase beta produced an improvement in the cellular uptake of the enzyme by fibroblasts. We performed in situ studies in 5 different primary cell lines obtained from adult male volunteers: wild-type (WT, control) representing normal α-gal A activity, 2 classic Fabry mutant fibroblasts (R301X and D136Y) with no α-gal A activity and 2 atypical variant Fabry mutants (A143T and R112H) with substantially lowered residual α-gal A activity. Fibroblasts were incubated with 0.5% DMSO (untreated) or either 4 (200 μM) or Gal-DNJ 8 (20 μM) (in blue), agalsidase beta (100 ng) or with a combination of both enzyme and stabilizing agent (in green) (Fig. 5A). After 24 h treatment, the cells were harvested and homogenized, and the intracellular α-gal A activity of the corresponding cell lysates was measured. The WT cell line presented normal α-gal A activity while untreated classic Fabry patients (R301X and D136Y) and variant mutation samples (A143T and R112H) showed reduced enzymatic activity. None of the cell lines, not even classical Fabry fibroblasts R301X and D136Y, showed a significant increase in α-gal A activity when incubated with 4 (200 μM) or 8 (20 μM) alone for 24 h. Of note, Gal-DNJ 8 is known to enhance α-gal A activity in R301Q lymphoblasts after in situ 4 day treatment of a 100 μM daily dose.34 Treatment with agalsidase beta showed a considerable increase in α-gal A activity in all the studied cell lines. This effect was improved in most cases with the combinatorial treatment of agalsidase beta and 4 or 8 after 24 h of incubation. We also measured α-gal A activity in media in order to confirm that the increase in α-gal A activity in cell lysates is due to stabilization of the enzyme (Fig. 5B). Thus, culture media were collected before harvesting the cells and α-gal A activity was measured after ConA purification. α-Gal A activity in media was at least double in all the cell lines treated with α-cyclosulfamidate 4 (200 μM) or Gal-DNJ 8 (20 μM), consistent with these compounds preventing α-gal A degradation during cell culture.Gb3 and lysoGb3 levels are corrected by α-cyclosulfamidate 4
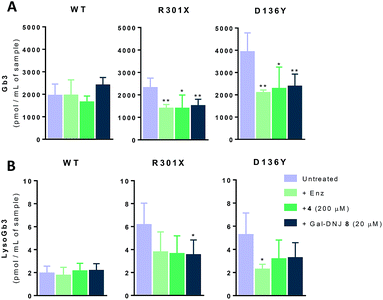
Generally, Fabry patients present elevated Gb3 which is further metabolized by acid ceramidase into lysoGb3 in lysosomes.35 LysoGb3 constitutes a signature of Fabry disease and allows diagnostic monitoring of disease progression,2,3,36–38 and has been linked to neuronopathic pain and renal failure through its effect on nociceptive neurons and podocytes.39–42 We investigated whether co-administration of α-cyclosulfamidate 4 and Gal-DNJ 8 with agalsidase beta would also have a positive effect in correcting these toxic metabolite levels. Gb3 and lysoGb3 levels from in situ treated cells were measured by LC-MS/MS (Fig. 6). Normal Gb3 and lysoGb3 levels observed in wild-type cells are in the range of around 2000 pmol mL−1 and 2 pmol mL−1 of Gb3 and lysoGb3, respectively. Cultured fibroblasts from classic Fabry patients (R301X and D136Y) treated with agalsidase beta resulted in a reduction of Gb3 and lysoGb3. This reduction was similar when fibroblasts were treated with the combination of α-cyclosulfamidate 4 (200 μM) or Gal-DNJ 8 (20 μM) and agalsidase beta. A variant Fabry A143T cell line presents normal Gb3 and lysoGb3 basal levels, whereas in R112H fibroblasts, these metabolites are increased and not corrected by agalsidase beta itself or inhibitor–agalsidase beta combination treatment (Fig. S6†).In situ 4 day treatment of cultured fibroblasts: increased α-gal A activity and Gb3 metabolite correction by α-cyclosulfamidate 4
