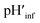Open Access Article
Open Access ArticleDetermination of proton concentration at cardiolipin-containing membrane interfaces and its relation with the peroxidase activity of cytochrome c†
Partha Pratim
Parui
 *ab,
Yeasmin
Sarakar
a,
Rini
Majumder
a,
Sanju
Das
ac,
Hongxu
Yang
b,
Kazuma
Yasuhara
b and
Shun
Hirota
*ab,
Yeasmin
Sarakar
a,
Rini
Majumder
a,
Sanju
Das
ac,
Hongxu
Yang
b,
Kazuma
Yasuhara
b and
Shun
Hirota
 *b
*b
aDepartment of Chemistry, Jadavpur University, Kolkata 700032, India. E-mail: parthaparui@yahoo.com; Fax: +91-33-24146223; Tel: +91-9433490492
bDivision of Materials Science, Nara Institute of Science and Technology, Nara 630-0192, Japan
cDepartment of Chemistry, Maulana Azad College, Kolkata 700013, India
First published on 6th August 2019
Abstract
The activities of biomolecules are affected by the proton concentrations at biological membranes. Here, we succeeded in evaluating the interface proton concentration (−log[H+] defined as pH′) of cardiolipin (CL)-enriched membrane models of the inner mitochondrial membrane (IMM) using a spiro-rhodamine-glucose molecule (RHG). According to fluorescence microscopy and 1H-NMR studies, RHG interacted with the Stern layer of the membrane. The acid/base equilibrium of RHG between its protonated open form (o-RHG) and deprotonated closed spiro-form (c-RHG) at the membrane interface was monitored with UV-vis absorption and fluorescence spectra. The interface pH′ of 25% cardiolipin (CL)-containing large unilamellar vesicles (LUVs), which possess similar lipid properties to those of the IMM, was estimated to be ∼3.9, when the bulk pH was similar to the mitochondrial intermembrane space pH (6.8). However, for the membranes containing mono-anionic lipids, the interface pH′ was estimated to be ∼5.3 at bulk pH 6.8, indicating that the local negative charges of the lipid headgroups in the lipid membranes are responsible for the deviation of the interface pH′ from the bulk pH. The peroxidase activity of cyt c increased 5–7 fold upon lowering the pH to 3.9–4.3 or adding CL-containing (10–25% of total lipids) LUVs compared to that at bulk pH 6.8, indicating that the pH′ decrease at the IMM interface from the bulk pH enhances the peroxidase activity of cyt c. The peroxidase activity of cyt c at the membrane interface of tetraoleoyl CL (TOCL)-enriched (50% of total lipids) LUVs was higher than that estimated from the interface pH′, while the peroxidase activity was similar to that estimated from the interface pH′ for tetramyristoyl CL (TMCL)-enriched LUVs, supporting the hypothesis that when interacting with TOCL (not TMCL), cyt c opens the heme crevice to substrates. The present simple methodology allows us to estimate the interface proton concentrations of complex biological membranes.
Introduction
Biological membranes separate cellular organelles from the exterior of cells. The water-exposed interfaces of bilayer membranes act as barriers keeping ions, proteins, and other molecules where they are needed, and preventing them from diffusing into areas where they should not be.1 Selective cellular uptake and refusal of various molecules by channels and other proteins at membrane interfaces frequently control biological processes, such as protein and ion transport,2 cell apoptosis,3 cell signaling,4 membrane trafficking,5etc. Membrane interface H+ concentration, which is affected by the membrane lipid composition, may also control the biological processes at the membrane interfaces. Although determination of the membrane interface H+ concentration is indispensable for evaluating the biomolecular activities at the membrane interface, the interface H+ concentration and its relationship with biological processes are often elusive.The proton gradient across the inner mitochondrial membrane (IMM) makes the IMM acidic (pH ∼ 6.8) and the matrix alkaline (pH ∼ 7.7) in isolated mitochondria.6 It has been reported that the pH change and proton gradient at the mitochondrial membrane are induced by physiological processes, such as Ca2+ transport,7 glutamate transport,8 and glucose starvation or sorbic acid stress.9 The lateral pH-profile along the p-side of cristae has been measured in situ by attaching a ratiometric fluorescent pH-sensitive GFP variant to oxidative phosphorylation complex IV and the dimeric F0F1 ATP-synthase in the mitochondrial membrane, showing that the local pH (−log[H+]) at F0F1 ATP-synthase dimers is 0.3 unit less acidic than that of complex IV.10
Self-assemblies of amphiphilic lipids and surfactants have been utilized to investigate the properties of biological membranes11 and their effects on protein activities,12 where the deviations in the interface −log[H+] (defined as pH′) from the bulk pH have been determined from the shifts in the acid/base pKa of small organic molecules upon interaction with self-assemblies.13 For example, the interface pH′ has been estimated from the shift in the acid/base pKa between the self-assembly interface and bulk by heterodyne-detected electronic sum frequency generation (HD-ESFG) spectroscopy.14 Various anionic amphiphilic self-assemblies, including inner and outer mitochondrial membranes, interact electrostatically with cationic rhodamine derivatives.15 Recently, we have introduced an interface pH′ detection method for various amphiphilic self-assemblies by exploiting the acid/base equilibrium of a H+ concentration probe (RHG), which is a glucose derivative of a spiro-rhodamine molecule.16
Cytochrome c (cyt c) contains 104 amino acid residues and is a positively charged protein at neutral pH. It is bound to the outer interface of the IMM, mainly by electrostatic interactions with anionic cardiolipins (CLs) in the membrane,17 and triggers apoptosis by its release from mitochondria.18 The peroxidase activity of cyt c increases upon interaction with CL, resulting in CL oxidation and subsequent apoptosis execution via cyt c permeabilization to the cytosol.19 Recently, it has been suggested that protein activities are affected by the pH gradient across biological membranes.20 The peroxidase activity of yeast cyt c adsorbed onto kaolinite was enhanced remarkably by decreasing the pH value below 4.21 In this study, by monitoring the acid/base equilibrium of RHG, we showed that the pH′ at the interfaces of large unilamellar vesicles (LUVs) containing monounsaturated tetraoleoyl CL (TOCL) or saturated tetramyristoyl CL (TMCL) (10–50% CL of total lipids) decreases ∼2.5–3.2 units from the neutral bulk pH. The peroxidase activity of cyt c was found to increase 5–7 fold at the LUV interface, due to the decrease in the interface pH′, while that at the interface of TOCL-containing LUVs was enhanced more than that estimated from the decrease in the interface pH′, apparently due to a cyt c structural change which has been previously reported.22
Results and discussion
Interaction of RHG with the membrane interface
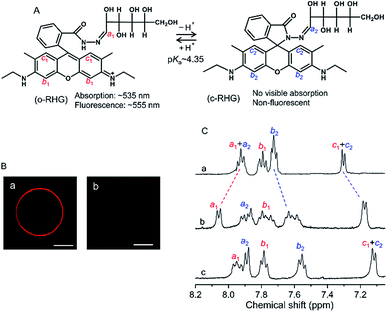
RHG shows a pH-dependent equilibrium between a protonated open form (o-RHG) and a deprotonated closed spiro-form (c-RHG), with an interconversion pKa of ∼4.35 (Fig. 1A).16c RHG comprises a hydrophobic rhodamine moiety (a cationic moiety for o-RHG) and a hydrophilic glucose moiety; thus, it binds to an anionic membrane. The difference in the o-RHG/c-RHG equilibrium at the membrane interface and in the bulk can be monitored by examining the UV-vis absorption and fluorescence spectra of RHG (Fig. 1A).16cThe pH of the intermembrane space is 6.8–6.9,23 while high concentrations of di-anionic CL are unique for mitochondrial membranes.24 Giant unilamellar vesicles (GUVs) (diameter, 1–10 μm; Fig. S1†) and LUVs (diameter, ∼100 nm) with a lipid composition of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)/TOCL = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were constructed to reproduce the major lipid components of the IMM24 (zwitterionic phosphatidylcholine (PC), ∼40%; zwitterionic phosphatidylethanolamine (PE), ∼30%; TOCL, ∼25%). The fluorescence of the o-RHG form of RHG (0.3 μM) was observed at the DOPC/DOPE/TOCL (2
1 were constructed to reproduce the major lipid components of the IMM24 (zwitterionic phosphatidylcholine (PC), ∼40%; zwitterionic phosphatidylethanolamine (PE), ∼30%; TOCL, ∼25%). The fluorescence of the o-RHG form of RHG (0.3 μM) was observed at the DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) GUV (total lipid, 500 μM) surface but not in the bulk medium (Fig. 1Ba), indicating that all the RHG molecules interacted with the GUV interface and the interface was more acidic than the bulk. However, for the interaction of RHG with a DOPC/1,2-dioleoyl-sn-glycero-3-phosphoglycerol (DOPG) (1
1) GUV (total lipid, 500 μM) surface but not in the bulk medium (Fig. 1Ba), indicating that all the RHG molecules interacted with the GUV interface and the interface was more acidic than the bulk. However, for the interaction of RHG with a DOPC/1,2-dioleoyl-sn-glycero-3-phosphoglycerol (DOPG) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) GUV containing mono-anionic lipids, the RHG fluorescence intensity decreased significantly compared to that for the interaction with the DOPC/DOPE/TOCL (2
2) GUV containing mono-anionic lipids, the RHG fluorescence intensity decreased significantly compared to that for the interaction with the DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) GUV under the same RHG and total lipid concentrations (Fig. 1Bb), apparently due to the decrease in H+ concentration and the o-RHG/RHG ratio at the DOPG GUV interface.
1) GUV under the same RHG and total lipid concentrations (Fig. 1Bb), apparently due to the decrease in H+ concentration and the o-RHG/RHG ratio at the DOPG GUV interface.
We obtained the absorption spectra of RHG in the presence of LUVs by subtracting the absorption spectrum of the LUV solution from that of the solution containing RHG and LUVs, although the LUV solution exhibited light scattering at the excitation (∼530 nm) and emission (∼560 nm) wavelengths of RHG (Fig. S2†). The absorbances of the DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUV solution (total lipid, 1 mM) were 0.080–0.145 and 0.071–0.130 at 530 and 560 nm, respectively, and those of the DOPG LUV solution were 0.051–0.097 and 0.045–0.088 at 530 and 560 nm, respectively, at pH 4.0–7.0 (Fig. S2A†). To identify the inner filter effect (IFE) in the RHG fluorescence intensity due to the LUV light scattering, we increased the concentration of DOPC/DOPE/TOCL (2
1) LUV solution (total lipid, 1 mM) were 0.080–0.145 and 0.071–0.130 at 530 and 560 nm, respectively, and those of the DOPG LUV solution were 0.051–0.097 and 0.045–0.088 at 530 and 560 nm, respectively, at pH 4.0–7.0 (Fig. S2A†). To identify the inner filter effect (IFE) in the RHG fluorescence intensity due to the LUV light scattering, we increased the concentration of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs at pH 4.5 and DOPG LUVs at pH 4.0 from 0.2 to 1 mM (total lipid) under a constant RHG concentration (0.2 μM), where the RHG fluorescence intensity was saturated (against LUV concentration) for all LUV concentrations used. The fluorescence intensity remained identical for the LUV concentrations studied (Fig. S3†), revealing that the IFE due to LUV light scattering was negligibly small, as reported previously.25 A linear correlation between the RHG concentration (keeping RHG
1) LUVs at pH 4.5 and DOPG LUVs at pH 4.0 from 0.2 to 1 mM (total lipid) under a constant RHG concentration (0.2 μM), where the RHG fluorescence intensity was saturated (against LUV concentration) for all LUV concentrations used. The fluorescence intensity remained identical for the LUV concentrations studied (Fig. S3†), revealing that the IFE due to LUV light scattering was negligibly small, as reported previously.25 A linear correlation between the RHG concentration (keeping RHG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lipid = 1
lipid = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 constant) and fluorescence intensity at 560 nm was observed up to 1.0 μM RHG at pH 5.5 and 6.0 (Fig. S4A†). The fluorescence intensity of the RHG (1 μM) solution was also measured in the presence of DOPC/DOPE/TOCL LUVs (DOPC
1000 constant) and fluorescence intensity at 560 nm was observed up to 1.0 μM RHG at pH 5.5 and 6.0 (Fig. S4A†). The fluorescence intensity of the RHG (1 μM) solution was also measured in the presence of DOPC/DOPE/TOCL LUVs (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at pH 5.5 and 6.0 under different concentrations of TOCL by changing the LUV concentration. A linear behaviour between the TOCL concentration and fluorescence intensity was observed up to 50 μM TOCL (Fig. S5B†). All these results demonstrate that there was no significant interference from light scattering on the fluorescence intensity. The absorbance and fluorescence intensity of RHG (1.0–2.0 μM) at pH 5.5 gradually increased upon increasing the amount of DOPC/DOPE/TOCL (2
1) at pH 5.5 and 6.0 under different concentrations of TOCL by changing the LUV concentration. A linear behaviour between the TOCL concentration and fluorescence intensity was observed up to 50 μM TOCL (Fig. S5B†). All these results demonstrate that there was no significant interference from light scattering on the fluorescence intensity. The absorbance and fluorescence intensity of RHG (1.0–2.0 μM) at pH 5.5 gradually increased upon increasing the amount of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) or DOPG LUVs (Fig. S5†), indicating that RHG interacted with LUVs and the o-RHG/RHG ratio increased due to the interaction. Thus, LUV-binding saturation conditions with high LUV concentrations were used at all pH for further experiments.