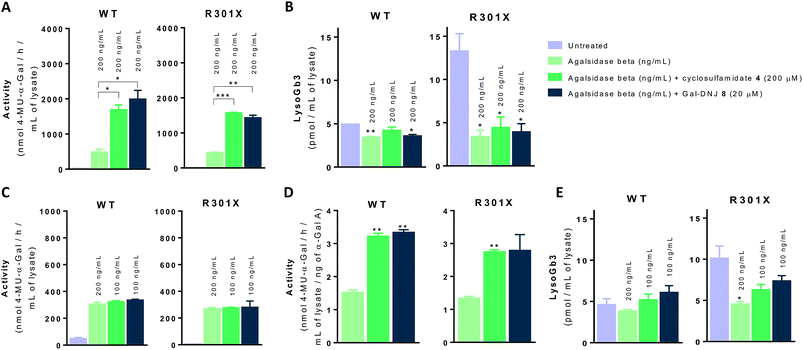
We next investigated whether the beneficial effect could be potentiated by extended incubation treatments. Thus, WT and classic Fabry (R301X) fibroblasts were treated with agalsidase beta (200 ng mL−1) or with a combination of enzyme (200 ng mL−1) and α-cyclosulfamidate 4 (200 μM) or Gal-DNJ 8 (20 μM) for 4 days. Fibroblasts were treated every 24 h with new medium and/or enzyme with or without inhibitor, and medium samples were collected for α-gal A activity quantification (see ESI Fig. S7†). α-Gal A activity was 3–4 times higher in fibroblasts treated with the combination of recombinant α-gal A and α-cyclosulfamidate 4 (200 μM) or Gal-DNJ 8 (20 μM) than those treated with agalsidase beta alone (Fig. 7A). This increase in α-gal A activity correlates with the reduction of lyso-Gb3 from ∼14 pmol mL−1 to ∼4 pmol mL−1 in the cell lysates of Fabry (classic R301X) fibroblasts (Fig. 7B). We finally studied whether the amount of required ERT could be decreased when stabilized with 4 or Gal-DNJ 8 and still produce a similar effect. WT and Fabry (classic R301X) fibroblasts were treated with agalsidase beta at 200 ng mL−1 and 100 ng mL−1. A reduction in toxic metabolites can be achieved in 4 days with half the concentration of enzyme (100 ng mL−1) when either α-cyclosulfamidate 4 (200 μM) or Gal-DNJ (20 μM) is added (Fig. 7C and D), with a similar reduction of toxic lyso-Gb3 from ∼10 pmol mL−1 to ∼5–6 pmol mL−1 in the cell lysates of Fabry (classic R301X) fibroblasts (Fig. 7E).Discussion
ERT with intravenous administration of recombinant human α-D-galactosidase (agalsidase beta, Fabrazyme® or agalsidase alpha, Replagal®) reduces the levels of Gb3 and lyso-Gb3 in some tissues of Fabry patients, but its clinical efficacy is still limited.5–7,43 The limited enzyme stability in plasma is a major drawback, and it is for this reason that enzyme active site binders that stabilize recombinant enzyme in circulation are pursued – with Gal-DNJ 8 (Migalastat®) currently in use in the clinic as the benchmark. Here we report the design and synthesis of the first-in-class conformational glycosidase inhibitor and α-gal A stabilizing agent, α-cyclosulfamidate 4. We show that this compound reversibly and selectively inhibits agalsidase beta with an IC50 value of 67 μM and a Ki of 110 μM. Ab initio metadynamics calculations and structural analysis of α-cyclosulfamidate 4 in complex with agalsidase beta show that this inhibitor binds in a 4C1 conformation mimicking the Michaelis complex conformation. We demonstrate that α-cyclosulfamidate 4 stabilizes recombinant human α-D-galactosidase (agalsidase beta, Fabrazyme®) in thermal stabilization assays and show that this prevents its degradation in cell culture medium. We further show that both α-gal-cyclosulfamidate 4 and Gal-DNJ 8 stabilize the enzyme more significantly at neutral pH than under acidic conditions (ΔTmmax difference of 8.1 °C for 4 and ΔTmmax difference of 12.2 °C for Gal-DNJ 8).To further study the stabilizing effect, we investigated whether α-gal-cyclosulfamidate 4 and Gal-DNJ 8 would stabilize α-gal A activity under in situ cell conditions. Treatment of fibroblasts (WT, classic and variant Fabry cell