1) or DOPG LUVs (Fig. S5†), indicating that RHG interacted with LUVs and the o-RHG/RHG ratio increased due to the interaction. Thus, LUV-binding saturation conditions with high LUV concentrations were used at all pH for further experiments.
The fluorescence quantum yield of RHG did not change upon varying the TOCL% (5–25% of total lipids) in DOPC/DOPE/TOCL LUVs at acidic pH 3.0, in which all of the c-RHG was converted to the o-RHG form (Fig. S6†). These results indicate that the fluorescence of RHG was not due to formation of a dimer, the formation of which has been reported for 10-N-nonyl acridine orange when it interacts with TOCL.26 To investigate the interaction of RHG with TOCL in more detail, the absorbance of RHG at 535 nm was measured for RHG solutions under various TOCL concentrations obtained by changing the concentration of DOPC/DOPE/TOCL LUVs (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; TOCL, 10 and 25% of total lipids) at pH 6.2. A gradual increase in the absorbance of RHG (2 μM) at 535 nm was observed upon increasing the TOCL concentration, and the absorbance saturated at ∼125 and ∼160 μM TOCL for LUVs containing 10 and 25% TOCL, respectively (Fig. S7†). Large [TOCL]/[RHG] ratios were required for the absorbance to saturate, suggesting a non-specific binding between RHG and TOCL. These results support the hypothesis that the dimer formation is not responsible for the increase in the fluorescence intensity of RHG.
1; TOCL, 10 and 25% of total lipids) at pH 6.2. A gradual increase in the absorbance of RHG (2 μM) at 535 nm was observed upon increasing the TOCL concentration, and the absorbance saturated at ∼125 and ∼160 μM TOCL for LUVs containing 10 and 25% TOCL, respectively (Fig. S7†). Large [TOCL]/[RHG] ratios were required for the absorbance to saturate, suggesting a non-specific binding between RHG and TOCL. These results support the hypothesis that the dimer formation is not responsible for the increase in the fluorescence intensity of RHG.
RHG binding to DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs was also investigated at pH 4.5–8.0 (Fig. S8†). The RHG (2 μM) solutions containing LUVs at various pHs from 4.5 to 8.0 were filtered using a 100 K molecular weight cut-off filter. The pH value of the filtrate was adjusted to 2.0, at which all of the RHG is converted to the o-RHG form and exhibits absorbance at 532 nm. For all the pH conditions (4.5–8.0) studied, the absorbance at 532 nm of the filtrate after adjusting the pH to 2.0 was less than 5% that of 2 μM RHG at pH 2.0. These results reveal that the binding of RHG to TOCL was more than 95% at pH 4.5–8.0.
1) LUVs was also investigated at pH 4.5–8.0 (Fig. S8†). The RHG (2 μM) solutions containing LUVs at various pHs from 4.5 to 8.0 were filtered using a 100 K molecular weight cut-off filter. The pH value of the filtrate was adjusted to 2.0, at which all of the RHG is converted to the o-RHG form and exhibits absorbance at 532 nm. For all the pH conditions (4.5–8.0) studied, the absorbance at 532 nm of the filtrate after adjusting the pH to 2.0 was less than 5% that of 2 μM RHG at pH 2.0. These results reveal that the binding of RHG to TOCL was more than 95% at pH 4.5–8.0.
We have previously reported that when RHG is located at the outer interface, the fluorescence of RHG can be selectively quenched by addition of Cu(ClO4)2/Na2S (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) solution containing a non-permeable Cu2+/S2− quencher.16c The amount of RHG localized at the inner interface can be estimated from the residual fluorescence intensity obtained after the addition of the quencher. DOPC/DOPE/TOCL (2
2) solution containing a non-permeable Cu2+/S2− quencher.16c The amount of RHG localized at the inner interface can be estimated from the residual fluorescence intensity obtained after the addition of the quencher. DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs (total lipid, 1 mM) were prepared in 10 mM cacodylate buffer, pH 6.0. The solution containing RHG (1 μM) and DOPC/DOPE/TOCL (2
1) LUVs (total lipid, 1 mM) were prepared in 10 mM cacodylate buffer, pH 6.0. The solution containing RHG (1 μM) and DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs (total lipid, 1 mM) in 10 mM cacodylate buffer, pH 6.0, was concentrated from 1 mL to ∼40 μM using a 100k Da molecular weight cut-off filter to separate unbound RHG from LUVs. Subsequently, we diluted the concentrated LUV solution to 1 mL with 10 mM HEPES buffer, pH 8.0, and the pH of the solution was adjusted to 8.0 with addition of ∼3 μL of 0.1 M NaOH. The fluorescence intensity of the pH-adjusted solution decreased to ∼9% compared to that of the solution at pH 6.0 before concentration (Fig. S9†). We also concentrated the solution containing RHG (1 μM) and DOPC/DOPE/TOCL (2
1) LUVs (total lipid, 1 mM) in 10 mM cacodylate buffer, pH 6.0, was concentrated from 1 mL to ∼40 μM using a 100k Da molecular weight cut-off filter to separate unbound RHG from LUVs. Subsequently, we diluted the concentrated LUV solution to 1 mL with 10 mM HEPES buffer, pH 8.0, and the pH of the solution was adjusted to 8.0 with addition of ∼3 μL of 0.1 M NaOH. The fluorescence intensity of the pH-adjusted solution decreased to ∼9% compared to that of the solution at pH 6.0 before concentration (Fig. S9†). We also concentrated the solution containing RHG (1 μM) and DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs (total lipid, 1 mM) in 10 mM cacodylate buffer, pH 6.0, and mixed the concentrated LUV solution with the filtrate. The fluorescence intensity of the mixture was similar to that of the solution before filtration (Fig. S9†). Upon addition of Cu(ClO4)2/Na2S (1
1) LUVs (total lipid, 1 mM) in 10 mM cacodylate buffer, pH 6.0, and mixed the concentrated LUV solution with the filtrate. The fluorescence intensity of the mixture was similar to that of the solution before filtration (Fig. S9†). Upon addition of Cu(ClO4)2/Na2S (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) (total salt, 2 mM) to the mixture, the fluorescence intensity decreased to a similar intensity (∼8%) to that observed when changing the pH from 6.0 to 8.0 with the concentration procedure (Fig. S9†). These results demonstrate that not only ∼8% RHG was incorporated into the inner LUV lumen during the concentration procedure but also RHG exhibited negligibly small fluorescence intensity in the presence of DOPC/DOPE/TOCL (2
2) (total salt, 2 mM) to the mixture, the fluorescence intensity decreased to a similar intensity (∼8%) to that observed when changing the pH from 6.0 to 8.0 with the concentration procedure (Fig. S9†). These results demonstrate that not only ∼8% RHG was incorporated into the inner LUV lumen during the concentration procedure but also RHG exhibited negligibly small fluorescence intensity in the presence of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs at pH 8.0.
1) LUVs at pH 8.0.
To identify the interface location of the two molecular forms of RHG (o-RHG and c-RHG), we performed 1H-NMR studies in D2O medium in the presence and absence of LUVs under the pH conditions at which o-RHG and c-RHG coexist: in the presence of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs at pH 6.5, in the presence of DOPG LUVs at pH 5.5, and in the absence of LUVs at pH 4.5 (Fig. 1C). In the absence of LUVs, 1H-NMR chemical shifts of the imine protons (H–C
1) LUVs at pH 6.5, in the presence of DOPG LUVs at pH 5.5, and in the absence of LUVs at pH 4.5 (Fig. 1C). In the absence of LUVs, 1H-NMR chemical shifts of the imine protons (H–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N: a1 and a2) were observed at 7.92 ppm for both o-RHG and c-RHG, and those of the aromatic protons c1 and c2 were observed at 7.30 ppm for o-RHG and c-RHG. However, the chemical shift of the aromatic proton b1 of o-RHG shifted downfield to 7.79 ppm compared to that of the aromatic proton b2 of c-RHG at 7.72 ppm, presumably due to the nearby positive charge field caused by the protonation of the amine moiety (Fig. 1A and C). The chemical shifts of c1 of o-RHG and c2 of c-RHG were both observed at the same chemical shifts of 7.17 and 7.11 ppm in the presence of DOPC/DOPE/TOCL (2
N: a1 and a2) were observed at 7.92 ppm for both o-RHG and c-RHG, and those of the aromatic protons c1 and c2 were observed at 7.30 ppm for o-RHG and c-RHG. However, the chemical shift of the aromatic proton b1 of o-RHG shifted downfield to 7.79 ppm compared to that of the aromatic proton b2 of c-RHG at 7.72 ppm, presumably due to the nearby positive charge field caused by the protonation of the amine moiety (Fig. 1A and C). The chemical shifts of c1 of o-RHG and c2 of c-RHG were both observed at the same chemical shifts of 7.17 and 7.11 ppm in the presence of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs and DOPG LUVs, respectively, and at 7.30 ppm in the absence of LUVs, indicating that the rhodamine unit was located in a strong negative charge field formed at the Stern layer of LUVs for both o-RHG and c-RHG. Similar upfield chemical shifts were observed for the b2 protons of o-RHG and c-RHG: from 7.72 to 7.60 ppm for DOPC/DOPE/TOCL (2
1) LUVs and DOPG LUVs, respectively, and at 7.30 ppm in the absence of LUVs, indicating that the rhodamine unit was located in a strong negative charge field formed at the Stern layer of LUVs for both o-RHG and c-RHG. Similar upfield chemical shifts were observed for the b2 protons of o-RHG and c-RHG: from 7.72 to 7.60 ppm for DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs and from 7.72 to 7.55 ppm for DOPG LUVs. Interestingly, the chemical shifts of the imine protons (a1 and a2) in the presence of LUVs were not the same for o-RHG and c-RHG; they differed relatively significantly for DOPC/DOPE/TOCL (2
1) LUVs and from 7.72 to 7.55 ppm for DOPG LUVs. Interestingly, the chemical shifts of the imine protons (a1 and a2) in the presence of LUVs were not the same for o-RHG and c-RHG; they differed relatively significantly for DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs (downfield from 7.92 to 8.06 ppm for a1 and upfield from 7.92 to 7.89 ppm for a2), suggesting positive and negative charge environments around the imine protons of o-RHG and c-RHG, respectively. When the cationic rhodamine moiety of o-RHG interacts with the anionic headgroup of TOCL as evaluated from the chemical shifts of the c1 protons, the imine-N—connecting the water-exposed glucose region and the rhodamine unit—may face the positive charge field, due to the increase in proton concentration around the interface compared to the bulk as identified in the microscopic observation (Fig. 1Ba). A relatively small downfield chemical shift from 7.92 to 7.94 for the imine proton of o-RHG was detected upon addition of DOPG LUVs (Fig. 1C), indicating that the difference in the proton concentration between the interface and bulk is relatively small for DOPG LUVs (Fig. 1Bb). However, irrespective of LUV compositions, a similar upfield chemical shift for the imine proton (a2) of c-RHG was observed at 7.89 ppm, suggesting a negative charge environment around the imine proton. The neutral hydrophobic rhodamine unit of c-RHG may move toward the hydrophobic acyl chain of the lipids, while the hydrophilic glucose region prefers to stay in the water phase, and eventually the imine-N connecting the two units will face a negative charge environment produced by the anionic lipid headgroups of the LUVs. All these results suggest that both o-RHG and c-RHG interact with the Stern layer of the LUVs, and are useful to estimate the interface H+ concentration.