lines) with only α-gal-cyclosulfamidate 4 (at 200 μM) and Gal-DNJ 8 (at 20 μM) for 24 h shows no effect on α-D-galactosidase activity. However, we observe an increased α-D-galactosidase activity in all cells treated with the combination of agalsidase beta and stabilizing agents (4 at 200 μM and 8 at 20 μM) when compared to the cells treated only with agalsidase beta. This result also correlates with an increased α-D-galactosidase activity in the cell medium of the cells treated with enzyme and 4 or 8. The stabilizing effect is more pronounced when cells are treated for longer time (4 days), suggesting that the agalsidase beta complexed with α-gal-cyclosulfamidate 4 or Gal-DNJ 8 is stabilized in the cell medium, internalized and dissociated from the reversible inhibitor in the lysosomes. Finally, co-administration of α-cyclosulfamidate 4 or Gal-DNJ 8 with agalsidase beta highlights a similar correction of toxic Gb3 and lyso-Gb3 metabolite levels as with the ERT alone. Importantly, similar α-gal A activity and correction of toxic metabolites is achieved with half the concentration of agalsidase beta when this is stabilized by α-cyclosulfamidate 4 or Gal-DNJ 8. The synergy between Gal-DNJ 8 and the human recombinant α-gal A in cultured fibroblasts from Fabry patients has recently been demonstrated both in agalsidase alpha and beta.16,19 This synergism, together with our agalsidase beta stabilization results, supports the idea that the efficacy of a combination treatment may be superior to ERT or PC alone for several reasons. Co-administration of ERT and the active site inhibitor may be effective in all Fabry patients, independent of mutations in their endogenous α-gal A. Furthermore, stabilization of the recombinant human α-gal A by a stabilizing agent may reduce the required enzyme dosages or extend IV injection intervals, and therefore improve the patient's lifestyle and reduce side effects and treatment costs.
Conclusions
In conclusion, we have developed a new class of α-D-galactosidase inhibitors based on cyclophellitol cyclosulfamidate as a conformational Michaelis complex isostere. Although cyclosulfamidate 4 is a 1000-fold weaker inhibitor of recombinant α-gal A compared to Gal-DNJ 8in vitro, it stabilizes α-gal A in cellulo at only 10 fold higher concentration, and we argue that non-basic, competitive glycosidase inhibitors are attractive starting points for clinical development as stabilizers of (recombinant) glycosidases in the context of lysosomal storage disorders. Also, compound 4 together with its structural isosteres (3 and 5) comprise a new class of competitive glycosidase inhibitors, and stabilizes agalsidase beta for the first time, not by the glycoside configurational mimicry and basic nature that is the hallmark of iminosugars (including Migalastat®), but by configurational and conformational mimicry of the Michaelis complex. Michaelis complex or product-like conformational competitive inhibitors have been reported to act on other glycosidases, for instance, thio-oligosaccharides44–46 and kifunensine.47,48 We believe that transferring the structural characteristics of our cyclosulfamidates to differently configured structural analogues may yield potent and selective competitive inhibitors targeting glycosidases and that these may have biological or biomedical value in their own right, be it as stabilizing agents or as classical enzyme inhibitors.Conflicts of interest