1) LUVs (downfield from 7.92 to 8.06 ppm for a1 and upfield from 7.92 to 7.89 ppm for a2), suggesting positive and negative charge environments around the imine protons of o-RHG and c-RHG, respectively. When the cationic rhodamine moiety of o-RHG interacts with the anionic headgroup of TOCL as evaluated from the chemical shifts of the c1 protons, the imine-N—connecting the water-exposed glucose region and the rhodamine unit—may face the positive charge field, due to the increase in proton concentration around the interface compared to the bulk as identified in the microscopic observation (Fig. 1Ba). A relatively small downfield chemical shift from 7.92 to 7.94 for the imine proton of o-RHG was detected upon addition of DOPG LUVs (Fig. 1C), indicating that the difference in the proton concentration between the interface and bulk is relatively small for DOPG LUVs (Fig. 1Bb). However, irrespective of LUV compositions, a similar upfield chemical shift for the imine proton (a2) of c-RHG was observed at 7.89 ppm, suggesting a negative charge environment around the imine proton. The neutral hydrophobic rhodamine unit of c-RHG may move toward the hydrophobic acyl chain of the lipids, while the hydrophilic glucose region prefers to stay in the water phase, and eventually the imine-N connecting the two units will face a negative charge environment produced by the anionic lipid headgroups of the LUVs. All these results suggest that both o-RHG and c-RHG interact with the Stern layer of the LUVs, and are useful to estimate the interface H+ concentration.
Red-shifts of 5–7 nm were observed in the wavelengths of absorption and fluorescence intensity maxima of RHG when it interacted with DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs at bulk pH 4.0–6.5 (Fig. 2 and S5†), presumably due to the decrease in dielectric constant at the interface compared to that of the bulk solution.15b,16c The absorption and fluorescence spectra of RHG (concentration: absorption, 2 μM; fluorescence, 1 μM) were measured in the presence of DOPC, DOPE, or DOPG LUVs (total lipid: absorption, 2 mM; fluorescence, 1 mM) and in the absence of LUVs. When RHG was attached to the DOPG LUVs, the maximum intensity wavelengths of absorption and fluorescence were observed at 539 and 560 nm, respectively, at pH 4.0–5.0, and they were also red-shifted 5–7 nm compared to the corresponding wavelengths in the absence of LUVs (Fig. S10†). However, the wavelengths and intensities of the absorption and fluorescence maxima in the RHG spectra did not change significantly when DOPC or DOPE LUV was used at pH 4.0–5.0, indicating that RHG interacts with DOPG LUV but not with DOPC or DOPE LUV.
1) LUVs at bulk pH 4.0–6.5 (Fig. 2 and S5†), presumably due to the decrease in dielectric constant at the interface compared to that of the bulk solution.15b,16c The absorption and fluorescence spectra of RHG (concentration: absorption, 2 μM; fluorescence, 1 μM) were measured in the presence of DOPC, DOPE, or DOPG LUVs (total lipid: absorption, 2 mM; fluorescence, 1 mM) and in the absence of LUVs. When RHG was attached to the DOPG LUVs, the maximum intensity wavelengths of absorption and fluorescence were observed at 539 and 560 nm, respectively, at pH 4.0–5.0, and they were also red-shifted 5–7 nm compared to the corresponding wavelengths in the absence of LUVs (Fig. S10†). However, the wavelengths and intensities of the absorption and fluorescence maxima in the RHG spectra did not change significantly when DOPC or DOPE LUV was used at pH 4.0–5.0, indicating that RHG interacts with DOPG LUV but not with DOPC or DOPE LUV.
There is a debate on the two pKa values of the phosphate groups of CL. It has been reported that the phosphate groups of CL have strongly disparate ionization behaviours (pK1 ∼ 2–4 and pK2 ∼ 7.5).27 In contrast, a recent study suggested that both of the phosphates ionize as strong acids with pKa values ranging between 1 and 1.5.28 RHG interacts with mono-anionic DOPG LUVs at bulk pHs 4.0–6.5 (Fig. 2 and S5†), but not with neutral DOPE LUVs at similar bulk pHs (4.0–5.0) (Fig. S10†), indicating that RHG may interact with CL even if it is monoionic under the acidic conditions used (pH 4.5). However, the concentration of RHG (1–2 μM) was considerably lower than that of the lipids (1–2 mM), and thus there was presumably no significant effect of RHG on the pKa of CL.
Estimation of H+ concentration at the LUV membrane interface
The absorbance and fluorescence intensity of RHG increased gradually upon decreasing the pH from 8.3 to 2.0 in the absence of lipids and in the presence of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) or DOPG LUV (RHG
1) or DOPG LUV (RHG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lipid = 1
lipid = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) under LUV-binding saturation conditions of RHG (Fig. S11†), indicating that c-RHG converted to o-RHG at low pH. The absorption and fluorescence intensity in the presence of DOPC/DOPE/TOCL (2
1000) under LUV-binding saturation conditions of RHG (Fig. S11†), indicating that c-RHG converted to o-RHG at low pH. The absorption and fluorescence intensity in the presence of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and DOPG LUVs saturated below pH ∼ 4.0 and ∼3.5, respectively, while they saturated below pH ∼ 2.0 in the absence of LUVs (Fig. S11†). However, the intensities of the saturated absorption and fluorescence spectra increased about 30% upon addition of DOPC/DOPE/TOCL (2
1) and DOPG LUVs saturated below pH ∼ 4.0 and ∼3.5, respectively, while they saturated below pH ∼ 2.0 in the absence of LUVs (Fig. S11†). However, the intensities of the saturated absorption and fluorescence spectra increased about 30% upon addition of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
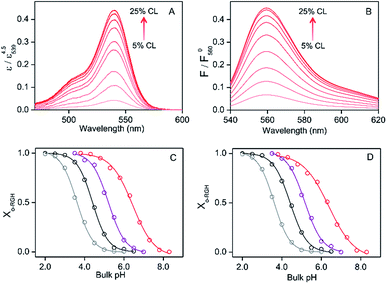
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs at pH 3.8 or DOPG LUVs at pH 3.5 (Fig. S11†), apparently due to changes in the UV-vis extinction coefficient and fluorescence quantum yield of o-RHG brought about by the change in polarity.16c The intensities of the absorption and fluorescence spectra were normalized by dividing the spectra by the maximum absorbance and fluorescence intensities, respectively, of the corresponding pH-saturated spectra, and the o-RHG/RHG ratios were calculated using the normalized spectra. The normalized absorption at 539 nm and fluorescence intensity at 560 nm of RHG increased gradually from 0.044 to 0.44 and 0.065 to 0.45, respectively, upon increasing TOCL from 5 to 25% in DOPC/DOPE/TOCL (DOPC
1) LUVs at pH 3.8 or DOPG LUVs at pH 3.5 (Fig. S11†), apparently due to changes in the UV-vis extinction coefficient and fluorescence quantum yield of o-RHG brought about by the change in polarity.16c The intensities of the absorption and fluorescence spectra were normalized by dividing the spectra by the maximum absorbance and fluorescence intensities, respectively, of the corresponding pH-saturated spectra, and the o-RHG/RHG ratios were calculated using the normalized spectra. The normalized absorption at 539 nm and fluorescence intensity at 560 nm of RHG increased gradually from 0.044 to 0.44 and 0.065 to 0.45, respectively, upon increasing TOCL from 5 to 25% in DOPC/DOPE/TOCL (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs at pH 6.5 (Fig. 2A and B), indicating the conversion of c-RHG to o-RHG due to the increase in acidity at the LUV interface.
1) LUVs at pH 6.5 (Fig. 2A and B), indicating the conversion of c-RHG to o-RHG due to the increase in acidity at the LUV interface.
The interface pH′ values of LUVs were estimated by measuring the difference in the o-RHG/RHG ratio at the interface and in the bulk (Fig. 2C and D). The apparent difference (Δ) between the pH′ at the LUV membrane interface and pH in the bulk medium is related to the difference in the o-RHG/c-RHG equilibrium at the interface and in the bulk. A similar value of Δ = ∼0.8 was obtained for DOPG LUVs by the absorption and fluorescence measurements under all bulk pH conditions measured (Fig. 2C and D, black and purple curves). For DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs, the value of Δ decreased from ∼2.1 to ∼1.5 and ∼1.2 according to absorption and fluorescence measurements, respectively, upon decreasing the bulk pH from 7.5 to 4.5 (Fig. 2C and D, black and red curves). However, the o-RHG/RHG ratio and thus Δ are affected by the polarity of the medium (Fig. S12†). The polarity contribution (δ) to Δ is estimated from the apparent pH shift caused by the polarity difference between the interface and the bulk. The LUV interface pH′
1) LUVs, the value of Δ decreased from ∼2.1 to ∼1.5 and ∼1.2 according to absorption and fluorescence measurements, respectively, upon decreasing the bulk pH from 7.5 to 4.5 (Fig. 2C and D, black and red curves). However, the o-RHG/RHG ratio and thus Δ are affected by the polarity of the medium (Fig. S12†). The polarity contribution (δ) to Δ is estimated from the apparent pH shift caused by the polarity difference between the interface and the bulk. The LUV interface pH′  is obtained from the bulk pH (pHbulk), Δ, and δ:16c
is obtained from the bulk pH (pHbulk), Δ, and δ:16c
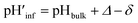 | (1) |
The interface dielectric constants of 45 and 44 were obtained for DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and DOPG LUVs, respectively, by utilizing an interface polarity detecting Schiff base molecule (2-((2-(pyridine-2-yl)ethylimino)methyl)-6-(hydroxymethyl)-4-methylphenol (PMP)) (Fig. S13†).29 The dielectric constant of the buffer solution was adjusted to 44–45 with addition of 58% (w/w) ethanol at pH 2.0–6.5,30 where the δ value was estimated to be ∼0.7 (Fig. 2C and D, gray and black curves). The pH′ values at DOPC/DOPE/TOCL (2
1) and DOPG LUVs, respectively, by utilizing an interface polarity detecting Schiff base molecule (2-((2-(pyridine-2-yl)ethylimino)methyl)-6-(hydroxymethyl)-4-methylphenol (PMP)) (Fig. S13†).29 The dielectric constant of the buffer solution was adjusted to 44–45 with addition of 58% (w/w) ethanol at pH 2.0–6.5,30 where the δ value was estimated to be ∼0.7 (Fig. 2C and D, gray and black curves). The pH′ values at DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and DOPG LUV interfaces under various bulk pHs were obtained from eqn (1) and are listed in Table 1. When the bulk pH was 7.0, the interface pH′ of the DOPC/DOPE/TOCL (2
1) and DOPG LUV interfaces under various bulk pHs were obtained from eqn (1) and are listed in Table 1. When the bulk pH was 7.0, the interface pH′ of the DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUV was ∼4.1, ∼2.9 units more acidic compared to the bulk pH. Considering these results, the interface pH′ of the IMM interface may be reduced to ∼3.9 when that of the mitochondrial intermembrane space is ∼6.8. However, the deviation between the interface pH′ and the bulk pH decreased gradually to ∼2.0 upon decreasing the bulk pH to ∼4.5 (Table 1), presumably due to protonation of one of the phosphate groups of TOCL. For the mono-anionic DOPG LUVs, a ∼1.5 unit decrease in interface pH′ from the bulk pH was detected under all bulk pH conditions investigated (pH 4.0–6.5) (Table 1).