There are no conflicts to declare.Author contributions
M. A., J. M. F. G. A., H. S. O. and G. J. D. conceived and designed the experiments. M. A., C. H. and A. S. carried out synthesis of inhibitors under supervision of G. A. v. d. M and J. D. C. C. L. R. performed ab initio metadynamics calculations under supervision of C. R. R. R. and L. W. carried out structural studies on enzyme–inhibitor complexes and thermostability assays under supervision of G. J. D. M. A., M. J. F and K. K. determined the IC50 values and kinetic parameters, and performed agalsidase beta stabilization studies in vitro and in cellulo, and lipid metabolite quantification. M. A., J. M. F. G. A., G. J. D and H. S. O. wrote the manuscript with input from all authors.Acknowledgements
We thank The Netherlands Organization for Scientific Research (NWO-CW, ChemThem grant to J. M. F. G. A. and H. S. O., and NWO TOP grant 2018-714.018.002 to H. S. O.), the European Research Council (ERC-2011-AdG-290836 “Chembiosphing”, to H. S. O., and ERC-2012-AdG-322942 “Glycopoise”, to G. J. D.), Sanofi Genzyme (research grant to J. M. F. G. A. and H. S. O. for financial support and postdoctoral contract to M. A.), the Spanish Ministry of Science, Innovation and Universities (MICINN) and the European Regional Development Fund (Fondo Europeo de Desarrollo Regional, FEDER) (CTQ2017-85496-P to C. R.), the Agency for Management of University and Research Grants of Generalitat de Catalunya (AGAUR) (2017SGR-1189 to C. R.) and the Spanish Structures of Excellence María de Maeztu (MDM-2017-0767 to C. R.). C. H. is supported by Villum Foundation (VKR023110). R. J. R. is supported by the BBSRC (BB/M011151/1). We thank the Diamond Light Source for access to beamline i02 and i04 (proposal number mx-13587) and the Barcelona Supercomputing Center for providing the computer resources at MareNostrum and the technical support (proposal RES-QCM-2017-2-0011). G. J. D. is supported by the Royal Society though the Ken Murray Research Professorship.References
- V. Lombard, H. Golaconda Ramulu, E. Drula, P. M. Coutinho and B. Henrissat, Nucleic Acids Res., 2014, 42, 490–495 CrossRef PubMed.
- J. M. Aerts, J. E. Groener, S. Kuiper, W. E. Donker-Koopman, A. Strijland, R. Ottenhoff, C. van Roomen, M. Mirzaian, F. A. Wijburg, G. E. Linthorst, A. C. Vedder, S. M. Rombach, J. Cox-Brinkman, P. Somerharju, R. G. Boot, C. E. Hollak, R. O. Brady and B. J. Poorthuis, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2812–2817 CrossRef CAS PubMed.
- H. Gold, M. Mirzaian, N. Dekker, M. J. Ferraz, J. Lugtenburg, J. D. C. Codée, G. A. Van Der Marel, H. S. Overkleeft, G. E. Linthorst, J. E. M. Groener, J. M. Aerts and B. J. H. M. Poorthuis, Clin. Chem., 2013, 59, 547–556 CrossRef CAS.
- R. J. Desnick, Y. A. Ioannou, C. M. Eng, α-Galactosidase A Deficiency: Fabry Disease, in The Online Metabolic and Molecular Bases of Inherited Diseases, ed. David Valle, et al., McGraw-Hill, New York, NY, 2014, DOI:10.1036/ommbid.181.
- M. Arends, M. Biegstraaten, D. A. Hughes, A. Mehta, P. M. Elliott, D. Oder, O. T. Watkinson, F. M. Vaz, A. B. P. Van Kuilenburg, C. Wanner and C. E. M. Hollak, PLoS One, 2017, 12, 1–17 CrossRef PubMed.
- M. Arends, F. A. Wijburg, C. Wanner, F. M. Vaz, A. B. P. van Kuilenburg, D. A. Hughes, M. Biegstraaten, A. Mehta, C. E. M. Hollak and M. Langeveld, Mol. Genet. Metab., 2017, 121, 157–161 CrossRef CAS.
- T. Alegra, F. Vairo, M. V de Souza, B. C. Krug and I. V. D. Schwartz, Genet. Mol. Biol., 2012, 35, 947–954 CrossRef CAS.
- A. Markham, Drugs, 2016, 76, 1147–1152 CrossRef PubMed.
- F. E. Mohamed, L. Al-Gazali, F. Al-Jasmi and B. R. Ali, Front. Pharmacol., 2017, 8, 448 CrossRef PubMed.
- S. Ishii, H.-H. Chang, K. Kawasaki, K. Yasuda, H.-L. Wu, S. C. Garman and J.-Q. Fan, Biochem. J., 2007, 406, 285–295 CrossRef CAS PubMed.
- E. R. Benjamin, J. J. Flanagan, A. Schilling, H. H. Chang, L. Agarwal, E. Katz, X. Wu, C. Pine, B. Wustman, R. J. Desnick, D. J. Lockhart and K. J. Valenzano, J. Inherited Metab. Dis., 2009, 32, 424–440 CrossRef CAS.
- D. A. Hughes, K. Nicholls, S. P. Shankar, G. Sunder-Plassmann, D. Koeller, K. Nedd, G. Vockley, T. Hamazaki, R. Lachmann, T. Ohashi, I. Olivotto, N. Sakai, P. Deegan, D. Dimmock, F. Eyskens, D. P. Germain, O. Goker-Alpan, E. Hachulla, A. Jovanovic, C. M. Lourenco, I. Narita, M. Thomas, W. R. Wilcox, D. G. Bichet, R. Schiffmann, E. Ludington, C. Viereck, J. Kirk, J. Yu, F. Johnson, P. Boudes, E. R. Benjamin, D. J. Lockhart, C. Barlow, N. Skuban, J. P. Castelli, J. Barth and U. Feldt-Rasmussen, J. Med. Genet., 2017, 54, 288–296 CrossRef CAS.
- G. Sunder-Plassmann, R. Schiffmann and K. Nicholls, Expert Opin. Orphan Drugs, 2018, 6, 301–309 CrossRef CAS.
- C. Porto, M. Cardone, F. Fontana, B. Rossi, M. R. Tuzzi, A. Tarallo, M. V. Barone, G. Andria and G. Parenti, Mol. Ther., 2009, 17, 964–971 CrossRef CAS.
- E. R. Benjamin, R. Khanna, A. Schilling, J. J. Flanagan, L. J. Pellegrino, N. Brignol, Y. Lun, D. Guillen, B. E. Ranes, M. Frascella, R. Soska, J. Feng, L. Dungan, B. Young, D. J. Lockhart and K. J. Valenzano, Mol. Ther., 2012, 20, 717–726 CrossRef CAS PubMed.
- C. Porto, A. Pisani, M. Rosa, E. Acampora, V. Avolio, M. R. Tuzzi, B. Visciano, C. Gagliardo, S. Materazzi, G. La Marca, G. Andria and G. Parenti, J. Inherited Metab. Dis., 2012, 35, 513–520 CrossRef CAS PubMed.
- D. G. Warnock, D. G. Bichet, M. Holida, O. Goker-Alpan, K. Nicholls, M. Thomas, F. Eyskens, S. Shankar, M. Adera, S. Sitaraman, R. Khanna, J. J. Flanagan, B. A. Wustman, J. Barth, C. Barlow, K. J. Valenzano, D. J. Lockhart, P. Boudes and F. K. Johnson, PLoS One, 2015, 10, 1–17 CrossRef.
- J. Castelli, S. Sitaraman, D. J. Lockhart, K. Nicholls, P. F. Boudes, R. Giugliani, D. P. Germain, D. A. Hughes, C. J. Jennette, A. Mehta, A. Bragat and L. Barisoni, Orphanet J. Rare Dis., 2012, 7, 91 CrossRef PubMed.
- A. Pisani, C. Porto, G. Andria and G. Parenti, J. Inherited Metab. Dis., 2014, 37, 145–146 CrossRef PubMed.