1) LUV was ∼4.1, ∼2.9 units more acidic compared to the bulk pH. Considering these results, the interface pH′ of the IMM interface may be reduced to ∼3.9 when that of the mitochondrial intermembrane space is ∼6.8. However, the deviation between the interface pH′ and the bulk pH decreased gradually to ∼2.0 upon decreasing the bulk pH to ∼4.5 (Table 1), presumably due to protonation of one of the phosphate groups of TOCL. For the mono-anionic DOPG LUVs, a ∼1.5 unit decrease in interface pH′ from the bulk pH was detected under all bulk pH conditions investigated (pH 4.0–6.5) (Table 1).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and DOPG LUVs at various bulk pHsa
1) and DOPG LUVs at various bulk pHsa
| pH | ||||
|---|---|---|---|---|
DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
DOPG | |||
| Abs | FL | Abs | FL | |
| a Interface pH′ values were estimated from the absorption (Abs) and fluorescence (FL) spectra of RHG at 25 °C. | ||||
| 7.50 | 4.59 ± 0.08 | 4.60 ± 0.05 | — | — |
| 7.00 | 4.10 ± 0.05 | 4.13 ± 0.04 | — | — |
| 6.50 | 3.65 ± 0.03 | 3.75 ± 0.02 | 4.92 ± 0.06 | 4.86 ± 0.05 |
| 6.00 | 3.33 ± 0.03 | 3.41 ± 0.02 | 4.44 ± 0.04 | 4.41 ± 0.03 |
| 5.50 | 2.99 ± 0.03 | 3.09 ± 0.02 | 3.93 ± 0.02 | 3.95 ± 0.02 |
| 5.00 | 2.68 ± 0.04 | 2.81 ± 0.03 | 3.43 ± 0.02 | 3.44 ± 0.02 |
| 4.50 | 2.44 ± 0.07 | 2.49 ± 0.05 | 2.97 ± 0.04 | 2.93 ± 0.03 |
| 4.00 | — | — | 2.55 ± 0.07 | 2.39 ± 0.06 |
We isolated mitoplasts from horse heart muscle, and tried to estimate the interface pH′ with RHG at pH 6.8. However, the mitoplast solution exhibited very large absorbances (∼1.5) at the excitation (530 nm) and emission (560 nm) wavelengths of RHG even at one order lower lipid concentration (∼0.1 mM) necessary for fluorescence saturation (Fig. S14†), not allowing us to measure the RHG absorption and fluorescence intensity under LUV-binding saturation conditions (lipid concentrations > ∼1 mM). Thus, we made LUVs with lipids extracted from mitochondrial membranes, where the interface pH′ of the LUVs made with extracted mitochondrial lipids was ∼4.5 at bulk pH 6.8 (Fig. S15†).
A similar amount of c-RHG-to-o-RHG conversion was obtained between DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and DOPC/TOCL (3
1) and DOPC/TOCL (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs for a wide range of pH (4.0–8.0) (Fig. S16†), showing that DOPE does not influence RHG binding to LUVs at pH 4.0–8.0. We obtained the fluorescence intensity of RHG (1 μM) under fluorescence saturation conditions with high LUV concentrations for all the measurements (total lipid, 1 mM; Fig. S17†). The interface pH′ decreased as the TOCL% in DOPC/DOPE/TOCL LUVs was increased at pH 5.5–7.0 (Fig. S18A†). However, using DOPC/DOPE/TOCL LUVs with constant DOPC and DOPE concentrations but different TOCL concentrations ([DOPC] = 360; [DOPE] = 180 μM; TOCL 60–290 μM) under saturated RHG fluorescence intensity conditions at bulk pH 6.5, the plots of the interface pH′ against the TOCL% in LUVs were similar to those obtained with a constant total lipid concentration (1 mM) (Fig. S18B†). These results indicate that the increase in the negative charges in LUVs causes a decrease in the interface pH′.
1) LUVs for a wide range of pH (4.0–8.0) (Fig. S16†), showing that DOPE does not influence RHG binding to LUVs at pH 4.0–8.0. We obtained the fluorescence intensity of RHG (1 μM) under fluorescence saturation conditions with high LUV concentrations for all the measurements (total lipid, 1 mM; Fig. S17†). The interface pH′ decreased as the TOCL% in DOPC/DOPE/TOCL LUVs was increased at pH 5.5–7.0 (Fig. S18A†). However, using DOPC/DOPE/TOCL LUVs with constant DOPC and DOPE concentrations but different TOCL concentrations ([DOPC] = 360; [DOPE] = 180 μM; TOCL 60–290 μM) under saturated RHG fluorescence intensity conditions at bulk pH 6.5, the plots of the interface pH′ against the TOCL% in LUVs were similar to those obtained with a constant total lipid concentration (1 mM) (Fig. S18B†). These results indicate that the increase in the negative charges in LUVs causes a decrease in the interface pH′.
It has been reported that the pH′ values at the anionic interfaces of amphiphilic self-assemblies are lower than the bulk pH.16c The o-RHG/RHG ratios at the interfaces of DOPC/DOPE/TOCL (DOPC/DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
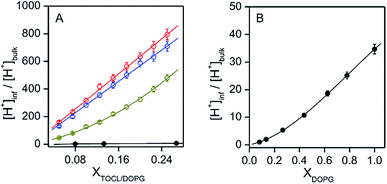
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; TOCL = 5–25%) and DOPC/DOPG (DOPG = 8–100%) LUVs increased for higher TOCL% and DOPG% (Fig. 2A and B, and S19†), strongly supporting the hypothesis that the negative charges of the anionic lipids at the interfaces are responsible for the decrease in pH′ at the interfaces compared to the bulk pH. [H+] values at the LUV interface, calculated from the interface pH′ (Fig. S18 and S19†), were higher than those in the bulk by ∼40- and ∼50-fold at pH 6.5 and 7.0, respectively, even for DOPC/DOPE/TOCL (DOPC
1; TOCL = 5–25%) and DOPC/DOPG (DOPG = 8–100%) LUVs increased for higher TOCL% and DOPG% (Fig. 2A and B, and S19†), strongly supporting the hypothesis that the negative charges of the anionic lipids at the interfaces are responsible for the decrease in pH′ at the interfaces compared to the bulk pH. [H+] values at the LUV interface, calculated from the interface pH′ (Fig. S18 and S19†), were higher than those in the bulk by ∼40- and ∼50-fold at pH 6.5 and 7.0, respectively, even for DOPC/DOPE/TOCL (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
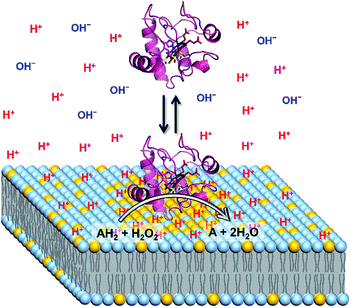
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs containing 5% TOCL (Fig. 3A). Upon increasing TOCL% from 5 to ∼25% in DOPC/DOPE/TOCL LUVs, the ratio of [H+] between the interface and the bulk increased linearly to ∼700 and ∼800 at bulk pH 6.5 and 7.0, respectively (Fig. 3A). For DOPC/DOPG LUVs containing 25% mono-anionic DOPG, the [H+] ratio between the interface and the bulk was ∼5 at bulk pH ∼ 5.0, and only a ∼35-fold increase was detected for DOPG LUVs at 100% DOPG (Fig. 3). For an anionic lipid membrane, the negatively charged headgroups of the lipids at the membrane interface may interact electrostatically with H+, whereas they repel OH−. [H+] and [OH−] may increase and decrease, respectively, at the interface compared to those in the bulk phase, while [H+] and [OH−] remain unchanged in the bulk (Fig. 4). However, a deviation (∼2.9) about twice as large between the interface pH′ and bulk pH was observed for DOPC/DOPE/TOCL (2
1) LUVs containing 5% TOCL (Fig. 3A). Upon increasing TOCL% from 5 to ∼25% in DOPC/DOPE/TOCL LUVs, the ratio of [H+] between the interface and the bulk increased linearly to ∼700 and ∼800 at bulk pH 6.5 and 7.0, respectively (Fig. 3A). For DOPC/DOPG LUVs containing 25% mono-anionic DOPG, the [H+] ratio between the interface and the bulk was ∼5 at bulk pH ∼ 5.0, and only a ∼35-fold increase was detected for DOPG LUVs at 100% DOPG (Fig. 3). For an anionic lipid membrane, the negatively charged headgroups of the lipids at the membrane interface may interact electrostatically with H+, whereas they repel OH−. [H+] and [OH−] may increase and decrease, respectively, at the interface compared to those in the bulk phase, while [H+] and [OH−] remain unchanged in the bulk (Fig. 4). However, a deviation (∼2.9) about twice as large between the interface pH′ and bulk pH was observed for DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUV compared to that (∼1.5) for DOPG LUV, although the DOPG ratio in DOPG LUV was four times higher than the TOCL ratio in DOPC/DOPE/TOCL LUV, showing that the local negative charges of the lipid headgroup affect the interface pH′ significantly (Fig. 4).
1) LUV compared to that (∼1.5) for DOPG LUV, although the DOPG ratio in DOPG LUV was four times higher than the TOCL ratio in DOPC/DOPE/TOCL LUV, showing that the local negative charges of the lipid headgroup affect the interface pH′ significantly (Fig. 4).
H+ concentration at the interface of the LUV membrane in the presence of cyt c
The effect of cyt c binding to TOCL on the interface pH′ was estimated at bulk pH 6.5 and 7.0 in the presence of oxidized horse cyt c (0.5–5.0) at a low TOCL concentration (10 μM) in DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
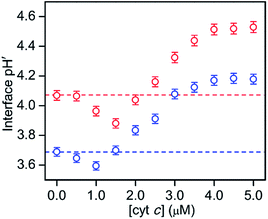
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs to avoid cyt c precipitation (Fig. 5 and S20†). A concentration of 0.05 μM was used for RHG to satisfy the fluorescence intensity saturation by LUV binding. The fluorescence intensity decreased a little, but less than 10%, for all the pH conditions studied (pH 2.5 to 5.3) (Fig. S21†). The fluorescence spectra of o-RHG in the presence of cyt c were calibrated taking into account of the quenching effect by cyt c (Fig. S20C and D†), and the interface pH′ at each concentration of cyt c was obtained from the calibrated spectra (Fig. 5). By increasing the cyt c concentration from 0 to 1.5 μM at pH 7.0, the interface pH′ decreased gradually from 4.07 to 3.86, followed by a gradual increase until it saturated at ∼4.51 at cyt c concentrations higher than 4.0 μM (Fig. 5). Although a similar trend was observed at pH 6.5, a smaller interface pH′ decrease from 3.69 to 3.59 was detected upon increasing the cyt c concentration from 0 to 1.0 μM (Fig. 5). Positively charged cyt c interacts electrostatically with negatively charged TOCL, forming TOCL-enriched lipid domains in LUVs (Fig. 4).31 When the concentration of TOCL is significantly higher than that of cyt c (TOCL concentration, 10 μM; cyt c concentration, <1.5 μM), the negative charge density at the TOCL-enriched domain may be higher compared to that of a homogeneous distribution domain, even though the negative charge density of the TOCL-enriched domain can be partially reduced by the positively charged cyt c. As a result, the increase in the negative charge density at the interface due to accumulation of TOCL may contribute to the decrease in the interface pH′ upon addition of 0.5 to 1.5 μM cyt c. At high cyt c concentrations (>1.5 μM), the effect of cyt c on the decrease in the negative charge density may supersede that by TOCL clustering, inducing an increase in the interface pH′ (Fig. 5). However, the interface pH′ saturated at ∼4.18 and ∼4.51 at bulk pH 6.5 and 7.0, respectively, upon addition of cyt c with concentrations higher than 4.0 μM, suggesting saturation of cyt c binding to LUVs.