- H. Brumer, P. F. G. Sims and M. L. Sinnott, Biochem. J., 1999, 339, 43–53 CrossRef CAS PubMed.
- A. I. Guce, N. E. Clark, E. N. Salgado, D. R. Ivanen, A. A. Kulminskaya, H. Brumer and S. C. Garman, J. Biol. Chem., 2010, 285, 3625–3632 CrossRef CAS PubMed.
- D. E. Koshland, Biol. Rev., 1953, 28, 416–436 CrossRef CAS.
- D. L. Zechel and S. G. Withers, Acc. Chem. Res., 2000, 33, 11–18 CrossRef CAS PubMed.
- G. Speciale, A. J. Thompson, G. J. Davies and S. J. Williams, Curr. Opin. Struct. Biol., 2014, 28, 1–13 CrossRef CAS PubMed.
- M. Artola, L. Wu, M. J. Ferraz, C. L. Kuo, L. Raich, I. Z. Breen, W. A. Offen, J. D. C. Codée, G. A. Van Der Marel, C. Rovira, J. M. F. G. Aerts, G. J. Davies and H. S. Overkleeft, ACS Cent. Sci., 2017, 3, 784–793 CrossRef CAS PubMed.
- R. J. Williams, J. Iglesias-Fernández, J. Stepper, A. Jackson, A. J. Thompson, E. C. Lowe, J. M. White, H. J. Gilbert, C. Rovira, G. J. Davies and S. J. Williams, Angew. Chem., Int. Ed., 2014, 53, 1087–1091 CrossRef CAS PubMed.
- L. I. Willems, T. J. M. Beenakker, B. Murray, S. Scheij, W. W. Kallemeijn, R. G. Boot, M. Verhoek, W. E. Donker-Koopman, M. J. Ferraz, E. R. Van Rijssel, B. I. Florea, J. D. C. Codée, G. A. Van Der Marel, J. M. F. G. Aerts and H. S. Overkleeft, J. Am. Chem. Soc., 2014, 136, 11622–11625 CrossRef CAS PubMed.
- G. Legler and S. Pohl, Carbohydr. Res., 1986, 155, 119–129 CrossRef CAS.
- N. F. Bráa, P. A. Fernandes and M. J. Ramos, J. Chem. Theory Comput., 2010, 6, 421–433 CrossRef PubMed.
- R. W. Wheatley, S. Lo, L. J. Jancewicz, M. L. Dugdale and R. E. Huber, J. Biol. Chem., 2013, 288, 12993–13005 CrossRef CAS PubMed.
- R. L. Lieberman, J. A. D'Aquino, D. Ringe and G. A. Petsko, Biochemistry, 2009, 48, 4816–4827 CrossRef CAS PubMed.
- J. Q. Fan, S. Ishii, N. Asano and Y. Suzuki, Nat. Med., 1999, 5, 112–115 CrossRef CAS PubMed.
- K. Kytidou, T. J. M. Beenakker, L. B. Westerhof, C. H. Hokke, G. F. Moolenaar, N. Goosen, M. Mirzaian, M. J. Ferraz, M. de Geus, W. W. Kallemeijn, H. S. Overkleeft, R. G. Boot, A. Schots, D. Bosch and J. M. F. G. Aerts, Front. Plant Sci., 2017, 8, 1026 CrossRef PubMed.
- N. Asano, S. Ishii, H. Kizu, K. Ikeda, K. Yasuda, A. Kato, O. R. Martin and J. Q. Fan, Eur. J. Biochem., 2000, 267, 4179–4186 CrossRef CAS PubMed.
- M. J. Ferraz, A. R. A. Marques, M. D. Appelman, M. Verhoek, A. Strijland, M. Mirzaian, S. Scheij, C. M. Ouairy, D. Lahav, P. Wisse, H. S. Overkleeft, R. G. Boot and J. M. Aerts, FEBS Lett., 2016, 590, 716–725 CrossRef CAS PubMed.