1) LUVs to avoid cyt c precipitation (Fig. 5 and S20†). A concentration of 0.05 μM was used for RHG to satisfy the fluorescence intensity saturation by LUV binding. The fluorescence intensity decreased a little, but less than 10%, for all the pH conditions studied (pH 2.5 to 5.3) (Fig. S21†). The fluorescence spectra of o-RHG in the presence of cyt c were calibrated taking into account of the quenching effect by cyt c (Fig. S20C and D†), and the interface pH′ at each concentration of cyt c was obtained from the calibrated spectra (Fig. 5). By increasing the cyt c concentration from 0 to 1.5 μM at pH 7.0, the interface pH′ decreased gradually from 4.07 to 3.86, followed by a gradual increase until it saturated at ∼4.51 at cyt c concentrations higher than 4.0 μM (Fig. 5). Although a similar trend was observed at pH 6.5, a smaller interface pH′ decrease from 3.69 to 3.59 was detected upon increasing the cyt c concentration from 0 to 1.0 μM (Fig. 5). Positively charged cyt c interacts electrostatically with negatively charged TOCL, forming TOCL-enriched lipid domains in LUVs (Fig. 4).31 When the concentration of TOCL is significantly higher than that of cyt c (TOCL concentration, 10 μM; cyt c concentration, <1.5 μM), the negative charge density at the TOCL-enriched domain may be higher compared to that of a homogeneous distribution domain, even though the negative charge density of the TOCL-enriched domain can be partially reduced by the positively charged cyt c. As a result, the increase in the negative charge density at the interface due to accumulation of TOCL may contribute to the decrease in the interface pH′ upon addition of 0.5 to 1.5 μM cyt c. At high cyt c concentrations (>1.5 μM), the effect of cyt c on the decrease in the negative charge density may supersede that by TOCL clustering, inducing an increase in the interface pH′ (Fig. 5). However, the interface pH′ saturated at ∼4.18 and ∼4.51 at bulk pH 6.5 and 7.0, respectively, upon addition of cyt c with concentrations higher than 4.0 μM, suggesting saturation of cyt c binding to LUVs.
Effect of pH′ decrease at the cardiolipin-containing LUV interface on the peroxidase activity of cyt c
Recently, theoretical calculations have shown that the charge density 2–3 nm away from the interface of a 100 nm diameter nanoparticle does not alter significantly.32 Cyt c is localized at the IMM interface and its diameter is 2–3 nm. Thus, cyt c may experience a pH′ close to ∼3.9 at the IMM interface (Fig. 4). It has been reported by NMR studies that only a certain number of cyt c molecules may exist in the TOCL-bound state in the presence of TOCL-containing LUVs, while the other cyt c molecules exist in the free state, indicating an equilibrium for cyt c binding to the TOCL-containing LUVs.33 However, the H+ concentration may affect the stability and function of cyt c at the IMM. At pH below 3.0, cyt c exists in the molten globule and denatured states in the presence of high and low salt concentrations, respectively, according to circular dichroism (CD), differential scanning calorimetry (DSC), and small angle X-ray scattering measurements.34 The structures of various partially unfolded intermediate states of cyt c have been determined during alkaline pH-dependent unfolding using the fluorometric photon counting histogram model.35 We obtained an approximate folded-to-unfolded transition temperature (Tm) of cyt c from the DSC thermogram as in other papers (Fig. S22†),36 although cyt c aggregation was detected in the DSC thermogram at all pH values studied (pH 3.9–6.8) at high temperatures (Fig. S22 and S23†). Tm decreased from 81 °C to 71 °C upon lowering the pH from 6.8 to 3.9, whereas it did not change significantly at pH 5.3 (∼80 °C) (Fig. S22†), indicating that the stability of cyt c decreases when it is bound to the IMM interface. Upon interaction with mono-anion-containing lipid membranes, cyt c may not destabilize significantly, owing to the pH′ at the membrane interface not changing appreciably from the bulk pH†). However, the peroxidase activity of cyt c increases when the Met80–heme iron coordination is cleaved or perturbed significantly.37 A decrease in the 695 nm absorbance, which is related to the Met80–heme iron coordination bond, was observed when decreasing the pH from 6.8 to 3.9 in the presence of H2O2 and 2-methoxyphenol, indicating that the Met80–heme iron bond was perturbed under high H+ concentration environments, such as the IMM interface (Fig. S24†).The product formation rates for 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) is frequently used as a model substrate in the oxidation reaction of heme proteins;22d,38 thus the oxidation of ABTS (40 μM) by cyt c (5 μM) in the presence of H2O2 (0−4 mM) was monitored at 730 nm (ε730 ∼ 14 mM−1 cm−1).39 The steady-state rate (kobs) increased linearly from 0.09 to 0.65 s−1 upon increasing the H2O2 concentration from 0.5 to 4 mM at pH 6.8, indicating that ABTS oxidation followed a bimolecular kinetics (Fig. 6). We estimated the effect of interface pH′ on the cyt c peroxidase activity for DOPG and DOPC/DOPE/TOCL LUVs (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; CL, 10−50% of total lipid) containing various mol% of a different CL, TOCL or TMCL. The product formation rate of the cyt c peroxidase reaction increased in the presence of DOPC/DOPE/TOCL (2
1; CL, 10−50% of total lipid) containing various mol% of a different CL, TOCL or TMCL. The product formation rate of the cyt c peroxidase reaction increased in the presence of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs upon increasing the TOCL concentration up to 0.2 mM and did not change further up to ∼0.28 mM, followed by a gradual decrease, apparently due to the degradation of cyt c (Fig. S25†).38b Thus, we used a constant concentration (0.25 mM) for TOCL and TMCL, exhibiting a relatively high cyt c peroxidase activity, and measured the cyt c peroxidase activity in the presence of LUVs with different CL ratios. DOPG LUVs (total lipid, 0.5 mM) were used as a reference by adjusting the total negative charge to that of DOPC/DOPE/TOCL LUVs (DOPC
1) LUVs upon increasing the TOCL concentration up to 0.2 mM and did not change further up to ∼0.28 mM, followed by a gradual decrease, apparently due to the degradation of cyt c (Fig. S25†).38b Thus, we used a constant concentration (0.25 mM) for TOCL and TMCL, exhibiting a relatively high cyt c peroxidase activity, and measured the cyt c peroxidase activity in the presence of LUVs with different CL ratios. DOPG LUVs (total lipid, 0.5 mM) were used as a reference by adjusting the total negative charge to that of DOPC/DOPE/TOCL LUVs (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). A similar pH dependence of the interface pH′ was observed for LUVs containing TMCL (pH′ 4.25, 3.95, and 3.55 for LUVs containing 10, 25, and 50% TMCL, respectively) and those containing TOCL (pH′ ∼ 4.32, 3.90, and 3.61 for LUVs containing 10, 25, and 50% TOCL, respectively) (Fig. S26†).
1). A similar pH dependence of the interface pH′ was observed for LUVs containing TMCL (pH′ 4.25, 3.95, and 3.55 for LUVs containing 10, 25, and 50% TMCL, respectively) and those containing TOCL (pH′ ∼ 4.32, 3.90, and 3.61 for LUVs containing 10, 25, and 50% TOCL, respectively) (Fig. S26†).
Similar to the Kitz–Wilson double-reciprocal plots,40 peroxidase activity constants, kcat and Km, were obtained from the intercept (1/kcat) and slope (Km/kcat) of the plots of the inverse of product formation rate (1/kobs) against inverse of H2O2 concentration, according to the following equation.
| 1/kobs = 1/kcat + (Km/kcat) × 1/[H2O2], | (2) |
In the presence of DOPC/DOPE/TMCL LUVs (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
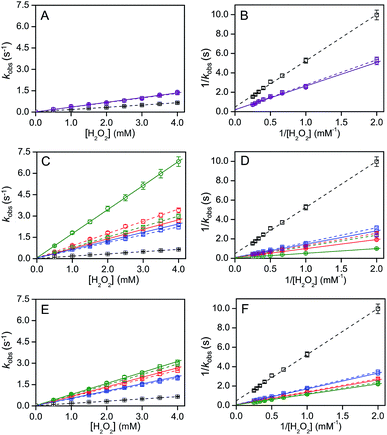
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the cyt c peroxidase activity increased 5 to 7 fold upon addition of LUVs depending on the TMCL% in the LUVs (Fig. 6 and Table 2). However, only a ∼2 fold increase in peroxidase activity was detected in the presence of DOPG LUVs (Fig. 6A and B). To evaluate the effect of pH decrease at the LUV interface on the peroxidase activity, we measured the peroxidase activity in the absence of LUVs at bulk pH identical to the interface pH′. Interestingly, the kinetic parameters (kcat and Km) at pH 6.8 in the presence of DOPC/DOPE/TOCL (DOPC
1), the cyt c peroxidase activity increased 5 to 7 fold upon addition of LUVs depending on the TMCL% in the LUVs (Fig. 6 and Table 2). However, only a ∼2 fold increase in peroxidase activity was detected in the presence of DOPG LUVs (Fig. 6A and B). To evaluate the effect of pH decrease at the LUV interface on the peroxidase activity, we measured the peroxidase activity in the absence of LUVs at bulk pH identical to the interface pH′. Interestingly, the kinetic parameters (kcat and Km) at pH 6.8 in the presence of DOPC/DOPE/TOCL (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, TOCL = 10−25%), DOPC/DOPE/TMCL (DOPC
1, TOCL = 10−25%), DOPC/DOPE/TMCL (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, TMCL = 10–50%), and DOPG LUVs were similar to the corresponding values obtained in the absence of LUVs at pH values corresponding to the interface pH′ (Fig. 6 and Table 2) (Fig. 6C–F and Table 2). However, the kcat in the presence of LUVs containing relatively high TOCL% (>50%) was ∼2-fold higher than that estimated by the pH decrease (Fig. 6C and D, and Table 2), indicating that other factors than the interface pH′ affect the peroxidase activity of cyt c. Related to this, protein modification of cyt c may also affect its peroxidase activity.41
1, TMCL = 10–50%), and DOPG LUVs were similar to the corresponding values obtained in the absence of LUVs at pH values corresponding to the interface pH′ (Fig. 6 and Table 2) (Fig. 6C–F and Table 2). However, the kcat in the presence of LUVs containing relatively high TOCL% (>50%) was ∼2-fold higher than that estimated by the pH decrease (Fig. 6C and D, and Table 2), indicating that other factors than the interface pH′ affect the peroxidase activity of cyt c. Related to this, protein modification of cyt c may also affect its peroxidase activity.41
| LUV | Bulk pH | Interface pH′ | k cat (s−1) | K m (μM) |
|---|---|---|---|---|
a DOPC/DOPE/TOCL LUVs (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2 DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, TOCL = 10, 25, and 50% of total lipids).
b DOPC/DOPE/TMCL LUVs (DOPC 1, TOCL = 10, 25, and 50% of total lipids).
b DOPC/DOPE/TMCL LUVs (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2 DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, TMCL = 10, 25, and 50% of total lipids). 1, TMCL = 10, 25, and 50% of total lipids).
|
||||
| None | 6.80 | 2.0 ± 0.1 | 10.0 ± 0.4 | |
| DOPG | 6.80 | 5.30 | 4.7 ± 0.2 | 11.8 ± 0.5 |
| None | 5.30 | 5.5 ± 0.2 | 12.3 ± 0.5 | |
| 10% TOCLa | 6.80 | 4.32 | 11.3 ± 0.4 | 12.7 ± 0.5 |
| None | 4.32 | — | 11.2 ± 0.4 | 14.0 ± 0.6 |
| 25% TOCLa | 6.80 | 3.90 | 15.5 ± 0.5 | 13.5 ± 0.5 |
| None | 3.90 | — | 14.4 ± 0.5 | 13.2 ± 0.6 |
| 50% TOCLa | 6.80 | 3.61 | 31.9 ± 1.0 | 14.0 ± 0.5 |
| None | 3.61 | 15.9 ± 0.5 | 14.1 ± 0.5 | |
| 10% TMCLb | 6.80 | 4.25 | 10.3 ± 0.4 | 12.1 ± 0.5 |
| None | 4.25 | 11.1 ± 0.4 | 11.9 ± 0.5 | |
| 25% TMCLb | 6.80 | 3.95 | 13.7 ± 0.5 | 13.5 ± 0.5 |
| None | 3.95 | 12.9 ± 0.5 | 13.3 ± 0.5 | |
| 50% TMCLb | 6.80 | 3.55 | 15.7 ± 0.5 | 13.9 ± 0.6 |
| None | 3.65 | 15.5 ± 0.5 | 13.7 ± 0.5 | |
It has been reported that the peroxidase activity of cyt c increases dramatically upon interaction with TOCL, due to the opening of the protein upon breaking of the Met80–heme iron bond and increase in ligand accessibility to the heme.22a–d Additionally, docking studies of cyt c with TOCL have shown that C11 of CL can bind to cyt c at a position adjacent to the heme.22f Full binding of cyt c to a membrane requires a cyt c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TOCL threshold ratio of 1
TOCL threshold ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 for cyt c to gain peroxidase activity.42 The structure of cyt c is perturbed significantly when it interacts strongly with TOCL-containing membranes.22e There are three possible TOCL binding sites of cyt c containing positive amino acid residues (Lys, His and Asp), and the heme crevice is opened to substrates by the simultaneous binding of two sites, at opposing sides to the heme, to the membrane.42 A ∼50-fold increase in the cyt c peroxidase activity has been reported for the reaction of cyt c (40 μM) with H2O2 (100 μM) and etoposide (700 μM) upon addition of DOPC/TOCL (1
5 for cyt c to gain peroxidase activity.42 The structure of cyt c is perturbed significantly when it interacts strongly with TOCL-containing membranes.22e There are three possible TOCL binding sites of cyt c containing positive amino acid residues (Lys, His and Asp), and the heme crevice is opened to substrates by the simultaneous binding of two sites, at opposing sides to the heme, to the membrane.42 A ∼50-fold increase in the cyt c peroxidase activity has been reported for the reaction of cyt c (40 μM) with H2O2 (100 μM) and etoposide (700 μM) upon addition of DOPC/TOCL (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs (total lipid, 400 μM), due to a change in the protein structure.22b,43 On the other hand, it has been reported that the peroxidase activity increases ∼15-fold for the reaction of cyt c (1 μM) with H2O2 (100 μM) upon addition of DOPC/TOCL (1
1) LUVs (total lipid, 400 μM), due to a change in the protein structure.22b,43 On the other hand, it has been reported that the peroxidase activity increases ∼15-fold for the reaction of cyt c (1 μM) with H2O2 (100 μM) upon addition of DOPC/TOCL (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs (total lipid, 250 μM).44 Although the peroxidase activity of cyt c at the membrane interface of TMCL-enriched LUVs (50% of total lipid) was similar to that estimated from the interface pH′, the cyt c peroxidase activity was higher than that estimated from the interface pH′ for TOCL-enriched LUVs (Fig. 6 and Table 2), supporting the hypothesis that cyt c opens the heme crevice to substrates when interacting with TOCL. We conclude that the peroxidase activity of cyt c increases due to both the pH decrease at the interface and the cyt c structural perturbation caused by the interaction with TOCL.
1) LUVs (total lipid, 250 μM).44 Although the peroxidase activity of cyt c at the membrane interface of TMCL-enriched LUVs (50% of total lipid) was similar to that estimated from the interface pH′, the cyt c peroxidase activity was higher than that estimated from the interface pH′ for TOCL-enriched LUVs (Fig. 6 and Table 2), supporting the hypothesis that cyt c opens the heme crevice to substrates when interacting with TOCL. We conclude that the peroxidase activity of cyt c increases due to both the pH decrease at the interface and the cyt c structural perturbation caused by the interaction with TOCL.
Conclusions
We demonstrate that RHG, an interface H+ concentration-sensing probe, can be used to measure the proton concentration (−log[H+], defined as pH′) at lipid membrane interfaces by monitoring the change in its acid/base equilibrium between the Stern layer and the bulk. The interface pH′ for CL-enriched membrane models of the IMM is evaluated to be ∼3.9, while the mitochondrial intermembrane space pH is ∼6.8. The large decrease in pH at the interfaces of the IMM model membranes compared to the bulk pH may enhance the peroxidase activity of cyt c by a factor of 5–7 fold. However, the peroxidase activity of cyt c increased not only because of the decrease in the interface pH′ but also due to the structural perturbation of cyt c when interacting with TOCL, whereas there was no additional increase in the peroxidase activity from that estimated from the interface pH′ when interacting with TMCL. Considering these results, we added information on interface pH′ to the peroxidase activity of cyt c at negatively charged membranes. These results also show that the inherent simplicity of our method for H+ concentration detection can be widely applied to various biological membrane interfaces.Experimental
Materials and general procedures
DOPC, DOPG, and DOPE were purchased from NOF Co. (Tokyo, Japan). TOCL and TMCL were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Horse cyt c, ABTS, and 2-methoxyphenol were purchased from Sigma-Aldrich (St. Louis, MO, USA). Cyt c was purified by gel filtration chromatography (Superdex 75, GE Healthcare, Sweden) before performing spectroscopic measurements. Different buffer compositions were used to obtain specific medium pH: pH 1.5–5.0, sodium citrate–phosphate buffer (a mixture of sodium phosphate and sodium citrate solutions); pH 5.0–6.0, sodium cacodylate buffer; pH 6.0–8.3, HEPES buffer. The pH value was adjusted by addition of 1.0 M NaOH or 1.0 M HCl, if necessary.Syntheses of a spiro-rhodamine-glucose molecule and a Schiff-base molecule
A spiro-rhodamine-glucose molecule (RHG)45 and a Schiff-base molecule (PMP)46 were prepared according to earlier procedures. Briefly, spiro-rhodamine 6G hydrazide was prepared by the reaction of rhodamine 6G hydrochloride with hydrazine in ethanol. The condensation reaction between rhodamine 6G hydrazide and glucose was performed in the presence of p-toluene sulfonic acid. The product was purified by silica gel column chromatography followed by rotary evaporation. Stock aqueous RHG solutions were used for all absorption and fluorescence experiments. The purity of RHG was confirmed by the 1H NMR spectrum in DMSO-d6 (Fig. S27†). PMP was synthesized by the condensation reaction between 2-hydroxy-3-(hydroxymethyl)-5-methylbenzaldehyde and 2-(2-aminoethyl)-pyridine.Purification of mitochondria and mitochondrial lipids
Horse heart meat chunks (200 g) were placed in 0.6 L of 10 mM sodium phosphate buffer, pH 8.0, containing 250 mM sucrose and 0.1 mM EDTA, and blended with a food processor (TK430, TESCOM) for 2 min. The blended sample was centrifuged at 900g for 10 min, and the supernatant was filtered with 2 layers of gauze to remove the fat. After adjusting the pH of the solution to 7.4 with 3 M NaOH, the solution was centrifuged at 7000g for 10 min. The precipitate was suspended in 25 mL of 10 mM sodium phosphate buffer, pH 7.4, containing 150 mM NaCl. The suspended solution (25 mL) was homogenized twice with a tissue grinder (WHEATON) and centrifuged at 7600g for 10 min. The supernatant was removed with a pipette, and the brown precipitate was suspended in 25 mL of 10 mM Tris–HCl buffer, pH 7.4, containing 70 mM sucrose and 210 mM mannitol. The suspended solution was homogenized twice with the tissue grinder and centrifuged at 7600g for 10 min. The supernatant was removed with a pipette, and the brown precipitate was suspended in 4 mL of 5 mM sodium phosphate buffer, pH 7.4, containing 10 mM KCl, 2 mM MgCl2, 70 mM sucrose, 0.2 mM EDTA, and 210 mM mannitol, obtaining mitochondria. To extract lipids from mitochondria, a mixture of chloroform (20 mL), methanol (40 mL), and pure water (12 mL) was added to the mitochondrial solution, and mixed for 2 min. Additional chloroform (20 mL) was added to the solution mixture, and the solution was mixed for 30 s. Subsequently, pure water (20 mL) was added to the sample solution, and the solution was mixed for another 30 s (the final proportion of chloroform, methanol, and water was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.8).47 The sample was filtered through filter paper on a Buchner funnel and transferred to a separatory funnel. After allowing a few minutes for complete separation and clarification of the solutions, the chloroform layer containing the mitochondrial lipids was collected.
1.8).47 The sample was filtered through filter paper on a Buchner funnel and transferred to a separatory funnel. After allowing a few minutes for complete separation and clarification of the solutions, the chloroform layer containing the mitochondrial lipids was collected.
Preparation of LUVs
Lipid components of DOPG, DOPC, DOPE, DOPC/DOPG (DOPG, 8–95%), DOPC/CL (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), DOPC/DOPE/CL (DOPC
1), DOPC/DOPE/CL (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; CL (TOCL/TMCL), 5–50% of total lipids), or lipids separated from mitochondria were dissolved in chloroform in a flask. The chloroform solvent was removed with a rotary evaporator at 40 °C, forming a thin lipid film on the wall of the flask. Residual chloroform in the thin lipid film was completely removed by drying in vacuo for 3 h. The lipids were hydrated by addition of a buffer with desired pH (2.0–8.3) at 40 °C. The lipid solution was mixed with a vortex mixer for ∼2 min for complete dissolution of the lipids. Seven cycles of freeze-and-thaw were performed at −196 and 50 °C to obtain multilamellar vesicles (MLVs). MLVs were extruded 15 times through two stacked polycarbonate membrane filters (pore size, 100 nm) equipped in a Liposo Fast mini extruder (Avestin, USA) to adjust the diameter of LUVs to 100 nm.
1; CL (TOCL/TMCL), 5–50% of total lipids), or lipids separated from mitochondria were dissolved in chloroform in a flask. The chloroform solvent was removed with a rotary evaporator at 40 °C, forming a thin lipid film on the wall of the flask. Residual chloroform in the thin lipid film was completely removed by drying in vacuo for 3 h. The lipids were hydrated by addition of a buffer with desired pH (2.0–8.3) at 40 °C. The lipid solution was mixed with a vortex mixer for ∼2 min for complete dissolution of the lipids. Seven cycles of freeze-and-thaw were performed at −196 and 50 °C to obtain multilamellar vesicles (MLVs). MLVs were extruded 15 times through two stacked polycarbonate membrane filters (pore size, 100 nm) equipped in a Liposo Fast mini extruder (Avestin, USA) to adjust the diameter of LUVs to 100 nm.