- H. Maruyama, K. Miyata, M. Mikame, A. Taguchi, C. Guili, M. Shimura, K. Murayama, T. Inoue, S. Yamamoto, K. Sugimura, K. Tamita, T. Kawasaki, J. Kajihara, A. Onishi, H. Sugiyama, T. Sakai, I. Murata, T. Oda, S. Toyoda, K. Hanawa, T. Fujimura, S. Ura, M. Matsumura, H. Takano, S. Yamashita, G. Matsukura, R. Tazawa, T. Shiga, M. Ebato, H. Satoh and S. Ishii, Genet. Med., 2019, 21, 44–52 CrossRef PubMed.
- M. Mirzaian, P. Wisse, M. J. Ferraz, A. R. A. Marques, P. Gaspar, S. V. Oussoren, K. Kytidou, J. D. C. Codée, G. van der Marel, H. S. Overkleeft and J. M. Aerts, Clin. Chim. Acta, 2017, 466, 178–184 CrossRef CAS PubMed.
- M. Mirzaian, P. Wisse, M. J. Ferraz, A. R. A. Marques, T. L. Gabriel, C. P. A. A. van Roomen, R. Ottenhoff, M. van Eijk, J. D. C. Codée, G. A. van der Marel, H. S. Overkleeft and J. M. Aerts, Clin. Chim. Acta, 2016, 459, 36–44 CrossRef CAS PubMed.
- L. Choi, J. Vernon, O. Kopach, M. S. Minett, K. Mills, P. T. Clayton, T. Meert and J. N. Wood, Neurosci. Lett., 2015, 594, 163–168 CrossRef CAS PubMed.
- M. D. Sanchez-Niño, D. Carpio, A. B. Sanz, M. Ruiz-Ortega, S. Mezzano and A. Ortiz, Hum. Mol. Genet., 2015, 24, 5720–5732 CrossRef PubMed.
- P. Colpart and S. Félix, Arch. Pathol. Lab. Med., 2017, 141, 1127–1131 CrossRef CAS PubMed.
- P. Rozenfeld and S. Feriozzi, Mol. Genet. Metab., 2017, 122, 19–27 CrossRef CAS PubMed.
- L. van Dussen, M. Biegstraaten, C. E. M. Hollak and M. G. W. Dijkgraaf, Orphanet J. Rare Dis., 2014, 9, 1–12 CrossRef PubMed.
- G. Sulzenbacher, H. Driguez, B. Henrissat, M. Schülein and G. J. Davies, Biochemistry, 1996, 35, 15280–15287 CrossRef CAS PubMed.
- A. J. Thompson, J. Dabin, J. Iglesias-Fernández, A. Ardèvol, Z. Dinev, S. J. Williams, O. Bande, A. Siriwardena, C. Moreland, T. C. Hu, D. K. Smith, H. J. Gilbert, C. Rovira and G. J. Davies, Angew. Chem., Int. Ed., 2012, 51, 10997–11001 CrossRef CAS PubMed.
- Y. Zhu, M. D. L. Suits, A. J. Thompson, S. Chavan, Z. Dinev, C. Dumon, N. Smith, K. W. Moremen, Y. Xiang, A. Siriwardena, S. J. Williams, H. J. Gilbert and G. J. Davies, Nat. Chem. Biol., 2010, 6, 125–132 CrossRef CAS PubMed.
- F. Vallée, K. Karaveg, A. Herscovics, K. W. Moremen and P. L. Howell, J. Biol. Chem., 2000, 275, 41287–41298 CrossRef PubMed.
- A. Males, L. Raich, S. J. Williams, C. Rovira and G. J. Davies, ChemBioChem, 2017, 18, 1496–1501 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary Fig. S1 to S7, Tables S1 and S2, materials and methods (biological and biochemical methods, DFT calculations, crystallography and chemical synthesis), and NMR spectra. See DOI: 10.1039/c9sc03342d |
| This journal is © The Royal Society of Chemistry 2019 |