Formation and microscopic observation of GUVs
A DOPC/DOPG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) or DOPC/DOPE/TOCL (2
2) or DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixture was dissolved in chloroform in a cylindrical container. A thin lipid film was prepared on the wall of the container as descried above. The lipid film was hydrated with 1 mM HEPES buffer, pH 6.5, containing 200 mM sucrose by placing the buffer gently on the lipid film at 30 °C and incubating overnight, resulting in the formation of GUVs (≥1 μm in diameter).
1) mixture was dissolved in chloroform in a cylindrical container. A thin lipid film was prepared on the wall of the container as descried above. The lipid film was hydrated with 1 mM HEPES buffer, pH 6.5, containing 200 mM sucrose by placing the buffer gently on the lipid film at 30 °C and incubating overnight, resulting in the formation of GUVs (≥1 μm in diameter).
RHG (0.3 μM) was added to GUVs (total lipid, 500 μM) in 1 mM HEPES buffer, pH 6.5. The solution containing RHG and GUVs was incubated for at least 30 min to obtain uniform fluorescence intensities among different GUV surfaces. GUVs were imaged at room temperature using an Olympus IX 71 microscope (Olympus, Center Valley, PA, USA). An Olympus 60×/1.4 NA Plan Apo oil immersion lens was used as an objective lens. Excitation light was obtained using an Hg lamp with a U-MWIY2 filter set (Olympus; excitation wavelength, 545–580 nm). Microscopic images were recorded using an Orca-Flash2.8 Scientific CMOS Camera (Hamamatsu, Japan).
1H-NMR measurements
1H-NMR spectra were measured in DMSO-d6 and D2O in the presence and absence of DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) or DOPG LUVs (total lipids, 15 mM) with a 300 MHz or 500 MHz NMR spectrophotometer (Bruker, USA) using tetramethylsilane (δ = 0) as a standard. The pD of the D2O medium in the presence and absence of LUVs was adjusted to 4.5–6.5 by addition of 0.01 M CF3COOH.
1) or DOPG LUVs (total lipids, 15 mM) with a 300 MHz or 500 MHz NMR spectrophotometer (Bruker, USA) using tetramethylsilane (δ = 0) as a standard. The pD of the D2O medium in the presence and absence of LUVs was adjusted to 4.5–6.5 by addition of 0.01 M CF3COOH.
Absorption and fluorescence measurements
The UV-vis optical absorption and fluorescence measurements were performed with a UV-2450 spectrophotometer (Shimadzu, Japan). Fluorescence measurements were performed with a PerkinElmer LS-55 spectrofluorimeter (PerkinElmer, USA) or a Fluoromax-4 spectrophotometer (HORIBA, USA). RHG (0.05–2.0 μM) was mixed with LUVs (total lipids, 0–2.0 mM), and the UV-vis absorption spectra (RHG, 2 μM) and fluorescence spectra (RHG, 0.05–1.0 μM) were measured at 25 °C in the presence and absence of cyt c (0.5–5.0 μM) with and without DOPC/DOPE/CL LUVs (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; CL (TOCL/TMCL), 5–50%). The spectra were also measured for DOPC/TOCL (3
1; CL (TOCL/TMCL), 5–50%). The spectra were also measured for DOPC/TOCL (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs and LUVs of mitochondrial lipids at pH 3.0–8.3, DOPC and DOPE LUVs at pH 3.5–5.0, DOPC/DOPG and DOPG LUVs (DOPG, 8–100%) at pH 2.0–7.0, and buffers containing ethanol (35–58% (w/w)) at pH 1.5–6.0. The fluorescence spectrum of RHG was corrected by subtracting the background intensity. The UV-vis absorption/fluorescence spectra were normalized by dividing them by the intensities of the corresponding spectra at 532/554 nm in the buffer medium and 539/560 nm in the buffer containing LUVs or ethanol, where the intensities were saturated by lowering the pH. The saturated spectra used for the calculations were measured at pH 3.0 for DOPC/DOPE/CL LUVs (DOPC
1) LUVs and LUVs of mitochondrial lipids at pH 3.0–8.3, DOPC and DOPE LUVs at pH 3.5–5.0, DOPC/DOPG and DOPG LUVs (DOPG, 8–100%) at pH 2.0–7.0, and buffers containing ethanol (35–58% (w/w)) at pH 1.5–6.0. The fluorescence spectrum of RHG was corrected by subtracting the background intensity. The UV-vis absorption/fluorescence spectra were normalized by dividing them by the intensities of the corresponding spectra at 532/554 nm in the buffer medium and 539/560 nm in the buffer containing LUVs or ethanol, where the intensities were saturated by lowering the pH. The saturated spectra used for the calculations were measured at pH 3.0 for DOPC/DOPE/CL LUVs (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE = 2
DOPE = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; CL (TOCL/TMCL), 5–50%) and LUVs of mitochondrial lipids, pH 2.0 for DOPC, DOPE, DOPG and DOPC/DOPG LUVs, and pH 1.5 for buffers containing ethanol. The plots of normalized intensities vs. bulk pH were fitted with sigmoidal-Boltzmann equations. The bulk and interface H+ concentrations were calculated from the bulk pH and interface pH′ values, respectively.
1; CL (TOCL/TMCL), 5–50%) and LUVs of mitochondrial lipids, pH 2.0 for DOPC, DOPE, DOPG and DOPC/DOPG LUVs, and pH 1.5 for buffers containing ethanol. The plots of normalized intensities vs. bulk pH were fitted with sigmoidal-Boltzmann equations. The bulk and interface H+ concentrations were calculated from the bulk pH and interface pH′ values, respectively.
The UV-vis absorption spectra of PMP were measured in the presence and absence of DOPC/DOPE/CL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), DOPC/DOPE (2
1), DOPC/DOPE (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), DOPC, or DOPG LUVs (total lipid, 3 mM) and LUVs of lipids from the mitochondrial membranes in 10 mM HEPES buffer, pH 6.5. The dielectric constant (D) at the LUV interface was estimated using the following relation as reported previously.29
1), DOPC, or DOPG LUVs (total lipid, 3 mM) and LUVs of lipids from the mitochondrial membranes in 10 mM HEPES buffer, pH 6.5. The dielectric constant (D) at the LUV interface was estimated using the following relation as reported previously.29
| ε420D/ε4208.0 = 0.42 × D − 1.8 | (3) |
The extinction coefficient at 420 nm for the buffer containing LUVs (ε420D) was divided by that for THF (ε4208.0), where D of THF is 8.0.
The UV-vis absorption spectra of cyt c (10–20 μM) in the 600–800 nm region were measured in the presence of H2O2 and 2-methoxyphenol (5 μM) at pH 3.9, 5.3, and 6.8 in citrate–phosphate buffer (a mixture of 10 mM sodium phosphate and 10 mM sodium citrate solutions), 10 mM cacodylate buffer, and 10 mM HEPES buffer, respectively, at 25 °C.
Determination of the fluorescence quantum yield
The fluorescence quantum yields of RHG (1 μM) in the presence of DOPC/DOPE/TOCL LUVs (total lipid, 1 mM) containing various TOCL% (5–25% of total lipids) were determined by adapting the procedure described previously.48 In brief, 9,10-diphenylanthracene in ethanol was used as the reference fluorophore with fluorescence quantum yield (ϕr) = 0.95. The fluorescence quantum yield of RHG (ϕs) in the presence of LUVs was measured by using the following equation:| ϕs = (ArFsns/AsFrnr) × ϕs | (4) |
Binding assay of RHG to LUVs
After mixing RHG (2.0 μM) with DOPC/DOPE/TOCL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUVs (total lipid, 2 mM) at pH 4.5–8.0, unbound RHG molecules were collected with a centrifugal filter (Amicon Ultra, Millipore; cut-off, 100k MW). The amount of RHG in the filtrate was calculated from the concentration of unbound RHG estimated by measuring the UV-vis absorption spectrum after adjusting the pH to 2.0.
1) LUVs (total lipid, 2 mM) at pH 4.5–8.0, unbound RHG molecules were collected with a centrifugal filter (Amicon Ultra, Millipore; cut-off, 100k MW). The amount of RHG in the filtrate was calculated from the concentration of unbound RHG estimated by measuring the UV-vis absorption spectrum after adjusting the pH to 2.0.
DSC measurements
DSC thermograms of oxidized horse cyt c (100 μM) at pH 3.9, 5.3, and 6.8 in citrate–phosphate buffer (a mixture of 10 mM sodium phosphate and 10 mM sodium citrate solutions), 10 mM cacodylate buffer, and 10 mM HEPES buffer, respectively, were measured with a VP-DSC calorimeter (MicroCal, GE Healthcare) at a scan rate of 1 °C min−1.Peroxidase activity measurements
The catalytic steady-state kinetics of ABTS oxidation was investigated with a UV-2450 spectrophotometer (Shimadzu) at 25 °C. The oxidation of ABTS (40 μM) with H2O2 (0–4 mM) was catalyzed by oxidized horse cyt c (5 μM) at pH 3.6–4.3, 5.3, and 6.8 in citrate–phosphate buffer (a mixture of 10 mM sodium phosphate and 10 mM sodium citrate solutions), 10 mM cacodylate buffer, and 10 mM HEPES buffer, respectively. The steady-state reaction rates were obtained by monitoring the absorbance at 730 nm using a molar absorption coefficient of 14 mM−1 cm−1 for ABTS oxidation.39 The reaction rate was determined from the initial reaction. Each experiment was repeated at least three times.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We thank Mr Leigh McDowell, NAIST, for his advice on manuscript preparation. This work was partially supported by Grants-in-Aid from JSPS for Scientific Research (Category B, no. JP18H02088 (SH) and Innovative Areas, no. JP16H00839 (SH)). PPP acknowledges UGC and the Government of West Bengal for financial support under the RUSA 2.0 scheme (no. 5400-F(Y)), and DST for a Special Grant to the Department of Chemistry, JU in the International Year of Chemistry 2011.Notes and references
-
(a) O. S. Andersen and R. E. Koeppe, Annu. Rev. Biophys. Biomol. Struct., 2007, 36, 107–130 CrossRef CAS PubMed
; (b) A. Finkelstein, J. Gen. Physiol., 1976, 68, 127–135 CrossRef CAS
.
- J. Kim, J. H. Jeon, H. J. Kim, H. Lim and I. K. Oh, ACS Nano, 2014, 8, 2986–2997 CrossRef CAS
.
- R. T. Uren, G. Dewson, C. Bonzon, T. Lithgow, D. D. Newmeyer and R. M. Kluck, J. Biol. Chem., 2005, 280, 2266–2274 CrossRef CAS
.
-
(a) F. Ramirez and D. B. Rifkin, Matrix Biol., 2003, 22, 101–107 CrossRef CAS
; (b) H. K. Kim, H. Wei, A. Kulkarni, R. M. Pogranichniy and D. H. Thompson, Biomacromolecules, 2012, 13, 636–644 CrossRef CAS PubMed
.
-
L. J. Earp, S. E. Delos, H. E. Park and J. M. White, in Membrane Trafficking in Viral Replication, ed. M. Marsh, Springer-Verlag, Heidelberg, 2005, vol. 285, pp. 25–66 Search PubMed
.
- J. Santo-Domingo and N. Demaurex, J. Gen. Physiol., 2012, 139, 415–423 CrossRef CAS PubMed
.
- D. Poburko, J. Santo-Domingo and N. Demaurex, J. Biol. Chem., 2011, 286, 11672–11684 CrossRef CAS PubMed
.
- G. Azarias, H. Perreten, S. Lengacher, D. Poburko, N. Demaurex, P. J. Magistretti and J. Y. Chatton, J. Neurosci., 2011, 31, 3550–3559 CrossRef CAS
.
- R. Orij, J. Postmus, A. Ter Beek, S. Brul and G. J. Smits, Microbiology, 2009, 155, 268–278 CrossRef CAS
.
- B. Rieger, W. Junge and K. B. Busch, Nat. Commun., 2014, 5, 3103 CrossRef PubMed
.
-
(a) E. London and D. A. Brown, Biochim. Biophys. Acta, 2000, 1508, 182–195 CrossRef CAS PubMed
; (b) K. Kuroda and G. A. Caputo, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 49–66 CAS
.
-
(a) A. Küchler, M. Yoshimoto, S. Luginbühl, F. Mavelli and P. Walde, Nat. Nanotechnol., 2016, 11, 409–420 CrossRef
; (b) T. Ravula, C. Barnaba, M. Mahajan, G. M. Anantharamaiah, S. C. Im, L. Waskell and A. Ramamoorthy, Chem. Commun., 2017, 53, 12798–12801 RSC
.
-
(a) C. Rottman and D. Avnir, J. Am. Chem. Soc., 2001, 123, 5730–5734 CrossRef CAS
; (b) H. Chakraborty, R. Banerjee and M. Sarkar, Biophys. Chem., 2003, 104, 315–325 CrossRef CAS
; (c) A. Chakrabarty, A. Mallick, B. Haldar, P. Purkayastha, P. Das and N. Chattopadhyay, Langmuir, 2007, 23, 4842–4848 CrossRef CAS PubMed
.
-
(a) S. Yamaguchi, K. Bhattacharyya and T. Tahara, J. Phys. Chem. C, 2011, 115, 4168–4173 CrossRef CAS
; (b) A. Kundu, S. Yamaguchi and T. Tahara, J. Phys. Chem. Lett., 2014, 5, 762–766 CrossRef CAS PubMed
.
-
(a) A. R. Gear, J. Biol. Chem., 1974, 249, 3628–3637 CAS
; (b) R. C. Scaduto and L. W. Grotyohann, Biophys. J., 1999, 76, 469–477 CrossRef CAS PubMed
; (c) M. Beija, C. A. M. Afonso and J. M. G. Martinho, Chem. Soc. Rev., 2009, 38, 2410–2433 RSC
.
-
(a) Y. Sarkar, S. Das, A. Ray, S. K. Jewrajka, S. Hirota and P. P. Parui, Analyst, 2016, 141, 2030–2039 RSC
; (b) R. Majumder, Y. Sarkar, S. Das, A. Ray and P. P. Parui, New J. Chem., 2017, 41, 8536–8545 RSC
; (c) Y. Sarkar, R. Majumder, S. Das, A. Ray and P. P. Parui, Langmuir, 2018, 34, 6271–6284 CrossRef CAS PubMed
.
- E. Kalanxhi and C. J. Wallace, Biochem. J., 2007, 407, 179–187 CrossRef CAS PubMed
.
-
(a) M. Ott, J. D. Robertson, V. Gogvadze, B. Zhivotovsky and S. Orrenius, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 1259–1263 CrossRef CAS PubMed
; (b) M. Huttemann, P. Pecina, M. Rainbolt, T. H. Sanderson, V. E. Kagan, L. Samavati, J. W. Doan and I. Lee, Mitochondrion, 2011, 11, 369–381 CrossRef PubMed
.
-
(a) X. Liu, C. N. Kim, J. Yang, R. Jemmerson and X. Wang, Cell, 1996, 86, 147–157 CrossRef CAS PubMed
; (b) X. Saelens, N. Festjens, L. Vande Walle, M. van Gurp, G. van Loo and P. Vandenabeele, Oncogene, 2004, 23, 2861–2874 CrossRef CAS PubMed
.
- P. R. Magalhaes, A. S. Oliveira, S. R. Campos, C. M. Soares and A. M. Baptista, J. Chem. Inf. Model., 2017, 57, 256–266 CrossRef CAS PubMed
.
- A. Ranieri, F. Bernini, C. A. Bortolotti, A. Bonifacio, V. Sergo and E. Castellini, Langmuir, 2011, 27, 10683–10690 CrossRef CAS PubMed
.
-
(a) Y. A. Vladimirov, E. V. Proskurnina, D. Y. Izmailov, A. A. Novikov, A. V. Brusnichkin, A. N. Osipov and V. E. Kagan, Biochemistry (Moscow)., 2006, 71, 998–1005 CrossRef CAS PubMed
; (b) N. A. Belikova, Y. A. Vladimirov, A. N. Osipov, A. A. Kapralov, V. A. Tyurin, M. V. Potapovich, L. V. Basova, J. Peterson, I. V. Kurnikov and V. E. Kagan, Biochemistry, 2006, 45, 4998–5009 CrossRef CAS PubMed
; (c) S. Hirota, Y. Hattori, S. Nagao, M. Taketa, H. Komori, H. Kamikubo, Z. Wang, I. Takahashi, S. Negi, Y. Sugiura, M. Kataoka and Y. Higuchi, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 12854–12859 CrossRef CAS PubMed
; (d) Z. Wang, T. Matsuo, S. Nagao and S. Hirota, Org. Biomol. Chem., 2011, 9, 4766–4769 RSC
; (e) J. Hanske, J. R. Toffey, A. M. Morenz, A. J. Bonilla, K. H. Schiavoni and E. V. Pletneva, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 125–130 CrossRef CAS PubMed
; (f) L. J. McClelland, H. B. Steele, F. G. Whitby, T. C. Mou, D. Holley, J. B. Ross, S. R. Sprang and B. E. Bowler, J. Am. Chem. Soc., 2016, 138, 16770–16778 CrossRef CAS PubMed
.
-
(a) A. M. Porcelli, A. Ghelli, C. Zanna, P. Pinton, R. Rizzuto and M. Rugolo, Biochem. Biophys. Res. Commun., 2005, 326, 799–804 CrossRef CAS PubMed
; (b) J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50–61 CrossRef CAS PubMed
.
- J. J. Krebs, H. Hauser and E. Carafoli, J. Biol. Chem., 1979, 254, 5308–5316 CAS
.
- J. X. Xu, B. C. N. Vithanage, S. A. Athukorale and D. M. Zhang, Analyst, 2018, 143, 3382–3389 RSC
.
- E. Mileykovskaya, W. Dowhan, R. L. Birke, D. H. Zheng, L. Lutterodt and T. H. Haines, FEBS Lett., 2001, 507, 187–190 CrossRef CAS
.
- G. Olofsson and E. Sparr, PLoS One, 2013, 8, e73040 CrossRef CAS PubMed
.
- M. Sathappa and N. N. Alder, Biochim. Biophys. Acta, 2016, 1858, 1362–1372 CrossRef CAS PubMed
.
- R. Majumder, Y. Sarkar, S. Das, S. K. Jewrajka, A. Ray and P. P. Parui, Analyst, 2016, 141, 3246–3250 RSC
.
- G. Akerlof, J. Am. Chem. Soc., 1932, 54, 4125–4139 CrossRef CAS
.
- P. A. Beales, C. L. Bergstrom, N. Geerts, J. T. Groves and T. K. Vanderlick, Langmuir, 2011, 27, 6107–6115 CrossRef CAS PubMed
.
- S. Atalay, M. Barisik, A. Beskok and S. Z. Qian, J. Phys. Chem. C, 2014, 118, 10927–10935 CrossRef CAS
.
- H. Kobayashi, S. Nagao and S. Hirota, Angew. Chem., Int. Ed., 2016, 55, 14019–14022 CrossRef CAS PubMed
.
-
(a) Y. Kuroda, S. Kidokoro and A. Wada, J. Mol. Biol., 1992, 223, 1139–1153 CrossRef CAS PubMed
; (b) S. Cinelli, F. Spinozzi, R. Itri, S. Finet, F. Carsughi, G. Onori and P. Mariani, Biophys. J., 2001, 81, 3522–3533 CrossRef CAS PubMed
.
- T. D. Perroud, M. P. Bokoch and R. N. Zare, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 17570–17575 CrossRef CAS PubMed
.
-
(a) J. Bagel'ova, M. Antalik and M. Bona, Biochem. J., 1994, 297(1), 99–101 CrossRef PubMed
; (b) L. Wang, E. V. Rivera, M. G. Benavides-Garcia and B. T. Nall, J. Mol. Biol., 2005, 353, 719–729 CrossRef CAS PubMed
.
- S. Papadopoulos, K. D. Jurgens and G. Gros, Biophys. J., 2000, 79, 2084–2094 CrossRef CAS PubMed
.
-
(a) L. Wan, M. B. Twitchett, L. D. Eltis, A. G. Mauk and M. Smith, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 12825–12831 CrossRef CAS PubMed
; (b) R. Radi, L. Thomson, H. Rubbo and E. Prodanov, Arch. Biochem. Biophys., 1991, 288, 112–117 CrossRef CAS PubMed
.
- T. Matsui, S. Ozaki, E. Liong, G. N. Phillips and Y. Watanabe, J. Biol. Chem., 1999, 274, 2838–2844 CrossRef CAS PubMed
.
-
(a) R. Kitz and I. B. Wilson, J. Biol. Chem., 1962, 237, 3245–3249 CAS
; (b) S. Prasad, N. C. Maiti, S. Mazumdar and S. Mitra, Biochim. Biophys. Acta, 2002, 1596, 63–75 CrossRef CAS PubMed
.
-
(a) Y. R. Chen, L. J. Deterding, B. E. Sturgeon, K. B. Tomer and R. P. Mason, J. Biol. Chem., 2002, 277, 29781–29791 CrossRef CAS PubMed
; (b) A. D. Nugraheni, C. Ren, Y. Matsumoto, S. Nagao, M. Yamanaka and S. Hirota, J. Inorg. Biochem., 2018, 182, 200–207 CrossRef CAS PubMed
.
- D. Mohammadyani, N. Yanamala, A. K. Samhan-Arias, A. A. Kapralov, G. Stepanov, N. Nuar, J. Planas-Iglesias, N. Sanghera, V. E. Kagan and J. Klein-Seetharaman, Biochim. Biophys. Acta, 2018, 1860, 1057–1068 CrossRef CAS PubMed
.
- L. V. Basova, I. V. Kurnikov, L. Wang, V. B. Ritov, N. A. Belikova, Vlasova, II, A. A. Pacheco, D. E. Winnica, J. Peterson, H. Bayir, D. H. Waldeck and V. E. Kagan, Biochemistry, 2007, 46, 3423–3434 CrossRef CAS PubMed
.
- M. Abe, R. Niibayashi, S. Koubori, I. Moriyama and H. Miyoshi, Biochemistry, 2011, 50, 8383–8391 CrossRef CAS PubMed
.
- W. Huang, P. Zhou, W. Yan, C. He, L. Xiong, F. Li and C. Duan, J. Environ. Monit., 2009, 11, 330–335 RSC
.
- R. Majumder, Y. Sarkar, S. Das, S. K. Jewrajka, A. Ray and P. P. Parui, Analyst, 2016, 141, 3246–3250 RSC
.
- E. G. Bligh and W. J. Dyer, Can. J. Biochem. Physiol., 1959, 37, 911–917 CrossRef CAS PubMed
.
- J. V. Morris, M. A. Mahaney and J. R. Huber, J. Phys. Chem., 1976, 80, 969–974 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available: Additional spectroscopic and data analysis. See DOI: 10.1039/c9sc02993a |
| This journal is © The Royal Society of Chemistry 2019 |